 Open Access Article
Open Access ArticleElectrochemical immunosensor based on mussel inspired coating for simultaneous detection and elimination of Staphylococcus aureus in drinks
Wenjin Wu†
a,
Yuping Yang†ad,
Lan Wanga,
Tingting Xu *b and
Rui Wang
*b and
Rui Wang *c
*c
aInstitute of Agricultural Products Processing and Nuclear-Agricultural Technology, Hubei Academy of Agricultural Sciences, Farm Products Processing Research Sub-Center of Hubei Innovation Center of Agriculture Science and Technology, Wuhan 430064, China
bJiangsu Co-Innovation Center of Efficient Processing and Utilization of Forest Resources, College of Light Industry and Food Engineering, Nanjing Forestry University, Nanjing 210037, China
cCollege of Food Science and Light Industry, State Key Laboratory of Materials-Oriented Chemical Engineering, Nanjing Tech University, Nanjing 211816, China. E-mail: ruiwang2013@njtech.edu.cn
dWuhan Institute for Drug and Medical Device Control, Wuhan 430075, Hubei, China
First published on 20th May 2021
Abstract
Staphylococcus aureus (S. aureus) is one of the most commonly isolated foodborne pathogens, and is considered as a major cause of foodborne illnesses worldwide. However, the development of smart and accurate analytical methods for the simultaneous detection and elimination of S. aureus in matrices of food or drinks remains challenging. In the present work, a mussel-inspired material, ε-poly-L-lysine-3,4-dihydroxy benzaldehyde (EPD), was designed and fabricated based on its Schiff base structure. Owing to the robust ability of the material to adhere onto wet electrode surfaces and the pH-responsive properties of EPD, the prepared immunosensor exhibited an excellent detection limit and linear range with on-demand antibacterial activity. In real milk samples, the average values obtained from the immunosensor were approximate to the standard results obtained from the plate count method, and the relative standard deviation was 3.16–6.54%, suggesting the good accuracy of the developed method. Moreover, it exhibited good selectivity, reproducibility, and stability, thus demonstrating the potential significant applications of the electrochemical immunosensor in drinks safety monitoring.
Introduction
Foodborne illness outbreaks caused by the consumption of food contaminated with harmful bacteria have drastically increased in the past decades.1 Therefore, the detection of harmful bacteria in food has become a pivotal factor for the diagnosis and prevention of problems associated with food safety and public health.2 Staphylococcus aureus (S. aureus) is one of the most common etiological agents in foodborne illness.3,4 S. aureus virulence factors such as toxins and proteases, often circulate in host blood vessels leading to life-threatening diseases.5 It mainly include infective endocarditis, toxic shock syndrome, scalded skin syndrome, or osteomyelitis. Even necrotizing fasciitis and necrotizing pneumonia were reported with S. aureus as the causative agent.6,7 Therefore, it is necessary to develop reliable methods for sensitive and rapid determination of S. aureus levels in complex food samples.Biosensors have been widely used in the food industry owing to their high sensitivity and rapid bacterial detection ability.1,8,9 Among different types of biosensors, electrochemical-based detection methods are the most prominent type of biosensors due to their well-known bio-interactions and inexpensive detection processes.10–12 However, electrochemical biosensors have certain limitations. The sensors are unable to work efficiently because of weak stability of the bio-recognition element on the surface of the electrode.13 The most commonly used methods to modify electrodes are drip coating and layer by layer self-assembly.14 However, the chemical stability achieved by these methods is unsatisfactory as the hydrous electrode surfaces hamper the immobilization of the functional molecules. Continuous efforts are being undertaken by researchers to enhance stable and firm absorption of the molecules on the electrode surfaces. Although the number of studies that focus on the rapid and precise detection of pathogenic bacterium have increased over the recent years, very little research has been conducted on the treatment of residual S. aureus in food, which makes it a “hidden danger”. Our group recently developed an electrochemical sensor for the detection and elimination of S. aureus in whole blood based on the hyaluronidase-responsive mechanism.15 However, to the best of our knowledge, there is no report of an electrochemical immunosensor technology that can detect and sterilize food or drinks simultaneously.
This is the first study that aimed to develop an immunosensor with dual capability of detection and elimination of S. aureus in drinks along with good selectivity, reproducibility, and stability. The immunosensor was constructed using a novel scaffold fabricated by robust binding of ε-poly-L-lysine-3,4-dihydroxy benzaldehyde (EPD) to polydopamine (PDA) pre-grafted gold electrode surfaces (Scheme 1). Mussels exhibit robust moisture-resistant adhesion properties owing to the byssus secretion that contains abundant adhesive mussel foot proteins (mfps), in particular, mfp-3 and mfp-5, which are rich in the catecholic amino acid 3,4-dihydroxyphenylalanine (DOPA) (10–20 and 25 mol%, respectively).16–18 DOPA, the main constituent in mfps, is believed to be the key factor responsible for the rapid and strong wet adhesion due to various types of interfacial interactions such as hydrogen bonding, metal chelation, π–π, and/or cation–π interactions with the tissue surface.19–22 In addition, the other constituents of mfps, e.g., cationic residues (lysine, K), also contributes to the wet adhesion.23–25 Here, a biomimetic polymer, EPD, was prepared and coated on the surface of a PDA-pretreated electrode to endow the immunosensor with good selectivity, reproducibility, and stability due to the robust binding of the antibody on the wet electrode surface. Moreover, the Schiff base structure in the EPD exhibits pH-responsive properties that allow on-demand ε-poly-L-lysine (ε-PL) delivery to eliminate S. aureus. It is known that bacterial growth produces protons and induces local acidification.26–29 The increased acidity will therefore cleave the Schiff base linkage to release ε-PL, which in turn kills the bacteria by targeting bacterial ribosomes and inhibiting protein synthesis.
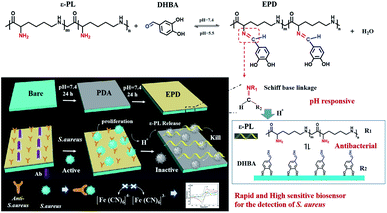 | ||
| Scheme 1 Schematic representation of fabrication of the immunosensor for detection and elimination of S. aureus. | ||
Experimental section
Materials
ε-Poly-L-lysine (ε-PL, molecular weight: 3500–5000 Da) was purchased from Nanjing Bioshineking Biotech Co., Ltd. Dopamine hydrochloride (DA) and 3,4-dihydroxy benzaldehyde (DBAH) were obtained from Aladdin Industrial Corporation (China). Methanol (absolute), triethylamine, sodium hydroxide (NaOH), potassium ferricyanide (K3[Fe(CN)6]), potassium ferrocyanide (K4Fe(CN)6·3H2O) and ethanol were provided by Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). All the solutions were prepared in Ultrapure water (18.2 MΩ, Milli-Q, Millipore). Anti-S. aureus antibody was obtained from Abcam Inc (Cambridge, UK), and BSA was purchased from Sigma (St. Louis, MO, USA). Milk was purchased from local markets.Synthesis and characterization of EPD
A mixture of ε-PL (1.5 mmol, 219 mg) and DBAH (1.5 mmol, 207 mg) in absolute methanol (36 mL) was stirred at room temperature under an argon atmosphere. After complete dissolution, triethylamine (1.5 mmol, 210 mL) was added gradually using a syringe. The reaction was left overnight under magnetic stirring, followed by methanol evaporation under reduced pressure. The solid was washed thrice with water (10 mL), filtered, and dried to obtain EPD. The chemical structure of EPD conjugates were characterized using 1H NMR (Varian, Palo, Alto, USA, 400 MHz) with D2O as the solvent.pH response of EPD monitored using 1H NMR
The pH response of EPD was tested by placing it in CBS (pH 5) for different time periods under magnetic stirring.30 After fixed periods of time, the respective samples were centrifuged and washed several times with water and EtOH. Then, the chemical structure of the samples was then characterized using 1H NMR (Varian, Palo, Alto, USA, 400 MHz) with D2O as the solvent.Preparation and characterization of the immunosensor
Fabrication and coating of the immunosensor comprises the following procedures. (a) The electrode was polished with 0.3 μm and 0.05 μm of aluminum oxide (Al2O3) powder and ultrasonically cleaned. (b) Twenty microliters of PDA solution (1.0 mg mL−1 in PBS) was injected over the electrode surface and fixed for 24 h. (c) Twenty microliters of activated EPD solution (1.0 mg mL−1) in PBS (pH 7.4) was injected and fixed for an additional 24 h. (d) Five microliters of anti-S. aureus antibody (0.1 mg mL−1) was added to the surface of the previously treated electrode, placed at 4 °C for 1 h, and gently rinsed with PBS to wash away unimmobilized antibodies. (e) Twenty microliters of BSA (1.0 mg mL−1) solution was added to the surface of the electrode to block non-specific reaction sites.The microscopic state of the electrode surfaces of different coatings was observed and photographed using an AFM. The WCA of the coating samples were recorded with a video-based contact angle measuring system. Pure water (2 μL) was dropped on the electrode surfaces and the WCA value for each sample was measured by calculating the average for a minimum of five data points at different locations for each sample.
Cytocompatibility assay
The cytocompatibility assay was carried out using 3T3 fibroblasts and cultured in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C, 5% CO2, and saturated humidity. For quantitative experiments, the cells were seeded in 96-well plates at a density of 2 × 104 cells per well. The cells were divided into two groups (control and EPD) with five wells per group. The plates were incubated overnight to allow the cells to attach, following which the medium was discarded and subjected to different treatments. Next, DMEM (100 μL) without BSA was added to each well, followed by 100 μL of PBS (pH 7.4) (control) and 100 μL of EPD (1 mg mL−1 in PBS, sterilized by filtration through a 0.22 μm filter). Cell viability was determined by MTT assay after 24 h of culturing under standard experimental conditions. For the live/dead staining experiment, cells were seeded in 24-well plates and grouped (grouping procedure same as the quantitative experiment). After 24 h of culture, the cells were stained with acridine orange (AO) and ethidium bromide (EB), observed under an inverted fluorescence microscope, and photographed.Hemolysis assay
Fresh blood was centrifuged (1500 rpm for 10 min) and the collected erythrocytes were washed three times with 10 mL PBS (2500 rpm centrifugation for 5 min each). The erythrocytes were resuspended in PBS at a concentration of 2% and EPD solution (1.0 mg mL−1) in PBS was added. The erythrocytes were and then incubated at 37 °C for 1 h. The EPD solution was replaced with PBS or water in the control sample. Thereafter, the mixture was centrifuged (1500 rpm for 10 min) and the absorbance of the supernatant was measured at 545 nm. The percentage of hemolysis was calculated using the following formula: hemolysis rate (%) = [(ODII − ODIII)/(ODI − ODIII)] × 100, where, ODI, ODII, and ODIII are the absorbance of PBS, EPD, and water, respectively.Electrochemical determination
All the electrochemical measurements were carried out using the electrochemical workstation at room temperature in 5 mM K3[Fe(CN)6]. The impedance was measured using electrochemical impedance spectroscopy (EIS) at a frequency ranging from 1 Hz to 100 kHz with 5 mV AC amplitude. Cyclic voltammetry (CV) was carried out over the potentials between −0.4 V and 1 V. Differential pulse voltammetry (DPV) measurements were performed at 0.05 V with a pulse width of 0.2 s.Actual sample test
Milk containing S. aureus was used as the test sample. Milk (1 mL) was diluted to 10 mL with PBS, divided into 5 portions, and different concentrations of S. aureus (2.2 × 105, 2.2 × 106, 2.2 × 107, 2.2 × 108 and 2.2 × 109 CFU per mL) was added. The above samples were separately detected by the immunosensor and the average of each sample, performed in triplicates, was calculated.Immunosensor performance analysis
The specificity of the sensor was determined by selecting E. coli and B. subtilis as the comparators. Here, the DPV tests were conducted for solutions containing E. coli, B. subtilis, and S. aureus (104 CFU per mL), respectively. The obtained peak current values were compared.For analysis of reproducibility, five immunosensors were prepared separately and the same concentration of S. aureus (5 × 105 CFU per mL) was added to the solution to compare the peak current values measured by the five sensors and the relative standard deviation (RSD) was calculated.
The prepared sensor was stored in a refrigerator at 4 °C for five weeks and the peak current values (S. aureus at 5 × 105 CFU per mL) were measured once a week. The data were compared and the RSD was calculated to analyze the stability of the sensor.
Antibacterial performance test
Milk containing S. aureus (2.20 × 106 CFU per mL) was used to test the antibacterial properties of the sensor. In order to calculate the bacteriostatic efficiency of the sensor, 100 μL of the milk was collected at 0, 30, and 60 min after the sensor was placed into the treated milk. The different concentrations of S. aureus were determined by plating 10-fold serial dilutions of the samples on agar plates. After incubation at 37 °C for 24 h, the plates were photographed and the survival rates of S. aureus were calculated. For the same time periods, 200 μL of milk was dropped onto a glass slide, fixed in 5 mL of glutaraldehyde solution (2.5%) for 4 h, followed by dehydration with different concentrations of ethanol (20–100% for 30 min), freeze-dried, and subsequently characterized by SEM analysis.Results and discussion
EPD synthesis and pH response monitored by H1 NMR
The schematic representation of the fabrication strategy of the immunosensor is shown in Scheme 1. It is pivotal to design a smart molecule as this sensor will automatically decompose and release ε-PL under acidic conditions. Under neutral conditions, EPD was synthesized by a condensation reaction between 3,4-dihydroxy benzaldehyde (DBAH) and ε-PL (Fig. 1a). Next, the pH response of EPD was tested by placing it in citrate buffer (CBS, pH 5). The 1H NMR spectra at pH 5 revealed that the signal of the imino proton (7.7 ppm) decreased with time, which was consistent with cleavage-decomposition of the Schiff base in EPD (Fig. 1b).30 This acid-responsive cleavage releases the ε-PL connected to the electrode, thereby killing S. aureus. This demonstrates the potential application of this smart response in the prevention of foodborne diseases. | ||
| Fig. 1 (a) Synthesis of EPD by condensation reaction between DBAH and ε-PL. (b) 1H NMR of EPD in citrus buffer (pH = 5) at fixed time periods. | ||
Surface morphology characterization of the immunosensor
For exploring the surface structure of the coatings, the PDA and EPD coatings deposited on electrode surface were scanned using an atomic force microscope (AFM). As shown in Fig. 2a, the PDA coating shows a rougher surface (Rq = 8.66 ± 0.12 nm) compared to the roughness parameter of the PDA sub-layer (Rq = 1.82 ± 0.07 nm).31 Furthermore, the surface density exhibited a significant increase after the deposition of EPD (Rq = 13.9 ± 0.09 nm). The rough morphology of EPD coating can be explained by strong interactions between DBAH and PDA, thus indicating that the polymer coatings were formed successfully on all the substrates.In addition, the substrates showed significant changes in static water contact angles (WCA) after the deposition of PDA and EPD coatings on the modified surfaces (Fig. 2b). Compared with the WCA values of the bare substrate (34.17 ± 5°) and PDA-coated substrates (27.5 ± 7°), the EPD-coated substrate (20.85 ± 8°) demonstrate slightly increased hydrophilicity.
Biocompatibility testing of EPD
EPD solution was co-cultured with 3T3 fibroblasts to assess its biological safety.32 As seen in Fig. 3a, there is no statistical difference in the cell viability between the EPD group and control group, which means that the presence of EPD does not inhibit cell proliferation or cause cell death. Furthermore, examination of the cell morphology (Fig. 3b) revealed that the cells co-cultured with EPD showed adherent and healthy spindle-shaped morphology. | ||
| Fig. 3 (a) Cell survival rate after a 24 h co-culture with EPD. ns: not significant. (b) Dead and live staining of cells from the 24 h co-culture with EPD. (c and d) Hemolysis assay of EPD. | ||
In addition, although EPD is used to detect and kill S. aureus in food, we assessed the hemocompatibility of EPD by calculating the hemolysis rate.33 The results (Fig. 3c and d) showed that even when EPD concentration reached 1.0 mg mL−1, no significant hemolysis was observed and the hemolysis rate was less than 3%, which further indicated the biosecurity of EPD.
Electrochemical characterization of the immunosensor
Fig. 4a shows the changes in the cyclic voltammetry (CV) signals at different stages of assembly of the immunosensor. The bare MGCE (curve a) showed a pair of well-defined redox peaks. Upon further modification of the bioactive molecules, the current response decreased significantly (curve b–d) due to the immobilized bioactive molecules that act as a barrier and prevent electron transfer between the electrode and the probe. The results from the electrochemical impedance spectroscopy (EIS) followed the same trend, thus confirming the results (Fig. 4b).The bare MGCE (curve a) showed lower resistance, while the resistance of the EPD-modified MGCE (curve b) increased significantly, which could be attributed to the polymer fixed on the electrode, thereby hindering the transfer of electrons.34 As expected, the resistance further increased with the addition of bovine serum albumin (BSA) and anti-S. aureus antibody.
Next, the relationship between S. aureus concentration and peak current was investigated using differential pulse voltammetry (DPV). The results suggested that as the concentration of S. aureus increased from 1 × 104 CFU per mL to 1 × 1010 CFU per mL, the amperometric response of the electrode decreased (Fig. 4c and d). The regression equation was as follows: ΔI (10−6 A) = −0.6729log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (CFU per mL) + 9.0222, (R2 = 0.9661).
C (CFU per mL) + 9.0222, (R2 = 0.9661).
In order to calculate the detection limit of the immunosensor, ten groups of parallel DPV tests for blank samples were carried out (Fig. 4e). The detection limit was estimated to be 28.55 CFU per mL with a signal-to-noise ratio of 3, which was similar to those reported in several studies.35–37
Performance characterization of the immunosensor
E. coli and B. subtilis were used as comparators to determine the specificity of the sensor. When the DPV tests were conducted for the solutions containing E. coli and B. subtilis (104 CFU per mL), respectively, the peak current values obtained were similar to those in the solutions without bacteria (Fig. 5a). When the DPV test was conducted for the S. aureus-containing solution (104 CFU per mL), the peak current values decreased significantly due to the specificity of the antibody for the antigen. Therefore, these results indicate the efficacy and feasibility of the immunosensor to detect S. aureus. | ||
| Fig. 5 (a) Analysis of specificity of the immunosensor to different microorganisms. (b) Analysis of accuracy and stability of the immunosensor. | ||
The accuracy and stability of the sensor were further evaluated using five immunosensors with the same concentration of S. aureus (105 CFU per mL) (Fig. 5b). The relative standard deviation (RSD) of the five sensors was 3.8%, indicating a considerable detection accuracy. Additionally, the EPD biosensor showed adequate long-term stability (RSD = 6.8%) for up to five weeks at room temperature. The above results thus demonstrate the acceptable and satisfactory performance of the developed immunosensor.
Analytical application of the immunosensor in real samples
We investigated the ability of the electrochemical immunosensor to detect S. aureus in actual samples. The results obtained are shown in Table 1. The concentration of S. aureus detected by the sensor were consistent with the actual added amount with a RSD between 3.16% and 6.54%. Besides, the detection time was shortened to less than 1 min. This indicated the good precision and fast speed of the immunosensor and its potential application in the detection of S. aureus in actual samples.| Samples | The concentration of S. aureus (CFU per mL) | SD | RSD (%) | |||
|---|---|---|---|---|---|---|
| Containing | Added | Immunosensor | Plate count | |||
| 1 | ND | 2.20 × 105 | 1.99 × 105 | 2.14 × 105 | 1.30 × 104 | 6.54 |
| 2 | ND | 2.20 × 106 | 2.04 × 106 | 2.07 × 106 | 1.09 × 105 | 5.32 |
| 3 | ND | 2.20 × 107 | 2.27 × 107 | 2.13 × 107 | 7.17 × 105 | 3.16 |
| 4 | ND | 2.20 × 108 | 2.11 × 108 | 2.10 × 108 | 8.60 × 106 | 3.91 |
| 5 | ND | 2.20 × 109 | 2.24 × 109 | 2.09 × 109 | 1.08 × 105 | 4.83 |
Antibacterial performance test
The developed biosensor proved its ability to detect S. aureus in phosphate-buffered saline (PBS). Next, we evaluated its potential to detect S. aureus in contaminated food products. The accumulation of S. aureus results in an increase in the acidity of the food, creating an acidic condition, which in turn breaks the Schiff base structure in EPD, thereby releasing ε-PL to kill S. aureus (Fig. 6a). Milk samples containing S. aureus (2.20 × 106 CFU per mL) were used to evaluate the antibacterial effect of the sensor (Fig. 6b and c). We observed that the total number of colonies, when compared with the initial state (0 min), significantly reduced after the sensor was placed in the solution, along with significant surface roughness and bacterial deformation. In addition, the antibacterial efficiency of the sensor at different time periods was calculated. The bacteriostatic efficiency reached 65% at 30 min and up to 98% at 60 min, thus indicating the high specificity of the immunosensor for S. aureus.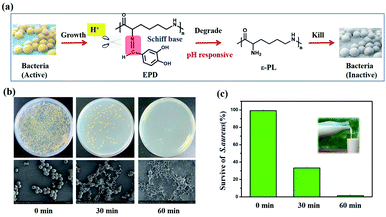 | ||
| Fig. 6 (a) Antibacterial mechanism of the immunosensor. (b and c) Antibacterial tests with S. aureus. | ||
Conclusions
In summary, this was the first study to develop an immunosensor with dual capability of detection and elimination of S. aureus. It displayed good biocompatibility and controlled release of ε-PL under acidic conditions, thus demonstrating its ability to kill S. aureus in food. EPD-modified MGCE exhibited a good electrochemical response to S. aureus in the range of 1 × 104 to 1 × 1010 CFU per mL, with high selectivity, stability, and repeatability. This response was confirmed using real food and reflected good bactericidal performance. Therefore, the developed immunosensor is an effective device for accurately detecting and effectively killing S. aureus in food.Moreover, it may have a promising application in solid food detection even polluted by other bacteria, fungi or viruses. Future work will be focused on determining the broad-spectrum utility of immunosensor based on EPD.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (2019YFD0901805), Jiangsu Agricultural Science and Technology Innovation Fund (CX(19)3115), the China Postdoctoral Science Foundation (2019M661814), the University Natural Science Research Project of Jiangsu Province (164105008), and the Nanjing Forestry University 2018 Scientific Research Start-up Funds (163105042).Notes and references
- M. Rubab, H. M. Shahbaz, A. N. Olaimat and D.-H. Oh, Biosens. Bioelectron., 2018, 105, 49–57 CrossRef CAS PubMed.
- R. L. Scharff, J. Food Prot., 2012, 75, 123–131 CrossRef PubMed.
- C. H. Wang, K. Y. Lien, J. J. Wu and G. B. Lee, Lab Chip, 2011, 11, 1521–1531 RSC.
- G. A. Suaifan, S. Alhogail and M. Zourob, Biosens. Bioelectron., 2017, 90, 230–237 CrossRef CAS PubMed.
- M. Otto, Curr. Opin. Microbiol., 2014, 17, 32–37 CrossRef CAS PubMed.
- J. S. Francis, M. C. Doherty, U. Lopatin, C. P. Johnston, G. Sinha, T. Ross, M. Cai, N. N. Hansel, T. Perl and J. R. Ticehurst, et al., Clin. Infect. Dis., 2005, 40, 100–107 CrossRef PubMed.
- L. G. Miller, F. Perdreau-Remington, G. Rieg, S. Mehdi, J. Perlroth, A. S. Bayer, A. W. Tang, T. O. Phung and B. Spellberg, N. Engl. J. Med., 2005, 352, 1445–1453 CrossRef CAS PubMed.
- S. Dai, S. Wu, N. Duan, J. Chen, Z. Zheng and Z. Wang, Biosens. Bioelectron., 2017, 91, 538–544 CrossRef CAS PubMed.
- S. Wu, N. Duan, C. Zhu, X. Ma, M. Wang and Z. Wang, Biosens. Bioelectron., 2011, 30(1), 35–42 CrossRef CAS PubMed.
- E. B. Setterington and E. C. Alocilja, Biosensors, 2012, 2, 15–31 CrossRef CAS PubMed.
- F. Gao, T. Fan, S. Ou, J. Wu, X. Zhang, J. Luo and D. Geng, Biosens. Bioelectron., 2018, 99, 201–208 CrossRef CAS PubMed.
- F. Gao, T. Fan, J. Wu, S. Liu, Y. Du, Y. Yao and D. Geng, Biosens. Bioelectron., 2017, 96, 62–67 CrossRef CAS PubMed.
- Y. W. Jung, H. J. Kang, J. M. Lee, S. O. Jung, W. S. Yun, S. J. Chung and B. H. Chung, Anal. Biochem., 2008, 374, 99–105 CrossRef CAS PubMed.
- C. Vericat, M. E. Vela, G. Benitez, P. Carro and R. C. Salvarezza, Chem. Soc. Rev., 2010, 39, 1805–1834 RSC.
- T. Xu, J. Li, S. Zhang, Y. Jin and R. Wang, Biosens. Bioelectron., 2019, 142, 111507 CrossRef CAS PubMed.
- R. Wang, J. Z. Li, W. Chen, T. T. Xu, S. F. Yun, Z. Xu, Z. Q. Xu, T. Sato, B. Chi and H. Xu, Funct. Mater., 2017, 27(8), 1604894 CrossRef.
- A. B. Kollbe, D. Saurabh, L. Roscoe, K. Yair and N. R. Martinez-Rodriguez, Nat. Commun., 2015, 6, 8663 CrossRef PubMed.
- B. Yang, N. Ayyadurai, H. Yun, Y. S. Choi and B. H. Hwang, Angew. Chem., Int. Ed., 2014, 53, 13360–13364 CrossRef CAS PubMed.
- B. P. Lee, P. B. Messersmith, J. N. Israelachvili and J. H. Waite, Annu. Rev. Mater. Res., 2011, 41, 99 CrossRef CAS PubMed.
- H. Lee, B. P. Lee and P. B. Messersmith, Nature, 2007, 448, 338 CrossRef CAS PubMed.
- J. H. Ryu, S. Hong and H. Lee, Acta Biomater., 2015, 27, 101–115 CrossRef CAS PubMed.
- L. Li, W. Smitthipong and H. Zeng, Polym. Chem., 2015, 6, 353–358 RSC.
- B. J. Kim, D. X. Oh, S. Kim, J. H. Seo and S. H. Dong, Biomacromolecules, 2014, 15, 1579–1585 CrossRef CAS PubMed.
- G. P. Maier, M. V. Rapp, J. H. Waite, J. N. Israelachvili and A. Butler, Science, 2015, 349, 628–632 CrossRef CAS PubMed.
- S. Seo, S. Das, P. J. Zalicki, R. Mirshafian, C. D. Eisenbach, J. N. Israelachvili, J. H. Waite and B. K. Ahn, J. Am. Chem. Soc., 2015, 137, 9214–9217 CrossRef CAS PubMed.
- J. J. Hu, Y. C. Quan, Y. P. Lai, Z. Zheng, Z. Q. Hu, X. Y. Wang, T. J. Dai, Q. Zhang and Y. Y. Cheng, J. Controlled Release, 2017, 247, 145–152 CrossRef CAS PubMed.
- A. Francesko, M. M. Fernandes, K. Ivanova, S. Amorim, R. L. Reis, I. Pashkuleva, E. Mendoza, A. Pfeifer, T. Heinze and T. Tzanov, Acta Biomater., 2016, 33, 203–212 CrossRef CAS PubMed.
- F. Hizal, I. Zhuk, S. Sukhishvili, H. J. Busscher, H. C. Van der Mei and C.-H. Choi, ACS Appl. Mater. Interfaces, 2015, 7(36), 20304–20313 CrossRef CAS PubMed.
- H.-S. Lee, S. S. Dastgheyb, N. J. Hickok, D. M. Eckmann and R. J. Composto, Biomacromolecules, 2015, 16(2), 650–659 CrossRef CAS PubMed.
- F. Nador, F. Novio and D. Ruiz-Molina, Chem. Commun., 2014, 50(93), 14570–14572 RSC.
- C. M. Xing, F. N. Meng, M. Quan, K. Ding, Y. Dang and Y. K. Gong, Acta Biomater., 2017, 59, 129–138 CrossRef CAS PubMed.
- R. Wang, X. X. Wang, Y. J. Zhan, Z. Xu, Z. Q. Xu, X. H. Feng, S. Li and H. Xu, ACS Appl. Mater. Interfaces, 2019, 11(41), 37502–37512 CrossRef CAS PubMed.
- T. Yu, A. Malugin and H. Ghandehari, ACS Nano, 2011, 5, 5717–5728 CrossRef CAS PubMed.
- S. J. Guo, D. Wen, Y. M. Zhai, S. J. Dongd and E. K. Wang, ACS Nano, 2010, 4, 3959–3968 CrossRef CAS PubMed.
- N. Tawil, F. Mouawad, S. Lévesque, E. Sacher, R. Mandeville and M. Meunier, Biosens. Bioelectron., 2013, 49, 334–340 CrossRef CAS PubMed.
- M. Braiek, K. B. Rokbani, A. Chrouda, B. Mrabet, A. Bakhrouf, A. Maaref and N. Jaffrezic-Renault, Biosensors, 2012, 2, 417–426 CrossRef CAS PubMed.
- T. Majumdar, R. Chakraborty and U. Raychaudhuri, Food Biosci., 2013, 4, 38–45 CrossRef CAS.
Footnote |
| † W. J. Wu. and Y. P. Yang contributed equally to this work. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. |
| This journal is © The Royal Society of Chemistry 2021 |


