Open Access Article
Open Access ArticleRestricted binding of a model protein on C3N4 nanosheets suggests an adequate biocompatibility of the nanomaterial†
Zonglin Gu *a,
Jose Manuel Perez-Aguilar
*a,
Jose Manuel Perez-Aguilar *b and
Qiwen Shaoc
*b and
Qiwen Shaoc
aCollege of Physical Science and Technology, Yangzhou University, Jiangsu 225009, China. E-mail: guzonglin@yzu.edu.cn; Tel: +86-0514-87975358
bSchool of Chemical Sciences, Meritorious Autonomous University of Puebla (BUAP), University City, Puebla 72570, Mexico. E-mail: jmanuel.perez@correo.buap.mx; Tel: +52-22-22-51-26-05
cInstitute of Quantitative Biology, Department of Physics, College of Life Science, Zhejiang University, Hangzhou 310027, China
First published on 12th February 2021
Abstract
Recently, C3N4, a carbon nitride nanomaterial, has attracted great attention in many scientific fields due to its outstanding properties. Specifically, this nanomaterial has displayed non- or low-toxicity in biological systems suggesting its excellent biocompatibility and biosafety. Nevertheless, few studies address the structural consequences from the direct interaction between C3N4 and biomolecules that could imply the physical origin of its bio-effect, particularly from the molecular level. Herein, we explored the interaction of a C3N4 nanosheet and a model protein, the λ-repressor protein. We found that the C3N4 nanosheet has a limited influence on the structure of the λ-repressor protein, which substantiates the outstanding biocompatibility of the nanomaterial. Detailed analyses showed that upon absorption on the C3N4 nanosheet, the λ-repressor protein remains located in a relatively fixed position without compromising the structural integrity of the protein. Furthermore, the protein-nanomaterial interaction is mediated by positively charged residues located on the surface of the protein and by the regional negatively charged center on the C3N4 nanosheet (i.e., N-rich defects). These findings provide further molecular-level insights into the good biocompatibility of the C3N4 nanomaterial and also suggest its potential usage as a protein drug delivery platform.
Introduction
The rapid development of carbon-based nanomaterials (CBNs) has inspired tremendous interests in biomedical applications,1,2 including nanotherapeutics,3–5 optical imaging,6 and gene delivery,7 because of their outstanding optical, mechanical, and electrical properties.8–10 Recently, ordered carbon nitrides (e.g. C2N,11 C3N,12,13 and C3N4 (ref. 14)), a new family class of CBNs, have also greatly stimulated research enthusiasm, due to their inherent electronic and optoelectronic properties. Based on their surprising electronic conductivity and optical properties, carbon nitrides were successfully utilized to produce various devices, such as photoelectrical devices, sensors, field-effect transistor devices, and so on.12,15,16 Carbon nitrides also exhibited interesting catalytic activity for the H2 evolution and oxygen reduction reaction.17–19 Moreover, some carbon nitrides exhibited promising potential in biomedical applications. For instance, ultrathin graphitic-phase C3N4 (g-C3N4) nanosheets, prepared by a “green” liquid exfoliation pathway from bulk g-C3N4 in water, displayed outstanding properties including (i) good stability in both acidic and alkaline solvents, (ii) intensive photoabsorption and photoresponse, and (iii) excellent biocompatibility.20 The g-C3N4 has also been utilized as a platform carrying up-conversion nanoparticles for cancer photodynamic therapy.21 Meanwhile, g-C3N4 was considered an excellent biosensor, due to its metal-free, high fluorescence quantum yield, easy preparation, high stability as well as adequate biocompatibility.22,23 For example, a g-C3N4 single-layer quantum dot was introduced for fluorescence imaging of the cellular nucleus.24 Through hybridization with gold nanoparticles, the g-C3N4 material was exploited as an electrochemiluminescence immunosensor.25 The ultrathin g-C3N4 nanosheet was developed for the detection of alkaline phosphatase exhibiting label-free high sensitivity in biological systems.26 The protonated g-C3N4 nanosheets without metal/label functionalization were established as a sensing platform for highly sensitive and selective detection of heparin.27 Although hybridized with some components, the g-C3N4 material was also exploited for biosensing of other molecules, e.g., hybridized with MOF for amyloid-β protein sensing,28 hybridized with MnSe for H2O2 and glucose sensing,29 hybridized with molecularly imprinted polymers for H2O2 sensing,22 etc.30–32Despite the widespread biomedical applications, thus far, there is very limited knowledge regarding the interaction of the C3N4 nanomaterial with biomolecules. Detailed knowledge of the biomolecule-C3N4 (bio-nano) interaction is central to support the desired biocompatibility of C3N4. Recently, Zhao et al. investigated the interaction between amino acid residues and a C3N4 nanosheet by employing density functional theory (DFT) and molecular dynamics simulations.33 They found that C3N4 preferentially attracted the backbone amino (NH2) group, yet this group appeared infrequently in any protein due to the dehydration synthesis from amino acids to protein. However, the positively charged side-chain groups from lysine and arginine may follow similar behavior regarding their preference to contact the C3N4. In this study, we probed the binding of a model protein, the λ-repressor protein, to a C3N4 nanosheet using all-atomic molecular dynamics (MD) simulations. We observed that the transverse migration of the λ-repressor protein on the C3N4 plane was significantly restricted, while the same protein on the graphene plane moved relatively free and randomly. Also, the protein structure exhibited little change throughout the binding process, suggesting excellent biocompatibility of the C3N4 nanomaterial. In addition, the positively charged residues were robustly attracted by the N-rich defects on the C3N4 surface, which was critical to the confined combination. Our works revealed the interaction basis at the molecular level of the C3N4 nanosheet and a model protein, which might be directly correlated with the nanomaterial's adequate biocompatibility that is central for its utility in biomedical devices.
Results
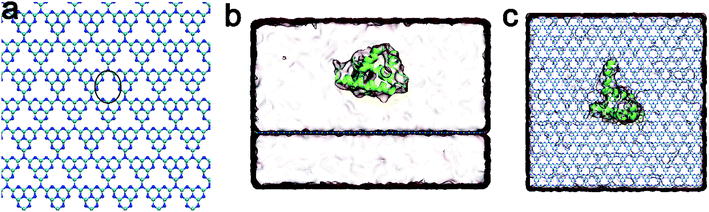
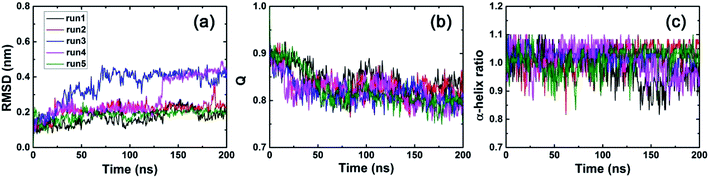
In order to investigate the binding of the λ-repressor protein to the C3N4 surface, we built an all-atom system consisting of a single biomolecule (the λ-repressor protein) and the C3N4 nanosheet as shown in Fig. 1. The system was investigated by unbiased MD simulations and five independent 200 ns-long trajectories were generated (see Methods for details). To assess the stability of the secondary and tertiary structures of the protein upon surface absorption, we performed analyses of several parameters along the different simulations, including the root mean square deviations (RMSDs) of Cα atoms, the fraction of native contacts (Q values) and the α-helix ratio (Fig. 2). Herein, a direct contact of the protein with the C3N4 surface was considered when any heavy atom of the λ-repressor protein was within 0.6 nm of any atom of C3N4 nanomaterial. Using the same direct contact criterion for the cut-off distance, the fraction of native contacts of protein over time t, Q(t), was defined as the ratio of the total number of native contacts at time t relative to that at time zero (i.e., the total number of native contacts as observed in the X-ray crystal structure); for this analysis, only residue pairs apart by at least 3 consecutive residues from each other were considered. The analysis indicates that the protein conserved its structural integrity in the five independent MD trajectories. Specifically, the RMSD values for the protein Cα atoms remained lower than 0.5 nm and even maintained values of ∼0.2 nm in three out of five trajectories, suggesting that the C3N4 nanosheet had little influence on the overall protein structure (i.e., the nanomaterial displays adequate biocompatibility). Moreover, both the Q values and the α-helix ratios also indicated a minor impact on the conformations sampled by the protein during the entire adsorption process, which substantiates the aforementioned conclusion regarding the protein structure. The mild effect of the C3N4 nanosheet on the protein tertiary structure substantially emerges from the particular molecular features of the surface (see detailed discussion below), which varies from that of pristine graphene.34–36 Fig. 3a illustrated the distributions of the center of mass (CoM) of the λ-repressor protein mapping onto the X–Y plane in the five trajectories. Clearly, during the entire simulation time, the CoMs of the protein was restricted on the C3N4 nanosheet in all five trajectories, showing a localized binding pattern. For comparison, we also performed another simulation in which the C3N4 nanosheet was replaced by a graphene surface (Fig. 3b). We similarly tracked the CoM of the λ-repressor protein on the graphene surface and found a different situation: the same protein moved quickly and randomly on the graphene plane. This difference was fundamentally caused by the ordered charge distribution and abundant defects on the C3N4 nanosheet which is in stark contrast to graphene's zero charge and uniform surface.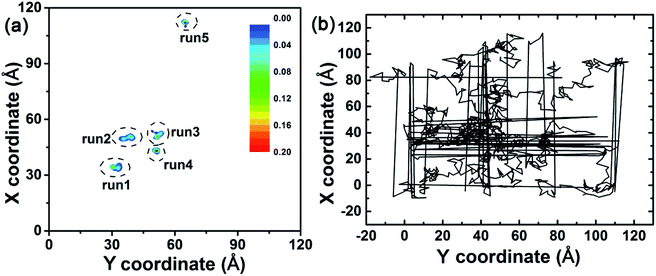 | ||
| Fig. 3 (a) Center of mass (CoM) distributions of the λ-repressor protein mapping onto the X–Y plane in five trajectories. (b) CoM track of the λ-repressor protein when bound to a graphene surface. | ||
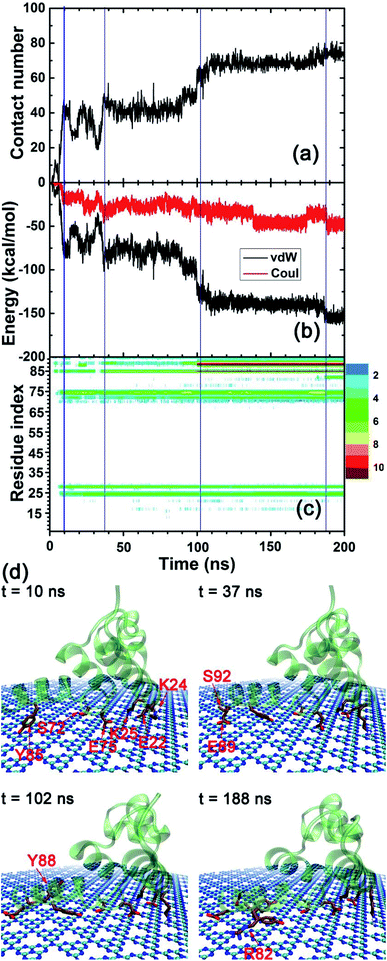
In order to explore the adsorption molecular determinants and interaction mechanism of the λ-repressor protein binding to the C3N4 nanosheet, we calculated the heavy atom contact number, the van der Waals (vdW), and coulombic energy contributions between the protein and the nanosheet, as well as the residue-dependent heavy atom contact number map (Fig. 4a–c). Additionally, according to their tendencies, we described in detail the adsorption process by plotting some snapshots at selected key time points along the entire trajectory (Fig. 4d). Specifically, at early stages, t = 10 ns, the λ-repressor protein approached the C3N4 nanosheet, with some residues directly contacting the surface, i.e., residues E22, K24, K25, S72, E75, and Y85. As expected, the values for the heavy atom contact number sharply increased to a value of around 43. Meanwhile, the vdW energy and the Coulombic energy decreased their values to −72.46 kcal mol−1 and −17.13 kcal mol−1, respectively. The larger decrement of its magnitude from the former energy term suggests that the vdW interactions may drive the initial protein-surface interactions. From 10 ns to 37 ns, the binding profile fluctuated significantly. At around t = 37 ns, the binding seems to reach a metastable state with minor increments in the protein-surface contact values mediated mainly by the interaction of the E89 and S92 residues (heavy atom contact number: ∼47, vdW energy: ∼−88.66 kcal mol−1, coulombic energy: ∼−37.49 kcal mol−1). At around t = 102 ns, and after the packing of residues E89, Y88, and Y85 (Fig. 4c), the adsorption event was further strengthened, where the heavy atom contact number reached values of close to 61 while the vdW and coulombic energies reached close to −122.29 and −31.10 kcal mol−1, respectively. Around t = 188 ns, the protein contact intensified slightly after the adsorption of the R82 residue. Even though the C3N4 nanomaterial comprised various negatively charged defects in its structure, during the absorption event it is the vdW interactions, and not the coulombic interactions, that seem to dominate the binding process. Yet interestingly, the positively charged residues account for around 30% of all the contact residues.
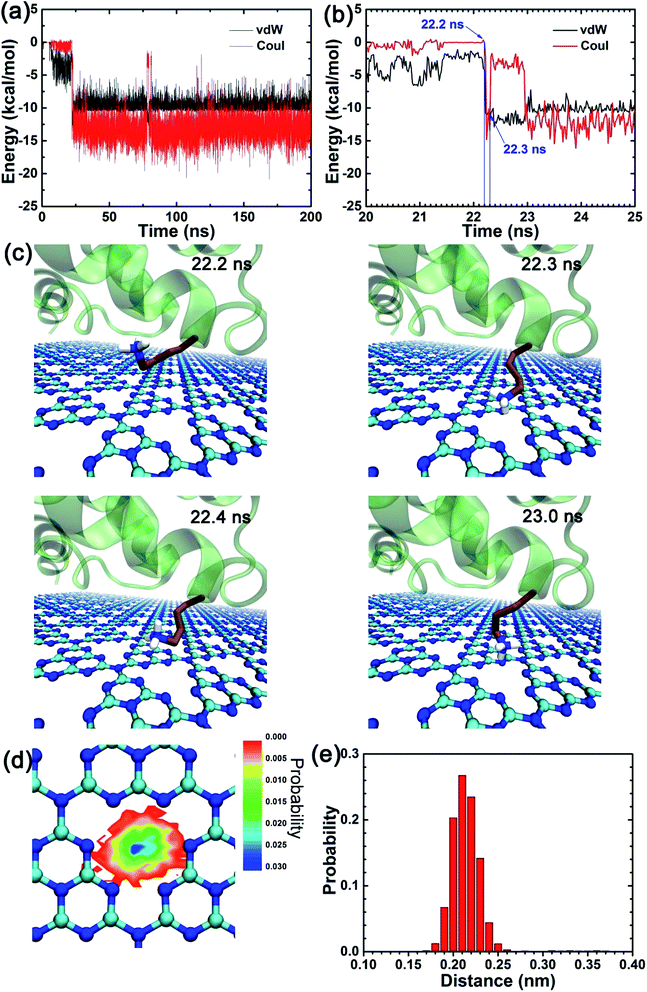
Due to the particular chemical topology of the C3N4 surface, the defects on the C3N4 nanosheet are surrounded by highly-concentrated negative charges (i.e., nitrogen atoms, as indicated in Fig. 1a) which enabled a specific attraction of these sites to the positively charged residues via electrostatic attraction. Therefore, we next explored the binding character of positively charged residues (choosing residue K24 as an example) as shown in Fig. 5. We first calculated the interaction energies between the K24 sidechain and the C3N4 nanosheet. Interestingly, both the vdW and the coulombic energies showed a sharp decline in the initial value of the contact number (∼10 ns). To interpret the absorption process of the K24 residue in the proximity of the C3N4 surface, we described in detail some snapshots from the MD trajectory as shown in Fig. 5c. Clearly, from 22.2 ns to 22.3 ns, the positively charged amino group of K24 quickly approaches the N-encircling defect in the surface. This event seems to be driven by the conjunct contributions, vdW and coulombic; the coulombic energy values range from −0.05 kcal mol−1 to −13.43 kcal mol−1 while the vdW energy values range from −4.21 kcal mol−1 to −12.21 kcal mol−1. Yet, soon after, at t = 22.4 ns and due to minor conformational perturbations of the K24 sidechain, the binding strength weakened significantly with a major contribution by the coulombic energy, which suggests that small conformational changes of the lysine residue can cause a large change in the value of this energy term. At t = 23.0 ns, the conformation recovered completely. We identified that the conformations where the K24 amino sidechain is directly positioned on the C3N4 surface defect were the structurally most stable and energetically more favorable binding modes; where the main contributions arise from the coulombic energy term. Moreover, our previous study also found a similar interacting pattern by the HP35 protein binding onto the C2N surface, suggesting more general binding features by this type of nanomaterials.37 Next, we mapped the location of the nitrogen atom from the amino group of residue K24 during the last 100 ns trajectory, on the X–Y plane (same plane as the C3N4 surface) as well as on the Z direction (perpendicular to the C3N4 plane), respectively (Fig. 5d and e). Remarkably, the largest distribution is located at the center of the N-encircling defect. Moreover, along the perpendicular direction, the position of this atom is mostly maintained at a distance of 0.20–0.23 nm over the C3N4 plane. This finding demonstrates the significance of the specific binding of basic residues to the C3N4 defects.
To probe if the aforementioned electrostatic interaction played a role in the observed fixed binding, we conducted a simulation by changing all atomic charges of C3N4 to zero. Notably, by tracking the CoM of the protein (as shown in Fig. S2†), we still observed a restricted binding of the protein on the C3N4 nanostructure, which thereby suggests that the electrostatic interactions are not the main factor for the observed fixed binding pattern of the protein but that the vdW interactions alone can induce the determinants of the observed binding event. We have demonstrated that the protein can freely move on graphene (Fig. 3b), and since the only difference between the two 2D nanomaterials is the surface topological structure, we concluded that the vdW interactions are enough to restrain the movement of the adsorbed protein and the key reason for the observed fixed binding pattern is the presence of significant defects in the nanomaterial surface topology. These defects could generate high free energy barriers and would affect the free protein movement, resembling the case for the C2N nanosheet.37 Further analysis of the surface water distribution on the C3N4 structure (Fig. S3†) showed that the surface defects aggregate many water molecules (with one water located nearby a structural defect), which forms a specific water layer in the proximity of the C3N4 material. This water layer may also contribute to the fixed binding of protein by forming a steric hindrance to prevent the lateral displacement of the protein, which resembles the results from our previous study regarding the dsDNA binding to the 2D C2N nanosheet.38 To sum up, the restricted protein binding on the C3N4 nanosheet is mainly attributed to the inherent porous structure of the C3N4 sheet as well as the water aggregation in the proximity of the surface.
To further confirm if the observed restricted binding pattern occurred in other biomolecules upon binding to the C3N4 nanosheet, we conducted MD simulations where we replaced the λ-receptor protein with (i) a double-stranded DNA segment (dsDNA) and (ii) the HP35 protein. As shown in Fig. S4,† it is notable that, despite their different physicochemical properties, both the HP35 protein and the dsDNA can bind to the C3N4 surface. Also, upon binding, these two biomolecules explored restrained conformations on the C3N4 surface comparable to those observed in the case of the λ-receptor protein bound to the C3N4 nanosheet. These findings substantiate our conclusion that the observed fixed binding pattern of biomolecules on the C3N4 nanosheet is mainly attributed to the inherent porous structure of the C3N4 nanomaterial as well as the water distribution in the surface proximity.
Usually, under real experimental environmental conditions, most water-soluble proteins (e.g., serum proteins) present a significant amount of hydrophilic residues on their surface for stability reasons in the water environment, particularly residue with positively charged side chains. In our previous study,39 we found that solvent-exposed basic residues (Lys and Arg) played an important role in mediating the interactions at the bio-nano interface, e.g., in the serum proteins–graphene interaction. Therefore, an abundance of basic residues may strengthen the protein absorption on the C3N4 surface in a physiologically relevant environment, because of the potent attraction between basic residues and the C3N4 pores. Similarly, by using the combination of MD simulations and experiments, we discovered that this kind of specific interactions between positively charged groups of bacterial lipid head groups and the C3N4 pores, also weakened the C3N4 insertion into the bacterial membrane due to the strong binding interactions between the lipid head groups and the C3N4 nanomaterial, reflecting the weak antibacterial activity of pristine C3N4 (ref. 40). However, after nitrogen plasma treatment, the pores of C3N4 were sealed, endowing the C3N4 nanosheet with a more homogeneous surface that results in an increase in membrane insertion and the concomitant increase in the C3N4 antibacterial activity. These findings further demonstrate that the interaction between positively charged chemical moieties (groups/side chains) and the C3N4 pores was extremely relevant for the behavior of the C3N4 nanomaterial in real experiments. Furthermore, surface functionalizations and larger topological defects on the C3N4 nanosheet should also be taken into account, due to the frequent presence during nanomaterial chemical synthesis. We have demonstrated that the main factors leading to the observed fixed protein binding is the porous C3N4 surface structure and the specific water distribution in the proximity of the nanosheet (as shown in Fig. S3†), instead of the electrostatic interaction. The functionalizations on the C3N4 surface will not have a significant impact on the determinants observed in the protein binding to the C3N4 nanosheet. The larger defects generate higher free energy barriers for lateral movement (resembling C2N nanosheet),37 which in turn intensifies the fixed binding of biomolecules. Thereby, surface functionalizations may have a limited influence on the observed fixed protein binding pattern.
We also compared our findings with other works regarding the interactions between other 2D nanomaterials and biomolecules as seen in Table S1,† particularly focusing on the consequences on the biomolecular structural integrity. Notably, abundant surface defects, such as nitrogen-enriched pores on the C3N4 and C2N nanomaterials, maybe more biocompatible to biomolecules than more homogeneous materials because their inherent porous structure can significantly restrict the lateral movement of the biomolecules. The 2D nanomaterials without abundant surface pores, such as graphene, C3N, boron nitride, and MoS2, may have an influence on the tertiary structure of the systems, due to the potential fast lateral migration of biomolecules. Moreover, surface wrinkles (e.g., in phosphorene) and hydrophilic groups (e.g., in silicene) may contribute to protect the biomolecular integrity. However, if the pores on the 2D nanomaterial are distributed less close-packed (the separation distance among these pores was larger than the size of adsorbed biomolecule), the biomolecule may suffer partial or complete denaturation, e.g., the HP35 protein on carboxyl functionalized graphene defects.41 Furthermore, surface-specific atomic arrangements also impact the structural change of biomolecules, e.g., Pt nanocrystals with (111) and (100) facets. In general, the factors mediating the structural influence of biomolecules after binding to 2D nanomaterials are complex, but it may be highly correlated to the surface characters of these 2D nanomaterials.
Conclusion
In summary, we have performed unbiased all-atom MD simulations to investigate the binding of the λ-repressor protein to a C3N4 nanosheet. Upon the protein's adsorption onto the C3N4 surface, the λ-repressor protein retained its structural integrity, that is, the conformations sampled upon absorption do not change its secondary and tertiary structure. The main driving force involved in the protein's absorption event was dominated by the van der Waals (vdW) interactions while the coulombic energy term exhibits a more discreet contribution. Moreover, our analysis of the mobility of the center of mass (CoM) of the λ-repressor protein found that the protein remains relatively fixed on the C3N4 nanosheet. Detailed analyses showed that the positively charged residues were mainly responsible for this specific binding pattern in which both interactions, vdW and coulombic, guided the determinants of the adsorption process. Our findings demonstrated a special interface of the protein and the C3N4 nanosheet and revealed a potential molecular mechanism that substantiates the adequate biocompatibility of the C3N4 nanomaterial observed in previous experiments.Model and methods
The C3N4 nanosheet used here consists of 1344 nitrogen and 1792 carbon atoms with a dimension of 9.98 × 9.88 nm2 as shown in Fig. 1a. Notably, C3N4 was a porous 2D nanomaterial, in the sense that every pore is surrounded by six nitrogen atoms forming a negatively charged center (due to inherent charge transfer from the C atoms to the N atoms). The force field parameters for C3N4 were obtained from our previous study,40 and the details were shown in Fig. S1.† The λ-repressor protein (PDB code 1LMB42) was utilized as a protein model to investigate its interaction with the C3N4 nanomaterial, due to its previous use in investigating the interactions of biomolecules with graphene.43 Next, the C3N4 nanosheet and the λ-repressor protein were embedded in a box (size: 9.98 × 9.88 × 6.89 nm3) of 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 028 water molecules, with their minimum initial distance setting to 1.5 nm. Two sodium ions were added to neutralize the entire system. Additionally, some other simulation systems were also built to support our conclusion, such as the λ-repressor protein with non-charged C3N4, HP35/C3N4, dsDNA/C3N4.
028 water molecules, with their minimum initial distance setting to 1.5 nm. Two sodium ions were added to neutralize the entire system. Additionally, some other simulation systems were also built to support our conclusion, such as the λ-repressor protein with non-charged C3N4, HP35/C3N4, dsDNA/C3N4.
The MD simulation was carried out with the GROMACS software package (version: 4.6.6).44 The VMD software45 was used to visualize the MD trajectories and rendered the molecular snapshots. The CHARMM36 force field46 was utilized for the protein in this system. TIP3P47 water model was applied to describe the water molecules. The system's temperature was maintained at 300 K using the velocity-rescale48 method while its pressure was coupled at 1 atm using the Parrinello–Rahman49 algorithm with semiisotropic type (coupling along Z direction). Periodic boundary conditions (PBC) were applied in all directions. The C3N4 nanosheet was frozen throughout the simulation which was similar to the protocol used in our previous works.38,50 The particle mesh Ewald (PME)51,52 method was used to treat long-range electrostatic interaction (with a cut-off distance of 1.2 nm) and the van der Waals (vdW) interactions were computed with a cutoff distance of 1.2 nm. The LINCS53 and SETTLE54 algorithms were employed to constrain the solute bonds associated with hydrogen atoms to their equilibrium values and water geometry, respectively. During the simulations, the time step was set to 2.0 fs and the coordinates were saved every 20 ps. The system was investigated by unbiased MD simulations and performed five independent 200 ns-long trajectories, for a total aggregated simulation time of 1.0 μs.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Shengtang Liu for helping with the manuscript. This work is supported by Youth Hundred Talents Program of Yangzhou University. J. M. P-A. thanks to the Laboratorio Nacional de Supercómputo del Sureste de México (LNS-BUAP) of the CONACyT network of national laboratories, for the computer resources and support provided and the computing time granted by LANCAD and CONACYT on the supercomputer xiuhcoatl at CGSTIC CINVESTAV.References
- J. S. Lee, H.-A. Joung, M.-G. Kim and C. B. Park, Graphene-based chemiluminescence resonance energy transfer for homogeneous immunoassay, ACS Nano, 2012, 6(4), 2978–2983 CrossRef CAS.
- C. Y. Cha, S. R. Shin, N. Annabi, M. R. Dokmeci and A. Khademhosseini, Carbon-based nanomaterials: multifunctional materials for biomedical engineering, ACS Nano, 2013, 7(4), 2891–2897 CrossRef CAS.
- M. Li, X. Yang, J. Ren, K. Qu and X. Qu, Using graphene oxide high near-infrared absorbance for photothermal treatment of alzheimer's disease, Adv. Mater., 2012, 24(13), 1722–1728 CrossRef CAS.
- K. Yang, J. Wan, S. Zhang, B. Tian and Y. Zhang, et al., The influence of surface chemistry and size of nanoscale graphene oxide on photothermal therapy of cancer using ultra-low laser power, Biomaterials, 2012, 33(7), 2206–2214 CrossRef CAS.
- Z. Yang, S.-g. Kang and R. Zhou, Nanomedicine: de novo design of nanodrugs, Nanoscale, 2014, 6(2), 663–677 RSC.
- B. Li, Y. Cheng, J. Liu, C. Yi and A. S. Brown, et al., Direct optical imaging of graphene in vitro by nonlinear femtosecond laser spectral reshaping, Nano Lett., 2012, 12(11), 5936–5940 CrossRef CAS.
- H. Bao, Y. Pan, Y. Ping, N. G. Sahoo and T. Wu, et al., Chitosan-functionalized graphene oxide as a nanocarrier for drug and gene delivery, Small, 2011, 7(11), 1569–1578 CrossRef CAS.
- V. C. Sanchez, A. Jachak, R. H. Hurt and A. B. Kane, Biological interactions of graphene-family nanomaterials: an interdisciplinary review, Chem. Res. Toxicol., 2012, 25(1), 15–34 Search PubMed.
- A. K. Geim, Graphene: status and prospects, Science, 2009, 324(5934), 1530–1534 CrossRef CAS.
- L. Feng and Z. Liu, Graphene in biomedicine: opportunities and challenges, Nanomedicine, 2011, 6(2), 317–324 CrossRef CAS.
- J. Mahmood, E. K. Lee, M. Jung, D. Shin and I.-Y. Jeon, et al., Nitrogenated holey two-dimensional structures, Nat. Commun., 2015, 6, 6486 CrossRef CAS.
- S. W. Yang, W. Li, C. C. Ye, G. Wang and H. Tian, et al., C3N-a 2d crystalline, hole-free, tunable-narrow-bandgap semiconductor with ferromagnetic properties, Adv. Mater., 2017, 29(16), 1065625 Search PubMed.
- J. Mahmood, E. K. Lee, M. Jung, D. Shin and H. J. Choi, et al., Two-dimensional polyaniline (C3N) from carbonized organic single crystals in solid state, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(27), 7414–7419 CrossRef CAS.
- Z. X. Zhou, Y. F. Shen, Y. Li, A. R. Liu and S. Q. Liu, et al., Chemical cleavage of layered carbon nitride with enhanced photoluminescent performances and photoconduction, ACS Nano, 2015, 9(12), 12480–12487 CrossRef CAS.
- Y. Bu, Z. Chen and W. Li, Using electrochemical methods to study the promotion mechanism of the photoelectric conversion performance of ag-modified mesoporous g-C3N4 heterojunction material, Appl. Catal., B, 2014, 144, 622–630 CrossRef CAS.
- X.-L. Zhang, C. Zheng, S.-S. Guo, J. Li and H.-H. Yang, et al., Turn-on fluorescence sensor for intracellular imaging of glutathione using g-C3N4 nanosheet–MnO2 sandwich nanocomposite, Anal. Chem., 2014, 86(7), 3426–3434 CrossRef CAS.
- X. F. Chen, J. S. Zhang, X. Z. Fu, M. Antonietti and X. C. Wang, Fe-g-C3N4-catalyzed oxidation of benzene to phenol using hydrogen peroxide and visible light, J. Am. Chem. Soc., 2009, 131(33), 11658–11659 CrossRef CAS.
- J. Mahmood, S. M. Jung, S. J. Kim, J. Park and J. W. Yoo, et al., Cobalt oxide encapsulated in C2N-h2d network polymer as a catalyst for hydrogen evolution, Chem. Mater., 2015, 27(13), 4860–4864 CrossRef CAS.
- Y. Zheng, Y. Jiao, J. Chen, J. Liu and J. Liang, et al., Nanoporous graphitic-C3N4@carbon metal-free electrocatalysts for highly efficient oxygen reduction, J. Am. Chem. Soc., 2011, 133(50), 20116–20119 CrossRef CAS.
- X. D. Zhang, X. Xie, H. Wang, J. J. Zhang and B. C. Pan, et al., Enhanced photoresponsive ultrathin graphitic-phase C3N4 nanosheets for bioimaging, J. Am. Chem. Soc., 2013, 135(1), 18–21 CrossRef CAS.
- L. L. Feng, F. He, B. Liu, G. X. Yang and S. L. Gai, et al., G-C3N4 coated upconversion nanoparticles for 808 nm near-infrared light triggered phototherapy and multiple imaging, Chem. Mater., 2016, 28(21), 7935–7946 CrossRef CAS.
- Y. H. Wu, Q. Chen, S. Liu, H. Xiao and M. L. Zhang, et al., Surface molecular imprinting on g-C3N4 photooxidative nanozyme for improved colorimetric biosensing, Chin. Chem. Lett., 2019, 30(12), 2186–2190 CrossRef CAS.
- Z. X. Zhou, Y. Y. Zhang, Y. F. Shen, S. Q. Liu and Y. J. Zhang, Molecular engineering of polymeric carbon nitride: advancing applications from photocatalysis to biosensing and more, Chem. Soc. Rev., 2018, 47(7), 2298–2321 RSC.
- X. D. Zhang, H. X. Wang, H. Wang, Q. Zhang and J. F. Xie, et al., Single-layered graphitic-C3N4 quantum dots for two-photon fluorescence imaging of cellular nucleus, Adv. Mater., 2014, 26(26), 4438–4443 CrossRef CAS.
- L. C. Chen, X. T. Zeng, P. Si, Y. M. Chen and Y. W. Chi, et al., Gold nanoparticle-graphite-like C3N4 nanosheet nanohybrids used for electrochemiluminescent immunosensor, Anal. Chem., 2014, 86(9), 4188–4195 CrossRef CAS.
- M. H. Xiang, J. W. Liu, N. Li, H. Tang and R. Q. Yu, et al., A fluorescent graphitic carbon nitride nanosheet biosensor for highly sensitive, label-free detection of alkaline phosphatase, Nanoscale, 2016, 8(8), 4727–4732 RSC.
- T. Y. Ma, Y. Tang, S. Dai and S. Z. Qiao, Proton-functionalized two-dimensional graphitic carbon nitride nanosheet: an excellent metal-/label-free biosensing platform, Small, 2014, 10(12), 2382–2389 CrossRef CAS.
- Y. Wang, Y. Zhang, H. Sha, X. Xiong and N. Jia, Design and biosensing of a ratiometric electrochemiluminescence resonance energy transfer aptasensor between a g-C3N4 nanosheet and Ru@mof for amyloid-beta protein, ACS Appl. Mater. Interfaces, 2019, 11(40), 36299–36306 CrossRef CAS.
- F. M. Qiao, Q. Q. Qi, Z. Z. Wang, K. Xu and S. Y. Ai, MnSe-loaded g-C3N4 nanocomposite with synergistic peroxidase-like catalysis: synthesis and application toward colorimetric biosensing of H2O2 and glucose, Sens. Actuators, B, 2016, 229, 379–386 CrossRef CAS.
- Y. X. Dong, J. T. Cao, B. Wang, S. H. Ma and Y. M. Liu, Spatial-resolved photoelectrochemical biosensing array based on a cds@g-C3N4 heterojunction: a universal immunosensing platform for accurate detection, ACS Appl. Mater. Interfaces, 2018, 10(4), 3723–3731 CrossRef CAS.
- Y. F. Zhang, R. Xu, Q. Kang, Y. Zhang and Q. Wei, et al., Ultrasensitive photoelectrochemical biosensing platform for detecting n-terminal pro-brain natriuretic peptide based on SnO2/SnS2/mpg-C3N4 amplified by PbS/SiO2, ACS Appl. Mater. Interfaces, 2018, 10(37), 31080–31087 CrossRef CAS.
- X. L. Fu, F. Hou, F. R. Liu, S. W. Ren and J. T. Cao, et al., Electrochemiluminescence energy resonance transfer in 2d/2d heterostructured g-C3N4/MnO2 for glutathione detection, Biosens. Bioelectron., 2019, 129, 72–78 CrossRef CAS.
- X. F. Zhao, P. K. Panda, D. Singh, X. Y. Yang and Y. K. Mishra, et al., 2d g-C3N4 monolayer for amino acids sequencing, Appl. Surf. Sci., 2020, 528, 146609 CrossRef CAS.
- G. Zuo, X. Zhou, Q. Huang, H. Fang and R. Zhou, Adsorption of villin headpiece onto graphene, carbon nanotube, and C60: effect of contacting surface curvatures on binding affinity, J. Phys. Chem. C, 2011, 115, 23323–23328 CrossRef CAS.
- X. C. Zhao, Self-assembly of DNA segments on graphene and carbon nanotube arrays in aqueous solution: a molecular simulation study, J. Phys. Chem. C, 2011, 115(14), 6181–6189 CrossRef CAS.
- Y. Tu, M. Lv, P. Xiu, T. Huynh and M. Zhang, et al., Destructive extraction of phospholipids from Escherichia coli membranes by graphene nanosheets, Nat. Nanotechnol., 2013, 8(8), 594–601 CrossRef CAS.
- B. Li, W. Li, J. M. Perez-Aguilar and R. Zhou, Mild binding of protein to C2N monolayer reveals its suitable biocompatibility, Small, 2017, 13(12), 1603685 CrossRef.
- Z. Gu, L. Zhao, S. Liu, G. Duan and J. M. Perez-Aguilar, et al., Orientational binding of DNA guided by the C2N template, ACS Nano, 2017, 11(3), 3198–3206 CrossRef CAS.
- Z. L. Gu, Z. X. Yang, L. L. Wang, H. Zhou and C. A. Jimenez-Cruz, et al., The role of basic residues in the adsorption of blood proteins onto the graphene surface, Sci. Rep., 2015, 5, 10873 CrossRef CAS.
- H. Y. Cui, Z. L. Gu, X. C. Chen, L. Lin and Z. G. Wang, et al., Stimulating antibacterial activities of graphitic carbon nitride nanosheets with plasma treatment, Nanoscale, 2019, 11(39), 18416–18425 RSC.
- Z. Gu, W. Song, S. H. Chen, B. Y. Li and W. F. Li, et al., Defect-assisted protein hp35 denaturation on graphene, Nanoscale, 2019, 11(41), 19362–19369 RSC.
- L. J. Beamer and C. O. Pabo, Refined 1.8 crystal structure of the λ repressor-operator complex, J. Mol. Biol., 1992, 227(1), 177–196 CrossRef CAS.
- J. Guo, X. Yao, L. Ning, Q. Wang and H. Liu, The adsorption mechanism and induced conformational changes of three typical proteins with different secondary structural features on graphene, RSC Adv., 2014, 4(20), 9953–9962 RSC.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation, J. Chem. Theory Comput., 2008, 4(3), 435–447 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, Vmd: Visual molecular dynamics, J. Mol. Graphics Modell., 1996, 14(1), 33–38 CrossRef CAS.
- J. Huang and A. D. MacKerell Jr, Charmm36 all-atom additive protein force field: validation based on comparison to NMR data, J. Comput. Chem., 2013, 34(25), 2135–2145 CrossRef CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79(2), 926–935 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126(1), 014101 CrossRef.
- M. Parrinello and A. Rahman, Polymorphic transitions in single-crystals – a new molecular-dynamics method, J. Appl. Phys., 1981, 52(12), 7182–7190 CrossRef CAS.
- Z. Gu, J. Perez-Aguilar, L. Meng and R. Zhou, Partial denaturation of villin headpiece upon binding to a carbon nitride polyaniline (C3N) nanosheet, J. Phys. Chem. B, 2020, 124(35), 7557–7563 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, Particle mesh ewald - an n.Log(n) method for ewald sums in large systems, J. Chem. Phys., 1993, 98(12), 10089–10092 CrossRef CAS.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden and H. Lee, et al., A smooth particle mesh ewald method, J. Chem. Phys., 1995, 103(19), 8577–8593 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. Fraaije, Lincs: a linear constraint solver for molecular simulations, J. Comput. Chem., 1997, 18(12), 1463–1472 CrossRef CAS.
- S. Miyamoto and P. A. Kollman, Settle – an analytical version of the shake and rattle algorithm for rigid water models, J. Comput. Chem., 1992, 13(8), 952–962 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10125g |
| This journal is © The Royal Society of Chemistry 2021 |