Open Access Article
Open Access ArticleOccurrence of disinfectant-resistant bacteria in a fresh-cut vegetables processing facility and their role in protecting Salmonella enteritidis†
Jing-Guo Xu,
Jiao Meng,
Wen-Jing Bao,
Jia-Mu Kang,
Jing-Yu Chen and
Bei-Zhong Han *
*
Beijing Laboratory for Food Quality and Safety, College of Food Science & Nutritional Engineering, China Agricultural University, Beijing, 100083, China. E-mail: hbz@cau.edu.cn
First published on 10th March 2021
Abstract
Chemical disinfectants are widely used to control foodborne pathogen contamination in fresh-cut vegetables (FVs) processing facilities. In this study, we investigated the disinfectant-resistant bacteria in a FVs processing facility and evaluate the effects of these bacteria on Salmonella enteritidis biofilm formation and disinfectant resistance. The disinfectant-resistance profiles were determined using 0.02% sodium hypochlorite (NaClO), 0.2% benzalkonium bromide (BAB) and 2% hydrogen peroxide (H2O2) solutions. The results showed the high occurrence of disinfectant resistant bacteria in the FVs processing environment, especially in the clean area. All isolates showed planktonic susceptibility to H2O2 and BAB, while the Gram-positive isolates were specifically resistant to NaClO. Isolates with biofilm-forming ability showed resistance to tested disinfectants. Disinfectant resistance of S. enteritidis was not significantly enhanced in most of the mixed-species biofilms, except for Bacillus paramycoides B5 which not only increased the biomass but also enhanced the survival ability of the Salmonella under NaClO treatment. Increased biomass and compact biofilm structures were observed in mixed-species biofilms by scanning electron microscopy (SEM). This study provides new insights into the disinfectant-resistant bacteria from food processing facilities and highlights their relevance for foodborne pathogen contamination.
1. Introduction
Fresh-cut vegetables (FVs) are minimally processed products of high quality, with nutritional properties, and are usually consumed in their raw form without further processing and cooking. However, FVs are vulnerable to foodborne pathogens during processing, as the cutting, peeling or other processing steps undermine the natural protective barriers of vegetables and result in the leakage of inner nutrients with water, which exposes the tissue to microbial contaminants.1 Recent studies have confirmed that FVs are potential risks for foodborne illness.2,3 Salmonella are the most important foodborne pathogens in various FVs products, and S. enteritidis was reported as the dominant serovar in sprouts and leaf greens.4,5Chemical disinfectants including NaClO, benzalkonium bromide as well as H2O2 have been widely used in washing process to lower microbial load on fresh produce. However, instability and low effectiveness of H2O2 and NaClO restrained their widely usage in fresh produce disinfection.6 Disinfectant-resistant strains are reported to persist in food processing environment due to the improper usage of these chemicals (e.g., sublethal doses and long-term exposure).7,8 Bacterial intrinsic traits including thick membrane, secreted substances and the drug efflux pump are likely to associate with the disinfectant resistance.9,10 Besides, the resistance of bacteria to disinfection is also associated with the presence of biofilms on surfaces. Biofilm with extracellular polymeric substances (EPS) may restrain disinfectants infiltration or neutralize their antibacterial ability.8,11 Most importantly, disinfectant-resistant strains that shows low susceptibility to such commonly used chemical biocides have profound effects on food safety, and they potentially protect the inadvertently contaminated foodborne pathogens during processing.12
Actually, mixed-species biofilms are predominant in food industry and usually show higher resistance to disinfectants compared to single-species ones.13 Studies have highlighted the importance and potential risks of mixed-species biofilms formed in various food processing facilities, including food catering service, dairy, fresh produce, aquatic products and meat processing environments.14,15 The interactive behavior in mixed-species biofilms shapes the biofilm structure and critically influences their stress resistance.16 The survival of foodborne pathogens in biofilms under disinfection treatment could be highly enhanced in the presence of other species from food processing facilities, usually with increased EPS production or interspecies communication.17,18
The disinfectant-resistant bacteria in FVs processing facility and their specific interactions with foodborne pathogens are still rather unexplored. In this study, we investigated the bacteria disinfectant-resistant profiles in this processing setting, and evaluated the effects of these bacteria on biofilm formation and disinfectant resistance of S. enteritidis.
2. Materials and methods
2.1 Food processing environmental samples collection
Samples were collected from a FVs processing facility located in Beijing, China, on December 13, 2018. Air samples were collected from different processing steps, including sorting, disinfection & cleaning, cutting and packaging, and storage, using the sedimentation method on Luria-Bertani (LB, Aoboxing Bio-Tech, Beijing, China) agar plates. At least three plates were settled in per workshop and left open for 20 min. Direct and indirect-contact surfaces to FVs products were sampled using the 3 M swab-sampler with 10 mL D/E Neutralizing Broth (3 M, USA, cat. number RS96010DE). Each area (10 cm by 10 cm) of floor, wall, equipment, utensil, and board from different processing steps was vigorously swabbed and scrapped 10 times at the horizontal and vertical directions. The adjacent area was collected as the duplicates. All samples were stored in an ice cooler (around 4 °C) and transported to the laboratory for analysis within 4 h.2.2 Bacteria isolation and identification
Airborne microorganism samples were directly incubated at 37 °C for 24 h. Swab samples were mixed at the maximum speed for 2 min. The homogenized sample solutions were 10-fold serially diluted with sterile saline solution (0.85% NaCl), and plated on LB agar plates in duplicates and incubated at 37 °C for 24 h. Colonies with different morphology on each plate were collected and individually transferred to new LB agar plates to obtain the pure cultures.Bacterial strain identification was conducted through 16S rRNA sequencing. Briefly, one loopful pure culture was transferred from the plate to LB broth (Aoboxing Bio-Tech, Beijing, China) and incubated overnight at 37 °C with shaking at 180 rpm min−1. Cultures were further centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min, and the pellets were washed with sterile saline. Genomic DNA was extracted using the TIANamp bacteria DNA kit (Tiangen Biotech, Beijing, China) following the manufacturer's protocol. A pair of universal primers (27F/1492R) was used to amplify the 16S rRNA gene.19 The PCR amplifying products were electrophoresed on 1% agarose gel and imaged using the Bio-Rad Imaging System (Bio-Rad, USA). Sequencing was performed using Sanger's method (Beijing Genomic Institute, Beijing, China). The sequences were compared with published 16S rRNA sequences on EzBioCloud 16S database (Chun's Lab, https://www.ezbiocloud.net). The 16S rRNA sequences of bacteria identified in this study were deposited in GenBank under the accession numbers MT704506-MT704546.
000 × g for 5 min, and the pellets were washed with sterile saline. Genomic DNA was extracted using the TIANamp bacteria DNA kit (Tiangen Biotech, Beijing, China) following the manufacturer's protocol. A pair of universal primers (27F/1492R) was used to amplify the 16S rRNA gene.19 The PCR amplifying products were electrophoresed on 1% agarose gel and imaged using the Bio-Rad Imaging System (Bio-Rad, USA). Sequencing was performed using Sanger's method (Beijing Genomic Institute, Beijing, China). The sequences were compared with published 16S rRNA sequences on EzBioCloud 16S database (Chun's Lab, https://www.ezbiocloud.net). The 16S rRNA sequences of bacteria identified in this study were deposited in GenBank under the accession numbers MT704506-MT704546.
2.3 Inoculum preparation for disinfection test and biofilm formation
The Salmonella enteritidis S01 strain was isolated from vegetable salads, and kindly provided by Professor S. H. Cui from the National Institutes for Food and Drug Control of China. Bacterial isolates from the FVs processing plant and S. enteritidis S01 were applied to the disinfection challenges and mixed-species biofilms formation. Inoculum was prepared as following: one loop of pure bacteria colony was transferred into LB broth and incubated at 37 °C overnight. Cultures were centrifuged at 2500 × g for 10 min, and the pellets were diluted with 1/2 dilution of LB broth at the optical density (OD) of 600 nm to 0.5 using a UV-vis spectrophotometer (UNICO, USA). The bacterial inoculum contained approximately ∼107 CFU mL−1 populations of each strain.2.4 Biofilm formation in polystyrene microtiter plates
Biofilms were prepared in 96-well polystyrene plates (Corning, USA, cat. number REF3599) for biomass quantification and disinfectant challenges.20 For mono-species biofilms, inoculum was directly added into each well; while in mixed-species biofilms, the inoculum of each isolate was mixed with the equivalent volume of S. enteritidis S01. The 96-well plates containing inoculum were statically incubated at 25 °C for 48 h. Each experiment was performed in six independent wells, and the broth-only wells were used as negative control.2.5 Biofilm formation on stainless-steel coupons
The mixed-species biofilms were also established on 304 stainless-steel (SS) coupons (1 cm × 1 cm × 0.1 cm) to simulate the commonly-used contact surface in food processing facility.21 Before use, coupons were immersed overnight in acetone, then rinsed with water and autoclaved at 121 °C for 15 min.22 Coupons were immersed into the 48-well polystyrene plates (Corning, USA, cat. number REF3548) filled with mono- or mixed-species suspensions. For mono-species suspensions, inoculum was directly added into each well, while the mixture of equivalent volume of each strain inoculum was as the mixed-species suspensions. Coupons were statically incubated at 25 °C for 48 h. The broth-only wells were used as negative control. Each experiment was performed in triplicates.2.6 Disinfectant challenge test
Disinfectants of NaClO, BAB and H2O2 (Macklin, Shanghai, China) were dissolved in sterile water to the recommended concentrations: NaClO at 0.02% (200 ppm), BAB at 0.2%, and H2O2 at 2%. Bacterial isolates in planktonic and biofilm states were all subjected to disinfectant treatments.The planktonic suspension was mixed with an equal volume of each of the three disinfectant solutions and challenged for 20 min at 25 °C. Disinfectant activity was quenched with Dey-Engley (D/E) neutralizing broth (Solarbio, Beijing, China) for 10 min, and then suspensions were transferred into 96 wells plates (100 μL per well). An equivalent volume of LB broth was added into the wells and further incubated at 37 °C for 24 h. OD600 nm values were measured using the Multiskan plates reader (Thermal Fisher Scientific Inc, USA). Experiment without disinfection treatment was used as control. Each experiment was performed in triplicates. Strain with the OD600 nm values exceed three times than that of blank control was defined as resistant bacteria.
Disinfectant treatments for the biofilms were conducted as Chorianopoulos et al. with modifications.21 Firstly, biofilms were rinsed three times with saline and then immersed in disinfectant solutions and treated at 25 °C for 20 min. Solutions were removed and the biofilms were washed twice with saline. The disinfectant resistance of biofilms formed in 96-well plates were evaluated using the tetrazolium salt (MTT) assay.23 MTT Assay Kit M1020 (Solarbio, Beijing, China) was used and performed according to the manufacturer's instructions. The disinfectant resistance of biofilms on SS coupons were determined by colony enumeration. Each experiment was performed in triplicates.
2.7 Crystal violet staining
Biomass were evaluated by crystal violet (CV) staining method as described by Burmolle et al. with some modifications.19 Briefly, biofilms in 96-well plates were rinsed three times with saline and fixed with anhydrous methanol for 15 min. After that, the biofilms were stained with 0.1% CV solution (Solarbio, Beijing, China) for 10 min and rinsed carefully with tap water. Residual CV was dissolved in 33% acetic acid and measured using the Multiskan plates reader at OD595 nm.2.8 Colony enumeration
The presence of S. enteritidis S01 in the mixed-species biofilms cultured on SS coupons was confirmed by colony enumeration method. Ciprofloxacin (Solarbio, Beijing, China) at the concentration of 10 μg mL−1 was used for S. enteritidis S01 selective culture according to the antibiotic susceptibility testing. Briefly, biofilms on SS coupons were sampled using sterile swab. Swabs were immersed into 3 mL saline and vortexed for 2 min. Dilutions were plated on LB agar supplemented with 10 μg mL−1 ciprofloxacin and incubated at 37 °C for 24 h. The presumptive S. enteritidis S01 colonies were confirmed by PCR test for the Salmonella specific invA gene.242.9 Scanning electron microscopy (SEM) observation
The spatial biofilm structure of S. enteritidis S01 and Bacillus paramycoides B-5 was observed by SEM. Biofilms on SS coupons were rinsed and fixed with 2.5% glutaraldehyde (Solarbio, Beijing, China) overnight. After that, biofilms were progressively dehydrated with graded ethanol solutions (30%, 50%, 70%, 90%, and 100%) and air drying. Before observation, coupons containing biofilms were sputter-coated with gold (IXRF Systems-Model MSP-2S, USA). SEM observation was performed on a tabletop microscope device (Hitachi TM3030, Japan).252.10 Statistical analysis
Colony Forming Units (CFU) were converted to log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU per coupon. Data were normalized with the Z-score method and demonstrated as heatmap to facilitate the comparison between different treatments using the ClustVis online tools (https://biit.cs.ut.ee/clustvis/). Significant difference was determined as p < 0.05 with the Student's t-test (IBM SPSS Statistics).
CFU per coupon. Data were normalized with the Z-score method and demonstrated as heatmap to facilitate the comparison between different treatments using the ClustVis online tools (https://biit.cs.ut.ee/clustvis/). Significant difference was determined as p < 0.05 with the Student's t-test (IBM SPSS Statistics).
3. Results
3.1 Bacterial isolates from the processing environment
Forty bacteria were isolated and identified (Table 1). The isolates were classified into 17 genera, with 13 isolates from processing surfaces, 7 from air, and 5 in both the two sites. The prevalent genera were Bacillus spp. (9 isolates), Microbacterium spp. (6 isolates), and Cellulosimicrobium spp. (4 isolates). Bacillus were isolated from both air and contact surface samples, while Microbacterium and Cellulosimicrobium were only detected in contact surfaces. Processing steps of sorting and disinfection & washing had the most abundant strains, with 11 and 18 bacterial isolates respectively; while in cutting and packaging rooms, generally classified as clean area, there were reduced strains of bacteria.| Processing steps | Sampling location | Isolatesa | Most similar strainb | Similarity % |
|---|---|---|---|---|
| a “K” represents the isolates from air, and “B” represents the isolates from contact surfaces.b Sequences with similarity above 97% were identified as the most similar species. Sequences alignment was performed on EzBioCloud 16S database on Dec. 18, 2019. | ||||
| Sorting | Air | K6 | Bacillus tequilensis (KCTC 13429) | 99.93 |
| K7 | Exiguobacterium acetylicum (DSM 20416) | 99.71 | ||
| K8 | Fictibacillus arsenicus (Con a/3) | 100 | ||
| K13 | Rothia marina (JSM 078151) | 100 | ||
| K1 | Aerococcus urinaeequi (CCUG 28094) | 100 | ||
| Cutting board | B14 | Corynebacterium callunae (DSM 20147) | 100 | |
| B18 | Microbacterium arborescens (DSM 20754) | 100 | ||
| Knife | B19 | Microbacterium arborescens (DSM 20754) | 99.92 | |
| Trimming table | B7 | Buttiauxella ferragutiae (ATCC 51602) | 99.61 | |
| B13 | Cellulosimicrobium funkei (ATCC BAA-886) | 99.78 | ||
| B20 | Microbacterium arborescens (DSM 20754) | 99.85 | ||
| Disinfection & washing | Air | K2 | Bacillus altitudinis (41KF2b) | 100 |
| K4 | Bacillus halotolerans (ATCC 25096) | 100 | ||
| K9 | Kocuria carniphila (CCM 132) | 99.78 | ||
| K10 | Kocuria palustris (DSM 11925) | 100 | ||
| K12 | Pseudomonas kribbensis (46-2) | 99.93 | ||
| Dryer | B17 | Kocuria rosea (DSM 20447) | 99.85 | |
| Conveyor belt | B3 | Bacillus altitudinis (41KF2b) | 100 | |
| B4 | Bacillus paramycoides (NH24A2) | 99.93 | ||
| B10 | Cellulosimicrobium cellulans (LMG 16121) | 100 | ||
| B15 | Escherichia fergusonii (ATCC 35469) | 99.93 | ||
| Drain | B8 | Buttiauxella agrestis (ATCC 33320) | 99.61 | |
| B9 | Cellulomonas pakistanensis (JCM 18755) | 99.7 | ||
| Tools shelf | B6 | Bacillus tequilensis (KCTC 13429) | 99.93 | |
| B16 | Exiguobacterium sibiricum (255-15) | 99.85 | ||
| B27 | Planococcus rifietoensis (M8) | 100 | ||
| Washing sink | B11 | Cellulosimicrobium cellulans (LMG 16121) | 99.63 | |
| B24 | Pantoea pleuroti (DSM 3493) | 99.92 | ||
| B25 | Pantoea dispersa (LMG 2603) | 99.85 | ||
| Cutting | Air | K5 | Bacillus paralicheniformis (KJ-16) | 100 |
| K11 | Pantoea vagans (LMG 24199) | 98.95 | ||
| Conveyor belt | B23 | Microbacterium arborescens (DSM 20754) | 99.85 | |
| B28 | Serratia glossinae (C1) | 99.85 | ||
| Soaking bucket | B1 | Aeromonas media (CECT 4232) | 99.85 | |
| Basket | B5 | Bacillus paramycoides (NH24A2) | 100 | |
| Packaging | Air | K3 | Bacillus altitudinis (41KF2b) | 100 |
| Basket | B21 | Microbacterium arborescens (DSM 20754) | 99.92 | |
| Wall | B29 | Serratia plymuthica (DSM 4540) | 99.42 | |
| Conveyor belt | B12 | Cellulosimicrobium cellulans (LMG 16121) | 99.85 | |
| B22 | Microbacterium arborescens (DSM 20754) | 99.93 | ||
3.2 Disinfectant-resistance profiles of isolates
The disinfectant-resistance profiles of isolates in planktonic state are shown in Fig. 1. Results showed that all isolates were susceptible to BAB and H2O2, while 15 out of 40 isolates were resistant to NaClO. The occurrence of resistant bacteria in sorting (5/11) and disinfection & washing (7/18) were rather higher than cutting (2/6), and packaging room (1/5) had the lowest proportion of resistant bacteria. As for sampling location, isolates from air were much more resistant (8/13) than strains from direct (5/20) and indirect-contact (2/7) surfaces. The differences of disinfectant resistance between species were distinct. Strains belonging to Bacillus spp., such as B. altitudinis B3/K2, B. halotolerans K4, B. paramycoides B5, B. paralicheniformis K5, and B. tequilensis K6 were mostly resistant to NaClO. All Microbacterium spp. were susceptible to the tested disinfectants.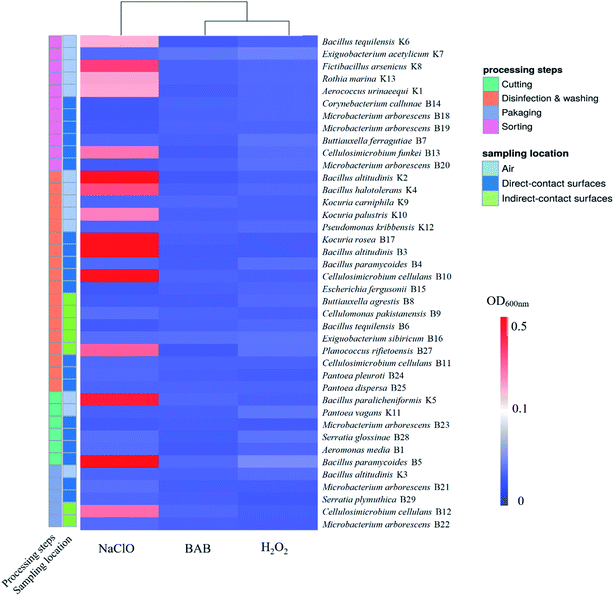 | ||
| Fig. 1 Disinfectant-resistance profiles of isolates in the planktonic state (n = 6). Each bacterial suspension was treated with disinfectant solutions (0.02% NaClO, 0.2% BAB, and 2% H2O2) for 20 min. | ||
The disinfectant-resistance profiles of the biofilm-forming strains are shown in Fig. 2. Seventeen isolates showed obvious biofilm-forming ability. E. fergusonii B15 biofilm cells were resistance to all tested disinfectants although its planktonic cells were susceptible (Fig. 1). The disinfectant resistance of biofilm bacteria was not directly related with their biofilm-forming ability. For example, R. marina K13 had the highest biofilm biomass but susceptible to disinfectant, while the weak biofilm-forming isolate E. fergusonii B15 was the most resistant strain with 43.58% residual metabolic ability after H2O2 treatment. The disinfectant resistance profiles were quite different for the same strain. B. agrestis B8, for example, was susceptible to BAB and H2O2 but resistant to NaClO. Overall, the disinfectant-resistance profiles of bacterial isolates were diverse in the fresh-cut vegetable processing facility.
3.3 Biomass and disinfectant resistance of mixed-species biofilms
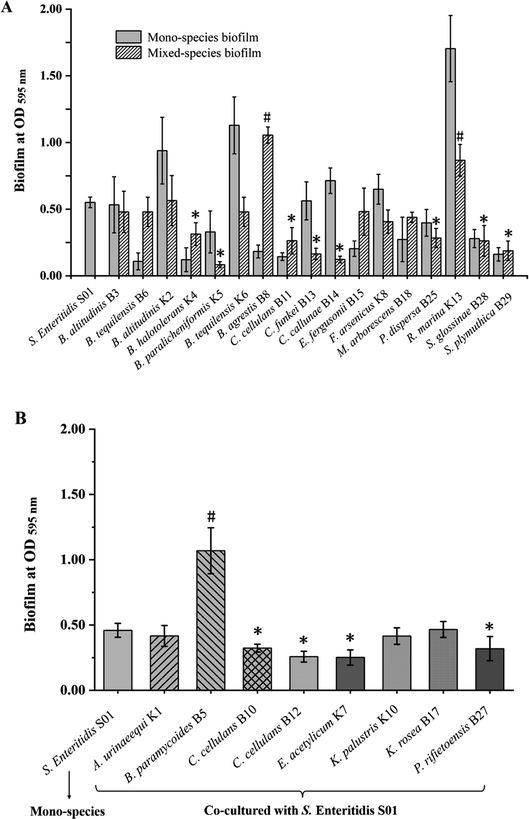
Mixed-species biofilms biomass formed by the biofilm-forming isolates and S. enteritidis S01 are shown in Fig. 3A. B. agrestis B8 and R. marina K13 showed the positive effects for biomass production, as the biomass of mixed-species biofilms were much higher (p < 0.05) than S. enteritidis S01 single-species biofilm. Isolates with weak biofilm-forming ability like B. halotolerans K4, B. paralicheniformis K5 and C. cellulans B11, had negative effects on mixed-species biofilms to S. enteritidis S01, as the biomass was significantly lower (p < 0.05) than that of the S. enteritidis S01 alone. In general, the disinfectant resistant abilities of mixed-species biofilm were impaired compared with their single-species counterparts (ESI Fig. S1†). The most resistant mixed-species biofilm was formed by S. plymuthica B29 and S. enteritidis S01 under BAB challenge, with approximately 5.5% of residual metabolic activity using MTT assay. However, S. enteritidis S01 was no detected Salmonella colonies (<2.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU per coupon) in most of the mixed-species biofilms under disinfectant treatment.
CFU per coupon) in most of the mixed-species biofilms under disinfectant treatment.
Moreover, biomass of mixed-species biofilms formed by the isolates with NaClO resistance but no biofilm-forming ability with S. enteritidis S01 were also tested. The results are shown in Fig. 3B. The mixed-species biofilm biomass of S. enteritidis S01 and B. paramycoides B5 was significantly higher (p < 0.05) than S. enteritidis S01 single-species biofilm, while C. cellulans B-10/B-12, E. acetylicum K7, and P. rifietoensis B27 had negative effects (p < 0.05) on mixed-species biofilm formation. Disinfectant resistance of S. enteritidis was not significantly enhanced in most of the mixed-species biofilms, except for Bacillus paramycoides B5 which significantly increased the Salmonella survival ability in mixed-species biofilm with the residue of 3.51 ± 0.22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU per coupon after 200 ppm NaClO treatment, while no detectable S. enteritidis S01 was found in its single-species biofilms under the same concentration of NaClO.
CFU per coupon after 200 ppm NaClO treatment, while no detectable S. enteritidis S01 was found in its single-species biofilms under the same concentration of NaClO.
3.4 Mixed-species biofilm formed by B. paramycoides B5 and S. enteritidis S01
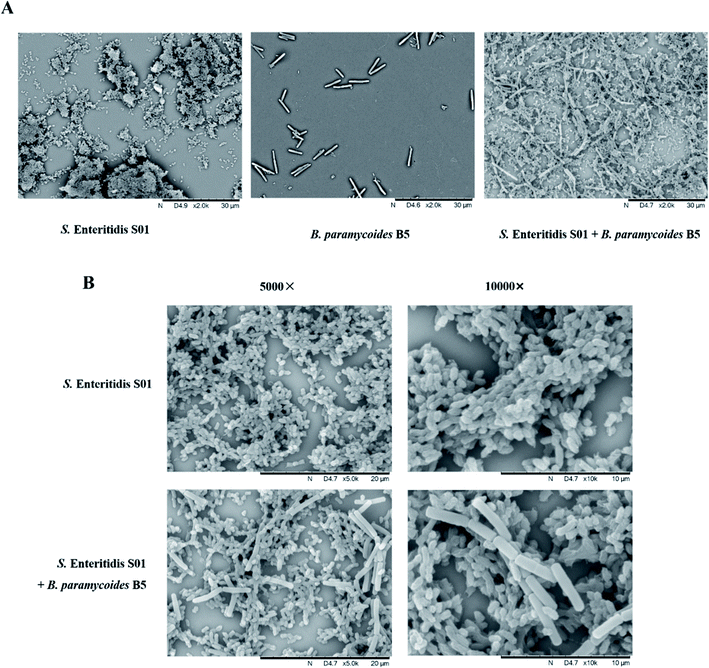
Scanning Electron Microscopy (SEM) showed that S. enteritidis S01 single-species culture formed a sparse and loose biofilm structure, while there were no detectable biofilm structures formed by B. paramycoides B5 (Fig. 4A). Mixed-species biofilms of S. enteritidis S01 and B. paramycoides B5 were widely distributed and covered the stainless steel with more compact biofilm structure than the Salmonella single-species biofilm. B. paramycoides B5 cells were directly interacted with S. enteritidis S01 by intercellular contact and embedded in the cell clusters (Fig. 4A). B. paramycoides B5 cell membrane remained intact after NaClO treatment. The colony enumeration method revealed the survival of Salmonella (3.51 ± 0.22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU per coupon) in mixed-species biofilm under NaClO treatment, although no intact S. enteritidis S01 cells were observed in the multi-species biofilms (Fig. 4B).
CFU per coupon) in mixed-species biofilm under NaClO treatment, although no intact S. enteritidis S01 cells were observed in the multi-species biofilms (Fig. 4B).
4. Discussion
Disinfection procedure in food processing facilities poses the selective pressures to resident or occasionally invaded pathogens by the sublethal doses and long-term exposure of these chemicals.7 Bacteria survived in these environments usually activated or evolved specific mechanisms to response the stresses.8–10 However, the disinfectant-resistance profiles of isolates especially those of the generally considered harmless microbiota in the food processing environment are scarcely reported. Notably, our results indicated the widely distribution of disinfectant resistant bacteria occurred in the processing facility and revealed the potential protection effects of these resistant bacteria to the commonly outbreak pathogen S. enteritidis.The pattern of disinfectant resistance of bacteria was quite different between processing steps, probably the different cleanliness requirements of these area. Processing steps of sorting and disinfection & washing had the most abundant strains, while in cutting and packaging steps which generally classified as clean area, there were reduced strains of bacteria. In this fresh-cut vegetable processing facility, chlorine-based disinfectants are commonly used for product contact surfaces, and high frequency of disinfection (1–2 times one day) and high dose of disinfectant (200 ppm) usually applied to the clean area. Although all isolates showed planktonic susceptibility to H2O2 and BAB, the NaClO resistance was more variable. Besides, contact surfaces from disinfection & washing are constantly interacted with washing water containing chlorine, strains survived in these surfaces are more likely to activate the intrinsic or evolve the new stress response mechanisms to disinfectants.12 Similarly, the occurrence of NaClO resistant bacteria was higher in sorting and disinfection & washing steps (12/29) than clean area (3/11). Importantly, strains belong to Microbacterium arborescens were observed in cutting board, knife and trimming table of sorting room, conveyor belt of cutting room, as well as basket and conveyor belt of packaging room. Cellulosimicrobium cellulans was proved to be the disinfectant resistant species, and found in both clean (cutting) and non-clean (disinfection & washing) area. These results indicated the potential transmission ability of some specific strains along the food processing lines. However, to date, transmission routes of microorganisms during food processing are still elusive and very hard to track or monitor and therefore more efforts are still to be made.
The pattern of bacteria disinfectant resistance was quite different between planktonic and biofilm states. Isolates of single species that belong to Gram-positive bacteria showed more stress resistance than Gram-negative ones in their planktonic state for their relatively thick cell walls that tolerate adverse stresses. Many studies demonstrated that Gram-negative bacteria were stress resistant and widely distributed in various environment.9,26,27 Besides, the efficacy of BAB and H2O2 against isolates was mostly compromised in the biofilm state, for example the biofilm cells of E. fergusonii B15 showed significant resistance to all tested disinfectants though its planktonic cells were disinfectant susceptible. The findings of biofilm-specific stress resistance are supported by numerous reports in the literature which demonstrated that the disinfectant resistance of bacteria is closely associated with their biofilm forming ability on surfaces.8,13 Unlike the planktonic state, bacteria cells in the biofilm state have various traits, including physiological status change, extracellular matrix, drug efflux pumps, and gene regulation, which can be associated with the biofilm-specific disinfectant resistance.28–31 However, the biofilm biomass were related but not the only factor that affects the bacteria disinfectant resistance. For example, R. marina K13 had the highest biofilm biomass but susceptible to disinfectant, while the weak biofilm-forming isolate E. fergusonii B15 was the most resistant strain (Fig. 2). These results are likely to be associated with the different development characteristics and biofilm physiologies of the various strains, though no further biofilm structure analysis were conducted in this study.
Bacterial microbiota in food processing environments usually involved multispecies and interspecies interactions.32,33 Ours results demonstrated that the biomass and disinfectant resistance were variable in mixed-species biofilms formed by resistant isolates and pathogen S. enteritidis (Fig. 3 and S1†). Specifically, the disinfectant resistant abilities of mixed-species biofilm were impaired compared with the disinfectant resistant single-species counterparts, but more resistant than S. enteritidis ones. The biomass and physiologies of the disinfectant resistant bacteria biofilm may be deteriorated by the introduction of S. enteritidis, indicating the antagonistic interactions between them. However, in the other hand, the findings also indicated that a mixed bacteria community provide some additional barrier for disinfectants diffusion that different from single-species biofilms, for which the survival of pathogen S. enteritidis was observed in mixed-species formed with B. paramycoides B5, indicating the synergistic interaction between these two species. The characteristics of morphologically distinct microcolonies were observed by SEM, B. paramycoides B5 and S. enteritidis cells were intercellular contacted in biofilm clusters and formed more biomass and compact biofilm structures than S. enteritidis single-species biofilm. This indicated that the increased NaClO resistance potentially due to the physiological change and intercellular interactions in mixed-species biofilm that is various from that of individual biofilm. Other studies have reported the enhanced stress resistance of Salmonella that induced by interspecies interactions.34,35
Overall, our results suggest that the high occurrence of disinfectant resistant bacteria in FVs processing environment, especially the clean area. Whether or not these resistant bacteria strains pose a health risk to humans, stringent disinfection measures should be made to prevent these microorganisms from becoming the potential reservoirs for drug resistance gene transfer and inadvertently contaminated foodborne pathogens contamination.
5. Conclusions
In summary, our results suggest the occurrence of disinfectant-resistant bacteria in the FVs processing facility. All isolates showed planktonic susceptibility to H2O2 and BAB, while the Gram-positive isolates were specifically resistant to NaClO. Isolates with biofilm-forming ability showed resistance to tested disinfectants. Besides, we prove that disinfectant resistant bacteria, for example B. paramycoides B5, involved in enhanced biofilm formation and disinfectant resistance of S. enteritidis in mixed-species biofilm.Author contributions
Jing-Guo Xu: conceptualization, formal analysis, writing-original draft, visualization. Jiao Meng: data curation, writing-review & editing. Wen-Jing Bao: investigation. Jia-mu Kang: writing-review & editing. Jing-Yu Chen: project administration, writing-review & editing. Bei-Zhong Han: funding acquisition, resources.Conflicts of interest
The authors declare that they have no competing conflicts of interest.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2017YFC1600806-1), Chinese Universities Scientific Fund (2020TC116) and Research Innovation Fund for Graduate Students of China Agricultural University (2020XYZC36A).References
- Sarengaowa, W. Z. Hu, K. Feng, Z. L. Xiu, A. L. Jiang and Y. Lao, Int. J. Food Sci. Technol., 2019, 54, 3128–3137 CrossRef CAS.
- C. K. Carstens, J. K. Salazar and C. Darkoh, Front. Microbiol., 2019, 10, 2667 CrossRef PubMed.
- Y. Wadamori, R. Gooneratne and M. A. Hussain, J. Sci. Food Agric., 2017, 97, 1396–1403 CrossRef CAS PubMed.
- A. N. Olaimat and R. A. Holley, Food Microbiol., 2012, 32, 1–19 CrossRef CAS PubMed.
- C. N. Berger, S. V. Sodha, R. K. Shaw, P. M. Griffin, D. Pink, P. Hand and G. Frankel, Environ. Microbiol., 2010, 12, 2385–2397 CrossRef PubMed.
- S. Van Haute, I. Tryland, A. Veys and I. Sampers, Food Control, 2015, 50, 173–183 CrossRef CAS.
- S. Wessels and H. Ingmer, Regul. Toxicol. Pharmacol., 2013, 67, 456–467 CrossRef CAS PubMed.
- A. Bridier, R. Briandet, V. Thomas and F. Dubois-Brissonnet, Biofouling, 2011, 27, 1017–1032 CrossRef CAS PubMed.
- A. Alonso-Hernando, C. Alonso-Calleja and R. Capita, Int. J. Food Microbiol., 2010, 137, 130–136 CrossRef CAS PubMed.
- J. A. Imlay, R. Sethu and S. K. Rohaun, Free Radical Biol. Med., 2019, 140, 4–13 CrossRef CAS PubMed.
- T. Beaudoin, L. Zhang, A. J. Hinz, C. J. Parr and T. F. Mah, J. Bacteriol., 2012, 194, 3128–3136 CrossRef CAS PubMed.
- G. Y. Gu, A. Ottesen, S. Bolten, L. Wang, Y. G. Luo, S. Rideout, S. Lyu and X. W. Nou, Int. J. Food Microbiol., 2019, 294, 31–41 CrossRef CAS PubMed.
- L. Yuan, M. F. Hansen, H. L. Roder, N. Wang, M. Burmolle and G. He, Crit. Rev. Food Sci. Nutr., 2020, 60, 2277–2293 CrossRef PubMed.
- G. S. Oliveira, D. R. G. Lopes, C. Andre, C. C. Silva, F. Bagliniere and M. C. D. Vanetti, Biofouling, 2019, 35, 819–831 CrossRef CAS PubMed.
- V. P. P. Alonso, A. M. M. Harada and D. Y. Kabuki, Front. Microbiol., 2020, 11, 177 CrossRef PubMed.
- C. D. Nadell, K. Drescher and K. R. Foster, Nat. Rev. Microbiol., 2016, 14, 589–600 CrossRef CAS PubMed.
- S. Elias and E. Banin, FEMS Microbiol. Rev., 2012, 36, 990–1004 CrossRef CAS PubMed.
- X. Y. Pang, Y. S. Yang and H. G. Yuk, J. Appl. Microbiol., 2017, 123, 651–660 CrossRef CAS PubMed.
- M. Burmolle, J. S. Webb, D. Rao, L. H. Hansen, S. J. Sorensen and S. Kjelleberg, Appl. Environ. Microbiol., 2006, 72, 3916–3923 CrossRef PubMed.
- R. Wang, N. Kalchayanand, J. W. Schmidt and D. M. Harhay, J. Food Prot., 2013, 76, 1513–1522 CrossRef PubMed.
- N. G. Chorianopoulos, E. D. Giaouris, P. N. Skandamis, S. A. Haroutounian and G. J. Nychas, J. Appl. Microbiol., 2008, 104, 1586–1596 CrossRef CAS PubMed.
- M. Kostaki, N. Chorianopoulos, E. Braxou, G.-J. Nychas and E. Giaouris, Appl. Environ. Microbiol., 2012, 78, 2586–2595 CrossRef CAS PubMed.
- E. A. Trafny, R. Lewandowski, I. Zawistowska-Marciniak and M. Stępińska, World J. Microbiol. Biotechnol., 2013, 29, 1635–1643 CrossRef CAS PubMed.
- K. Rahn, S. A. De Grandis, R. C. Clarke, S. A. McEwen, J. E. Galán, C. Ginocchio, R. Curtiss and C. L. Gyles, Mol. Cell. Probes, 1992, 6, 271–279 CrossRef CAS PubMed.
- F. Lopez-Galvez, M. I. Gil, P. Truchado, M. V. Selma and A. Allende, Food Microbiol., 2010, 27, 199–204 CrossRef CAS PubMed.
- A. G. Ríos-Castillo, F. F. Umaña and J. J. Rodríguez-Jerez, Food Control, 2018, 93, 219–225 CrossRef.
- M. A. Fernández-Fuentes, H. Abriouel, E. Ortega Morente, R. Pérez Pulido and A. Gálvez, Int. J. Food Microbiol., 2014, 172, 49–56 CrossRef PubMed.
- L. Lavilla Lerma, N. Benomar, A. Gálvez and H. Abriouel, Int. J. Food Microbiol., 2013, 161, 97–106 CrossRef CAS PubMed.
- A. Bridier, P. Sanchez-Vizuete, M. Guilbaud, J. C. Piard, M. Naitali and R. Briandet, Food Microbiol., 2015, 45, 167–178 CrossRef CAS PubMed.
- L. Luque-Sastre, E. M. Fox, K. Jordan and S. Fanning, J. Food Prot., 2018, 81, 1481–1490 CrossRef CAS PubMed.
- M. Cadena, T. Kelman, M. L. Marco and M. Pitesky, Foods, 2019, 8(7), 275 CrossRef CAS PubMed.
- H. L. Roder, P. K. Raghupathi, J. Herschend, A. Brejnrod, S. Knochel, S. J. Sorensen and M. Burmolle, Food Microbiol., 2015, 51, 18–24 CrossRef PubMed.
- S. Mitri, J. B. Xavier and K. R. Foster, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(suppl 2), 10839–10846 CrossRef CAS PubMed.
- M. Schwering, J. Song, M. Louie, R. J. Turner and H. Ceri, Biofouling, 2013, 29, 917–928 CrossRef CAS PubMed.
- E. Heir, T. Moretro, A. Simensen and S. Langsrud, Int. J. Food Microbiol., 2018, 275, 46–55 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09325d |
| This journal is © The Royal Society of Chemistry 2021 |