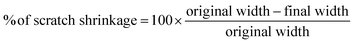Open Access Article
Open Access ArticleDevelopment of bactericidal spinel ferrite nanoparticles with effective biocompatibility for potential wound healing applications
Atiya Rabbania,
Reihaneh Haghniazb,
Taous Khanc,
Romana Khand,
Ayesha Khalida,
Syeda Sohaila Naze,
Mazhar Ul-Islamf,
Fereshteh Vajhading and
Fazli Wahid *ah
*ah
aDepartment of Biotechnology, COMSATS University Islamabad, Abbottabad Campus, Pakistan
bKhademhosseini's Laboratory, Center for Minimally Invasive Therapeutics (CMIT) California NanoSystems Institute, University of California Los Angles, Los Angles, USA
cDepartment of Pharmacy, COMSATS University Islamabad, Abbottabad Campus, Pakistan
dDepartment of Environmental Sciences, COMSATS University Islamabad, Abbottabad Campus, Pakistan
eDepartment of Nanosciences and Technology, National Centre for Physics, Islamabad, Pakistan
fDepartment of Chemical Engineering, College of Engineering, Dhofar University, Salalah, Oman
gDepartment of Chemistry, Faculty of Science, Yazd University, Iran
hDepartment of Biomedical Sciences, Pak-Austria Fachhochschule: Institute of Applied Sciences and Technology, Mang, Khanpur Road, Haripur, Pakistan. E-mail: fazli.wahid@fbse.paf-iast.edu.pk
First published on 6th January 2021
Abstract
The current study was devised to explore the antibacterial activity and underlying mechanism of spinel ferrite nanoparticles (NPs) along with their biocompatibility and wound healing potentials. In this regard, nickel ferrite and zinc/nickel ferrite NPs were synthesized via a modified co-precipitation method and were characterized by X-ray diffraction (XRD), scanning electron microscopy (SEM) and energy Energy-dispersive X-ray spectroscopy (EDX). The biocompatibility of the synthesized NPs with human dermal fibroblast (HDF) and red blood cells (RBCs) was assessed. The biocompatible concentrations of the NPs were used to investigate the antimicrobial activity against various pathogenic Gram-negative and Gram-positive bacteria. The mode of bactericidal action was also explored. In vitro scratch assay was performed to evaluate the wound healing potential of NPs. The SEM-EDX analysis showed that the average particles size of nickel ferrite and zinc/nickel ferrite were 49 and 46 nm, respectively, with appropriate elemental composition and homogenous distribution. The XRD pattern showed all the characteristic diffraction peaks of spinel ferrite NPs, which confirmed the synthesis of the pure phase cubic spinel structure. The biocompatible concentration of nickel ferrite and zinc/nickel ferrite NPs was found to be 250 and 125 μg ml−1, respectively. Both the NPs showed inhibition against all the selected strains in the concentration range of 50 to 1000 μg ml−1. Studies on the underlying antimicrobial mechanism revealed damage to the cell membrane, protein leakage, and intracellular reactive oxygen species production. The in vitro scratch assay confirmed the migration and proliferation of fibroblast with artificial wound shrinkage. This study shows that nickel ferrite and zinc/nickel ferrite NPs could be a strong candidate for antibacterial and wound healing nano-drugs.
1. Introduction
The rise of antibiotic resistance is becoming a global public health crisis.1 Although, the misuse of antibiotics for the treatment of human diseases is the major cause of the problem, the overuse of antimicrobial agents in veterinary medicines, agriculture and industry is also worsening the situation. The use of antibiotics in poultry and livestock industries for the prevention of fatal infections in chickens, cattle, pigs, sea food and farmed fishes is a common practice worldwide.2–4 This excessive use of antibiotics has led to mutations and possibly to resistance in bacterial strains.5 This emerging health threat has highlighted the urgent need for formulating innovative antimicrobial agents for the control of infections.6 In this context, the field of nanotechnology has rapidly evolved as a new approach to deal with the complexity of antibiotic resistance.7 Nanotechnology based materials such as nanoparticles (NPs) have displayed immense potential to combat the microbial infection more effectively as compared to conventional antibiotics.8 Among these materials, metal and metal oxide NPs are attracting much attention, as several studies suggested their superior activity toward resistant microorganisms.9 The most comprehensively studied particles having superior antimicrobial activity includes silver, gold, zinc oxide, copper, copper oxide, titanium dioxide and iron oxide NPs.10–13Spinel ferrite (SF) are ferromagnetic compounds that are usually oxides of various transition metals, containing iron e.g. ZnFe2O4, NiFe2O4, Zn/NiFe2O4, CoFe2O4, MnFe2O4, and MgFe2O4.13–15 These NPs have unique optical, electrical and magnetic properties and can easily be magnetized or demagnetized due to their soft and insulating nature. SF NPs are mostly used as magnetic, refractory and catalytic materials.16 Ferrites are used in diverse technological applications such as microelectronics, humidity sensors, microwave reflections, drug delivery system and magnetic resonance imaging (MRI).17 Moreover, ferrites are also utilized in high density data storage materials, catalyst, converters and antenna frames.14 Conjugation of ferrites and some divalent metallic ions dramatically enhance the properties of ferrites.18 The addition of metal ion to ferrite strengthens its property of coercivity.19 NiFe2O4 (NF) and Zn–NiFe2O4 (ZNF) NPs have gained much attention due to their diverse applications. NF NPs are applied in magnetic extraction, MRI, cell labeling, drug delivery and magnetic hyperthermia.20 Likewise, Zn substituted nickel SF NPs have gained much interest due to modifiable magnetic parameters, high electromagnetic performance, and good chemical stability. Similarly, ZNF NPs have been employed in magnetic hyperthermia, MRI, drug delivery and magnetic extraction.17
Although well-reported for above mentioned potentials, substantial efforts are needed regarding its medical applications and biological activities specifically the bactericidal mechanism, biocompatibility, and wound healing abilities. It has been proposed that the existing biological features can be improved by incorporating transition metals into ferrite NPs.21 Linking to the stated concept, we aimed to synthesize multipurpose NF and ZNF NPs and to observe their biocompatibility and detailed bactericidal mechanism. Biocompatibility was analyzed against human dermal fibroblast and red blood cells, whereas the mechanisms of bactericidal action were established against Gram-positive and Gram-negative strains through multiple analytical techniques. We hope that this study may provide a strong base for the use of NF and ZNF NPs as possible candidates for control of bacterial infections and improving the process of wound healing.
2. Experimental
2.1. Synthesis of nanoparticles
NF NPs were synthesized by previously reported method of co-precipitation with slight modification.22 Stoichiometric amounts of iron (10 mmol) and nickel (0.5 mmol) salts were added in 100 ml of deionized water under constant stirring until complete dissolution. Oleic acid was added to the solution at the rate of 1 drop per 25 ml. Sodium hydroxide (3 mol) solution was then added drop wise (2 ml min−1) under constant stirring, until the pH become alkaline (>12). Temperature of the solution was maintained at 80 °C throughout the reaction. After attaining the desired pH, the solution was centrifuged at 5000 rpm for 5 min. These precipitates were washed with enough distilled water and ethanol till neutralization, then dried at 80 °C and finally annealed at 500 °C to get the NF NPs.For the synthesis of ZNF NPs, the aqueous solution of nickel, zinc and ferric chloride was prepared in fixed molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 of Ni/Zn/Fe. As a surfactant, 2–3 drops of oleic acid were added to each 75 ml of solution. Under constant stirring, 1.5 mol NaOH solution was added drop wise at the rate of 2 ml min−1, until pH become >12. Temperature was maintained at 80 °C throughout the reaction. The solution was centrifuged at 6000 rpm and the obtained precipitates were washed with distilled water and ethanol several times until pH become neutral. In last, particles were dried at 70 °C to get powder form and finally annealed at 500 °C.23
2 of Ni/Zn/Fe. As a surfactant, 2–3 drops of oleic acid were added to each 75 ml of solution. Under constant stirring, 1.5 mol NaOH solution was added drop wise at the rate of 2 ml min−1, until pH become >12. Temperature was maintained at 80 °C throughout the reaction. The solution was centrifuged at 6000 rpm and the obtained precipitates were washed with distilled water and ethanol several times until pH become neutral. In last, particles were dried at 70 °C to get powder form and finally annealed at 500 °C.23
2.2. Characterization of NPs
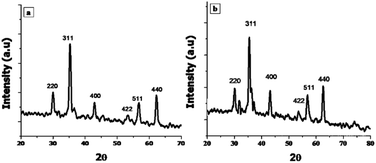
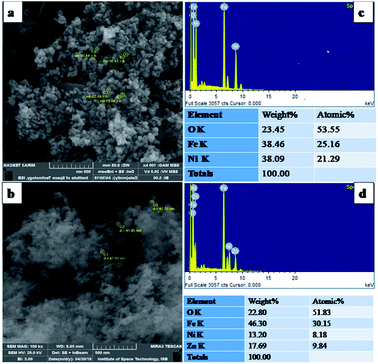
X-ray Diffraction (XRD) technique was used for the determination of crystallite size and phase identification of synthesized nanomaterials. These analyses were carried out by using Bruker, D8 Advanced at the scan rate of 1.2/min in 2θ range of 10–80°. Cu Kα (λ = 1.54056 Å) was used as radiation source and generated at 40 kV and 40 mA. Scanning electron microscopy (SEM) along with energy-dispersive X-ray spectroscopy (EDX) was used to confirm the surface morphology and elemental composition of NPs (SEM MAG: 25.0 kx, MIRA3 TESCAN Institute of space technology, ISB).242.3. Biocompatibility
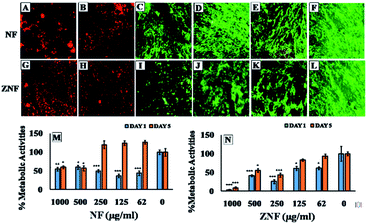
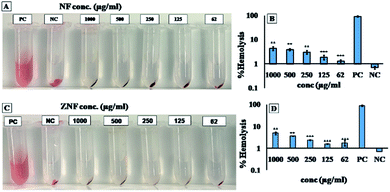
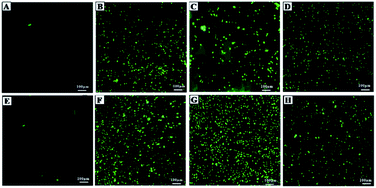
Biocompatibility studies were carried out to check the safety of NF and ZNF NPs. For this purpose, human dermal fibroblast cells (HDF) were maintained and cultured in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with 10% Fetal Bovine Serum (FBS) and 2% Penicillin–Streptomycin (Pen/Strep) under standard condition of 5% CO2 and 37 °C in a cell culture incubator (Thermo Fisher Scientific PA, USA). PrestoBlue assay was performed to evaluate the cell metabolic activity as per previously reported protocol.25 Briefly, the HDF cells were trypsinized with 0.5% trypsin–EDTA, followed by counting with hemocytometer, centrifuged and re-suspended the cell pellet in DMEM complete media. Afterwards, cells (5000 cells per ml) were seeded in 48 well culture plates for 24 h. Then, different concentrations (1000, 500, 250, 125 and 62 μg ml−1) of both NF and ZNF NPs were added and incubated for different time periods. PrestoBlue assay was performed on day 1 and day 5 and fluorescent intensity was measured using micro plate reader (excitation 530 nm, emission at 570 nm, BioTek UV/VIS synergy 2, VT, USA). Cell viability was assessed via live/dead fluorescence assay. Briefly, cells on day 5 of NPs treatment were incubated for 20 min with 1 ml of staining solution containing ethidium homodimer-1 (20 μl) and calcein AM (5 μl) in Dulbecco's Phosphate Buffer Saline (DPBS). Finally, the images were taken using fluorescence microscopy (Axio Observer 5, Zeiss Germany) with excitation/emission wavelength of 528/617 for ethidium homodimer-1 and 528/617 for calcein.Moreover, hemolysis assay was performed according to the ASTM E2524-08 standard protocol to evaluate the hemocompatibility of the NPs.26 For this purpose, whole human blood was acquired from ZenBio and used according to the institutionally (UCLA) approved safety protocols. Blood was stored in heparinized tubes, refrigerated, and used in the assay not later than 48 h. Concentration of the hemoglobin in the blood was estimated by Drabkin's reagent, using a standard curve generated on different concentrations of pure human hemoglobin (1000–62 μg ml−1). The blood was diluted with DPBS to adjust the hemoglobin concentration to 10 ± 2 mg ml−1. The required concentrations of NPs were added in Eppendorf tubes containing 800 μl DPBS, followed by adding 100 μl of the diluted blood. Polyethylene glycol (PEG) (4.4% v/v in DPBS) and triton X-100 (1% v/v in DPBS) treated lysed red blood cells were used as positive control and diluted blood served as negative control. All the samples were incubated in a water bath (37 °C) for 3 h ±15 min, followed by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The supernatant (100 μl) was transferred to a 96-well plate containing equal volume of Drabkin's reagent and was kept in dark on the shaker (100 rpm) for 15 min. The absorbance was recorded at 540 nm against the reagent blank by using a microplate reader (540 nm, BioTek UV/VIS Synergy 2, VT, USA). The concentration of cell-free hemoglobin in each sample was estimated using the standard curve. Eventually, hemolysis (%) was determined by the following equation;
000 rpm for 15 min. The supernatant (100 μl) was transferred to a 96-well plate containing equal volume of Drabkin's reagent and was kept in dark on the shaker (100 rpm) for 15 min. The absorbance was recorded at 540 nm against the reagent blank by using a microplate reader (540 nm, BioTek UV/VIS Synergy 2, VT, USA). The concentration of cell-free hemoglobin in each sample was estimated using the standard curve. Eventually, hemolysis (%) was determined by the following equation;
2.4. Antimicrobial activity
All the bacterial strains i.e. Escherichia coli (E. coli) (ATCC 15224), Pseudomonas aeruginosa (P. aeruginosa) (ATCC-15442), Klebsiella pneumoniae (K. pneumoniae) B5055, Salmonella typhi (S. typhi) (ATCC 14028) and Staphylococcus aureus (S. aureus) (ATCC 6538) were kindly provided by Department of Pharmacy, Quaid-i-Azam University, Pakistan. The methicillin-resistant Staphylococcus aureus (MRSA) was donated by Dr Shujaat Ali Khan, Department of Pharmacy, COMSATS University Islamabad, Abbottabad Campus, Pakistan. Various drug sensitivity tests were performed by Shujaat et al. to confirm that clinical isolate of MRSA is resistant to antibiotics.27The antimicrobial activity of synthesized NPs was performed using ager well diffusion method with some modifications.28–31 For this assay, inoculums of selected strain were used with optical density (OD) between 0.1–0.5 at 600 nm. The inoculum was spread evenly on nutrient agar plates, wells of 70 mm2 diameter were bored and selected concentrations of NPs solution was added into the well. Selected concentrations of NF and ZNF were 1000 μg ml−1, 600 μg ml−1, 300 μg ml−1, 100 μg ml−1 and 50 μg ml−1. The plates were incubated for 24 h at 37 °C. Silver sulfadiazine was used as positive control and dimethyl sulfoxide (DMSO) as a vehicle control. Zone of inhibitions were measured after 24 h of treatment.
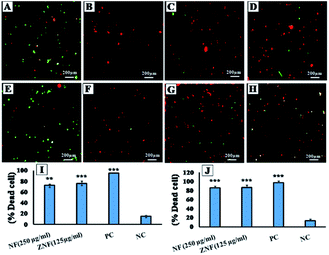
Moreover, live dead assay was performed with representative Gram-negative strain E. coli and Gram-positive S. aureus. Concentrations of 250 μg ml−1 for NF and 125 μg ml−1 for ZNF were used for live dead assay, as these concentrations are biocompatible with human cells. Live and dead bacterial cells assessment was carried out with live/dead back light L-7012 (Invitrogen, USA) using fluorescence light microscopy as previously reported with little modifications.32 Briefly, the overnight grown cultures of E. coli and S. aureus were adjusted to concentration of 1 × 105 CFU ml−1. The bacterial cells were treated with the selected dose of NF and ZNF NPs for 8 h. Tetracycline treated bacterial cell served as positive control. At the end, treated cells were collected and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min at 4 °C. Resultant pallet was washed with phosphate buffer saline (PBS) and stained with SYTO 9 and propidium iodide (PI) for 15 min under dark conditions. The stained bacterial cells were observed under fluorescence microscope to record images. Fluorescence from PI was detected using a filter with excitation wavelength of 540–580 nm and an emission filter of 600–660 nm. Fluorescence from SYTO9 was detected using a filter with excitation wavelength of 465–495 nm and an emission filter of 515–555 nm (Axio Observer 5, zeiss, Germany).
000 rpm for 5 min at 4 °C. Resultant pallet was washed with phosphate buffer saline (PBS) and stained with SYTO 9 and propidium iodide (PI) for 15 min under dark conditions. The stained bacterial cells were observed under fluorescence microscope to record images. Fluorescence from PI was detected using a filter with excitation wavelength of 540–580 nm and an emission filter of 600–660 nm. Fluorescence from SYTO9 was detected using a filter with excitation wavelength of 465–495 nm and an emission filter of 515–555 nm (Axio Observer 5, zeiss, Germany).
2.5. Mechanism of action of NF and ZNF
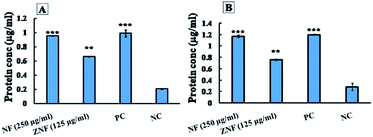
The mechanisms of bactericidal action of synthesized NPs were analyzed by fluorescence-activated cell sorting (FACS) analysis, Bio-Rad protein assay and dichlorofluorescin diacetate dye method (DCFDA). The selected doses for this analysis were same as that of live/dead assay i.e. 250 μg ml−1 for NF and 125 μg ml−1 for ZNF. The E. coli was selected as a representative strain from Gram-negative and S. aureus from Gram-positive group.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min) followed by re-suspending in saline. Suspensions of E. coli and S. aureus were treated separately with 250 μg ml−1 of NF and 125 μg ml−1 of ZNF NPs for 8 h. Tetracycline treated bacterial cells were used as positive control, while untreated bacterial cells were considered as negative control. After treatment, each bacterial suspension was centrifuged at 12
000 rpm for 15 min) followed by re-suspending in saline. Suspensions of E. coli and S. aureus were treated separately with 250 μg ml−1 of NF and 125 μg ml−1 of ZNF NPs for 8 h. Tetracycline treated bacterial cells were used as positive control, while untreated bacterial cells were considered as negative control. After treatment, each bacterial suspension was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min and the obtained supernatant was analysed for protein estimation using Bio-Rad Protein assay kit (Bradford method).37 The protein concentration was estimated from standard curve that was established using known concentrations of bovine serum albumin (BSA).
000 rpm for 15 min and the obtained supernatant was analysed for protein estimation using Bio-Rad Protein assay kit (Bradford method).37 The protein concentration was estimated from standard curve that was established using known concentrations of bovine serum albumin (BSA).2.6. In vitro scratch assay
In vitro scratch method was performed to evaluate the wound healing potentials of the NPs. For this, in vitro cell migration assay were performed as per reported protocol.39 Briefly, primary mouse embryonic fibroblast NIH-3T3 cells were seeded with DMEM containing 10% FBS and 1% pen/strep in 6-well plate at the cell density of 106 cell per ml and kept in CO2 incubator at 37 °C. When monolayer of cells covered the wells, a scratch was made with 200 μl pipette tips followed by washing cells with FBS free DMEM, twice. Next, the sterilized solution of required concentration of NF and ZNF NPs in DMEM (without FBS) were added to the wells. A control group was also run in which cells were cultured in DMEM (without FBS) and left untreated. After being cultured for 0 and 18 h, the images of cells were taken with fluorescence microscope in bright field (Axio Observer 5, zeiss, Germany). In addition, the original and final width of the scratch was measured with ImageJ software by using MRI wound healing tool and percentage of scratch shrinkage was calculated with the following formula:2.7. Statistical analysis
Where necessary, data was taken in triplicates and their mean standard deviation was measured with the standard deviation tool of Microsoft excel 2010. The p value was measured by using Student t-test, p ≤ 0.05 was considered significant.3. Results and discussion
3.1. Characterization of NPs
3.2. Biocompatibility
Despite of the antimicrobial potential and diverse biological applications of various NPs, their use as a therapeutic agent is limited because of their toxicity.45 In the current study, the biocompatibility of NF and ZNF NPs with HDF cells was analyzed via PrestoBlue assay. As the PrestoBlue reagent is a resazurin-based solution that utilizes the reducing power of the living cells to quantitatively measure cell metabolic activity. If the cells are alive, these maintain a reducing environment in cytosol.46 The results showed that NF and ZNF NPs were not cytotoxic to HDF cells at concentration of equal or below 250 and 125 μg ml−1, respectively as shown in Fig. 3C and J. It was also observed that at safer concentrations, the fibroblast cells proliferate significantly with the passage of time (Fig. 3C–E, J and K). To the best of the literature search, this is the first study to report the biocompatibility of NF and ZNF with HDF cells. However, earlier studies reported that Vero cells (monkey kidney epithelial cells) were more than 90% viable when treated with 250 μg ml−1 of NF NPs.47 The results also revealed that the cell viability significantly decreased at higher doses and cells seems to be almost dead (Fig. 3A, B, G and H). As more than 50% cells were died due to arrest of metabolic activities at higher concentration (Fig. 3M and N); therefore 250 μg ml−1 of NF and 125 μg ml−1 of ZNF NPs were considered the safer concentrations and chosen for antimicrobial and other experimentations.NPs induce effects on human erythrocytes through different mechanisms. The most common mechanisms include hemolysis (rapture of erythrocytes membrane) and generation of free radical that can further lead to erythrocytes apoptosis.48 Other effects of NPs on human erythrocytes include morphological changes, oxidative stress induction and alteration of enzymatic activities. Therefore, hemolytic activity of NF and ZNF NPs were evaluated at different concentrations (1000, 500, 250, 125 and 62 μg ml−1) to find out the best hemocompatible dose. Results showed that there was no significant hemolysis at any tested concentration. It can be seen in Fig. 4B and D that there is no significant difference between the negative control and NPs treated group at all concentrations. However, there is significant difference in positive control and treated groups. In Fig. 4A and B, no hemoglobin was observed in supernatant of NPs treated samples and negative control, while hemoglobin was present in supernatant of positive control, which indicated that no hemolysis was observed in any sample except for positive control. A previous study reported that NF NPs were biocompatible at 200 μg ml−1 but ZNF induced hemolysis at 200 μg ml−1.20 Some other studies reported that iron oxide NPs induced reduction in hemoglobin concentration in rat49 and produced hemolysis and oxidative stress in human50
3.3. Antibacterial activities of NF and ZNF
The antibacterial activities of the NF and ZNF NPs were evaluated against four Gram-negative bacteria including E. coli, P. aeruginosa, Klebsiella and S. typhi and two Gram-positive bacteria i.e., S. aureus and MRSA. As shown in Table 1, the NF and ZNF NPs exhibited dose dependent antibacterial activity against the selected microbial strains. The ZNF NP was more active against E. coli, Klebsiella, S. typhi and S. aureus. Similarly, NF was more potent against P. aeruginosa and MRSA. Previously, similar antimicrobial results were reported for NF NPs;43,51 however, there is no reported literature available for antimicrobial activity of ZNF. The structure and chemical composition of cell wall are different in Gram-negative and Gram-positive bacteria. The cell wall of Gram-negative bacteria (e.g. E. coli) mainly consists of lipid A, lipopolysaccharide and peptidoglycan; however, Gram-positive bacteria (e.g. S. aureus) cell wall comprises mainly of peptidoglycan.52 The difference in antibacterial activity of NF and ZNF NPs against various bacterial strains may be due to differences in cell wall composition and variation in interaction with membrane at molecular and cellular level. Moreover, particle size, crystal structure, morphology, surface area and charge also play a significant role in antibacterial activity.53–55| Conc. (μg ml−1) | E. coli | P. aeruginosa | K. pneumoniae | S. typhi | S. aureus | MRSA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF | ZNF | NF | ZNF | NF | ZNF | NF | ZNF | NF | ZNF | NF | ZNF | |
| a NF = NiFe2O4, ZNF = Zn–NiFe2O4, PC = positive control, VC = vehicle control. | ||||||||||||
| 1000 | 12 ± 0.3 | 12 ± 0.2 | 12 ± 0.5 | 12 ± 0.5 | 12 ± 0.4 | 13 ± 0.2 | 11 ± 0.1 | 12 ± 0.4 | 14 ± 0.2 | 14 ± 0.4 | 12 ± 0.5 | 13 ± 0.5 |
| 600 | 11 ± 0.4 | 12 ± 0.9 | 11 ± 0.1 | 11 ± 0.2 | 11 ± 0.3 | 12 ± 0.2 | 11 ± 0.5 | 12 ± 0.2 | 14 ± 0.3 | 14 ± 0.4 | 12 ± 0.2 | 12 ± 0.3 |
| 300 | 10 ± 0.5 | 11 ± 0.5 | 10 ± 0.1 | 9 ± 0.5 | 10 ± 0.3 | 10 ± 0.2 | 10 ± 0.4 | 11 ± 0.1 | 13 ± 0.1 | 11 ± 0.5 | 12 ± 0.5 | 12 ± 0.5 |
| 100 | 10 ± 0.3 | 9 ± 0.5 | 10 ± 0.1 | 9 ± 0.2 | 10 ± 0.3 | 9 ± 0.5 | 10 ± 0.6 | 10 ± 0.1 | 13 ± 0.4 | 10 ± 0.7 | 11 ± 0.2 | 11 ± 0.5 |
| 50 | 10 ± 0.2 | 9 ± 0.3 | 9 ± 0.2 | 8 ± 0.1 | 9 ± 0.1 | 9 ± 1.3 | 9 ± 0.4 | 8 ± 0.5 | 12 ± 0.9 | 10 ± 0.1 | 10 ± 0.1 | 10 ± 0.6 |
| PC | 13 ± 0.1 | 14 ± 0.5 | 16 ± 0.2 | 19 ± 2 | 13 ± 0.4 | 14 ± 0.5 | 17 ± 0.6 | 15 ± 0.4 | 18 ± 1.9 | 18 ± 1.2 | 14 ± 0.9 | 16 ± 1 |
| VC | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
To further confirm the antimicrobial activity, live dead assay was performed against the representative Gram-negative (E. coli) and Gram-positive (S. aureus) bacterial strains. The green fluorescent dye SYTO 9 can only bind with the viable and healthy bacterial cells, whereas, PI dye binds to damaged and/or dead cells, and hence, emit red fluorescence.56 In Fig. 5A (E. coli without any treatment) and Fig. 5E (S. aureus without treatment) serves as negative control. The intense green fluorescence shows the viable E. coli (Fig. 5A) and S. aureus (Fig. 5E) cells. Fig. 5B and F showed maximum dead cells, which represented the positive control of E. coli and S. aureus cells in the study. Whereas, in Fig. 5C red fluorescence show that majority of the E. coli cells treated with 250 μg ml−1 of NF NPs were dead. Same results were observed for S. aureus (Fig. 5G) when treated with the same concentration. Fig. 5D (E. coli) and Fig. 5H (S. aureus) also show that majority of the bacterial cells are dead when treated with 150 μg ml−1 of ZNF NPs. The observed results indicated the prominent antimicrobial activity of NF and ZNF NPs at biocompatible doses. These results are in accordance with the previous literature that SF NPs possess broad-spectrum antibacterial activities against Gram-positive and Gram-negative bacterial strains.55,57,58 In one such study, Elayakumar et al., investigated that Ce3+ doped CuFe2O4 caused inhibition of S. aureus and Klebsiella pneumonia.55 Another study revealed that ZnFe2O4 and Ag hybrid nanostructures effectively halted the growth of Candida albicans.59 Similarly, CoFe2O4 NPs efficiently killed Bacillus subtilis, S. aureus, E. coli and P. aeruginosa,57 while MgFe2O4 NPs exhibited good antibacterial property against P. aeruginosa, E. coli, S. aureus and Serratia marcescens.58 Other studies reported that NiFe2O4 based nanocomposite possesses prominent antibacterial activities against E. coli and S. aureus,60 while nickel substituted copper ferrite displayed better antimicrobial activity against E. coli, K. pneumonia, S. aureus, and B. subtilis.61
Furthermore, the size of NPs greatly influences their biomedical application.62 Previously, Elsabahy and Wooley suggested that intermediate sizes (20–200 nm) of NPs have the greater potential for biomedical applications.63 ZnO NPs with particle size of 50 ± 5 nm exhibited good antimicrobial activity against Escherichia coli (8 and 16 mg ml−1) and Staphylococcus aureus (4 and 8 mg ml−1), respectively.64 Likewise, ZnO NPs coated with crustacean immune molecule having a particle size of 20–50 nm restrained the growth of S. aureus and P. vulgaris.65 Similarly, our synthesised NF and ZNF NPs have a particle size of 49 ± 3.8 and 47 ± 2.5 nm, respectively, have potent antimicrobial activity even at a concentration of 50 μg ml−1. It was noticed that NF and ZNF NPs acted on both human and bacterial cells, but the toxicity to human cells was low or negligible in comparison to bacterial cells when the same concentration was used. A previous study also reported similar results.66 The different effects on human and bacterial cells might be due to the alterations in the cell membrane structures of mammalian and bacterial cells. The existence of endocytic machinery, and multiple intracellular compartments in human cells also prevent the entry of NPs within the cell. Thus, keeping the NPs away from the direct interaction with the important molecules present within the human cells. Furthermore, NF and ZNF NPs have negative surface charge at neutral pH that make them less toxic in comparison to positive charge NPs67
3.4. Mechanism of action of NF and ZNF
Similarly, ZNF NPs, at the dose 125 μg ml−1, showed protein leakage of 1.1 and 0.7 μg ml−1 from E. coli and S. aureus, respectively (Fig. 7A and B). The protein leakage of NPs treated groups was significantly higher as compared to control groups. Previously, similar results were reported for cobalt ferrite;80 however, no study reported the effects of NF and ZNF NPs on protein leakage. It is important to note that the difference in the protein leakage profile of Gram-positive and Gram-negative bacteria might be due to the thickness of peptidoglycan layer of bacterial cell wall. Peptidoglycan layer provide protection to bacteria against antibacterial agents like antibiotics, toxins, chemicals, enzymes and NPs.81 Similarly, Steffy et al., found that protein leakage was higher in Gram-negative bacteria compared to Gram-positive bacteria.82
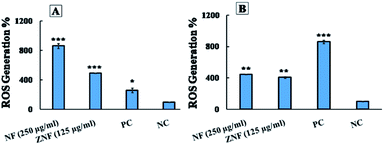
The H2O2 treatment was considered as positive control. It was interesting that NPs generated more ROS than H2O2 in E. coli and less in S. aureus. The group without treatment was considered as negative control group. A pervious study reported that Ni doped CoFe2O4 NPs also generate ROS in bacterial cell.85 Likewise, a study reported that ZnFe2O4 NPs generate ROS, which can be used in photodynamic therapy.86 Hence, it can be concluded that NF and ZNF NPs exert antimicrobial activity through free radicals (ROS) production that generate oxidative stress in bacteria. The oxidative stress causes breakdown of cell membrane with subsequent leakage of cytoplasmic material (protein) thus resulting in distortion of metabolic activities and consequently the cell death.
3.5. Scratch assay
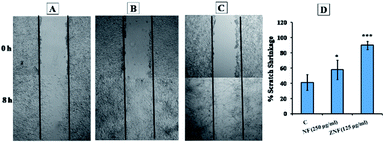
Scratch assay is the most convenient method to illustrate the wound healing potential of drug, molecule or NPs.87 Therefore, in vitro scratch assay was performed to find the wound healing potentials of NF and ZNF NPs. The results showed that the proliferation and migration of cells in artificial wound area was significantly higher in NF and ZNF NPs treatment as compare to control (Fig. 9A–C). Both the NPs promoted the migration of fibroblasts, but the restoration of cellular density was faster in ZNF NPs than NF NPs. Percent scratch shrinkage of both NF and ZNF NPs treated sample was significantly higher than control (untreated) as shown in Fig. 9D. Previous study reported similar results for silver NPs loaded collagen/chitosan scaffolds.87,88 It was observed that silver NPs loaded collagen/chitosan scaffolds increased the fibroblast migration using in vitro scratch assay. Previously, no data is available for NF and ZNF NPs wound healing potential; therefore further animal studies can be performed to fully investigate the effect of NF and ZNF NPs on wound healing process.4. Conclusions
It can be concluded from the results that the prepared NF and ZNF NPs were homogeneous with uniform elemental distribution. The prepared NPs were compatible with human dermal and blood cells, which suggested that these can be potential candidates for biomedical applications. The NPs were active against both Gram-positive and Gram-negative bacterial strain at biocompatible doses. It can be suggested that NF and ZNF NPs causes bacterial cell death via membranes disruption, protein leakage and ROS generation. The in vitro scratch assay revealed that these NPs have strong wound healing potentials. In summary, NF and ZNF NPs can act as potent antimicrobial nano-drugs against infectious organisms and may also be used in wound healing formulations.Conflicts of interest
There is no conflict to declare.Acknowledgements
We acknowledge the Higher Education Commission of Pakistan for providing International Research Support Initiate Program (IRSIP) facility. We would also like to thank Professor Ali Khademhosseini research group at California NanoSystems Institute, University of California Los Angeles, USA for providing cell culture lab facility.Notes and references
- M. Unemo, C. del Rio and W. M. Shafer, Microbiol. Spectrum, 2016, 10, 213–237 CAS.
- G. M. Steede, C. Meyers, N. Li, E. Irlbeck and S. Gearhart, J. Appl. Commun. Res., 2018, 102(4), 49–61 Search PubMed.
- V. Economou and P. Gousia, Infect. Drug Resist., 2015, 8, 49–61 CrossRef.
- B. M. Marshall and S. B. Levy, Clin. Microbiol. Rev., 2011, 24, 718–733 CAS.
- I. Levin-Reisman, I. Ronin, O. Gefen, I. Braniss, N. Shoresh and N. Q. Balaban, Science, 2017, 355, 826–830 CrossRef CAS.
- E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans and Y. Carmeli, Lancet Infect. Dis., 2018, 18, 318–327 CrossRef.
- Y. Liu, L. Shi, L. Su, H. C. van der Mei, P. C. Jutte, Y. Ren and H. J. Busscher, Chem. Soc. Rev., 2019, 48, 428–446 RSC.
- A. Rai, M. Comune and L. Ferreira, in Antibacterial Drug Discovery to Combat MDR, Springer, 2019, pp. 605–633 Search PubMed.
- N.-Y. Lee, P.-R. Hsueh and W.-C. Ko, Front. Pharmacol., 2019, 10, 1153 CAS.
- N. A. Villegas, S. Ravetti, J. M. Bermúdez, A. G. Cid, D. A. Allemandi and S. D. Palma, in Materials for Biomedical Engineering, Elsevier, 2019, pp. 383–407 Search PubMed.
- S. Rehman, R. Jermy, S. M. Asiri, M. A. Shah, R. Farooq, V. Ravinayagam, M. A. Ansari, Z. Alsalem, R. Al Jindan and Z. Reshi, RSC Adv., 2020, 10, 32137–32147 CAS.
- S. Rehman, R. Farooq, R. Jermy, S. M. Asiri, V. Ravinayagam, R. A. Jindan, Z. Alsalem, M. A. Shah, Z. Reshi and H. Sabit, Biomolecules, 2020, 10, 622 CAS.
- S. Rehman, S. M. Asiri, F. A. Khan, B. R. Jermy, V. Ravinayagam, Z. Alsalem, R. Al Jindan and A. Qurashi, Sci. Rep., 2020, 10, 1–9 CrossRef.
- K. K. Kefeni, T. A. Msagati, T. T. Nkambule and B. B. Mamba, Mater. Sci. Eng., C, 2019, 110314 Search PubMed.
- S. Rehman, M. A. Ansari, M. A. Alzohairy, M. N. Alomary, B. R. Jermy, R. Shahzad, N. Tashkandi and Z. H. Alsalem, Processes, 2019, 7, 714 CrossRef CAS.
- F. da Silva, J. Depeyrot, A. Campos, R. Aquino, D. Fiorani and D. Peddis, J. Nanosci. Nanotechnol., 2019, 19, 4888–4902 CrossRef CAS.
- H. Ghayour, M. Abdellahi, N. Ozada, S. Jabbrzare and A. Khandan, J. Phys. Chem. Solids, 2017, 111, 464–472 CrossRef CAS.
- A. Ashour, A. I. El-Batal, M. A. Maksoud, G. S. El-Sayyad, S. Labib, E. Abdeltwab and M. El-Okr, Particuology, 2018, 40, 141–151 CrossRef CAS.
- X. Zhao, A. Sun, W. Zhang, L. Yu, Z. Zuo, N. Suo, X. Pan and Y. Han, J. Mater. Sci.: Mater. Electron., 2019, 1–17 Search PubMed.
- N. L. Martínez-Rodríguez, S. Tavárez and Z. I. González-Sánchez, Toxicol. in Vitro, 2019, 57, 54–61 CrossRef.
- N. Sanpo, C. C. Berndt, C. Wen and J. Wang, Acta Biomater., 2013, 9, 5830–5837 CrossRef CAS.
- K. Maaz, S. Karim, A. Mumtaz, S. Hasanain, J. Liu and J. Duan, J. Magn. Magn. Mater., 2009, 321, 1838–1842 CrossRef CAS.
- I. Gul, W. Ahmed and A. Maqsood, J. Magn. Magn. Mater., 2008, 320, 270–275 CrossRef CAS.
- A. Madni, R. Khan, M. Ikram, S. Naz, T. Khan and F. Wahid, Int. J. Polym. Sci., 2019, 2019, 1–8 CrossRef.
- A. Sheikhi, J. de Rutte, R. Haghniaz, O. Akouissi, A. Sohrabi, D. Di Carlo and A. Khademhosseini, MethodsX, 2019, 6, 1747–1752 CrossRef.
- J. Laloy, H. Haguet, L. Alpan, D. Raichman, J.-M. Dogné and J.-P. Lellouche, Nano Converg., 2018, 5, 31 CrossRef.
- K. Ullah, S. A. Khan, A. Mannan, R. Khan, G. Murtaza and M. A. Yameen, Curr. Pharm. Biotechnol., 2020, 21, 948–954 CAS.
- T. Pranati, R. Anitha, S. Rajeshkumar and T. Lakshmi, Res. J. Pharm. Technol., 2019, 12, 2799–2803 CrossRef.
- L. L. Tshweu, M. A. Shemis, A. Abdelghany, A. Gouda, L. A. Pilcher, N. R. Sibuyi, M. Meyer, A. Dube and M. O. Balogun, RSC Adv., 2020, 10, 19770–19780 RSC.
- S. K. Sahni, Sustain. Humanosphere, 2020, 16, 316–319 Search PubMed.
- Y. N. Aung, Y. M. Aung and Z. Z. Yin, Journal of the Myanmar Academy of Arts and Science, 2020, 18, 159 Search PubMed.
- V. Gopinath, S. Priyadarshini, M. F. Loke, J. Arunkumar, E. Marsili, D. MubarakAli, P. Velusamy and J. Vadivelu, Arabian J. Chem., 2017, 10, 1107–1117 CrossRef CAS.
- K. Saritha, A. Rajesh, K. Manjulatha, O. H. Setty and S. Yenugu, Front. Microbiol., 2015, 6, 577 CrossRef.
- K. Saritha, A. Rajesh, K. Manjulatha, O. H. Setty and S. Yenugu, Front. Microbiol., 2015, 6 DOI:10.3389/fmicb.2015.00577.
- M. L. Mangoni, N. Papo, D. Barra, M. Simmaco, A. Bozzi, A. Di Giulio and A. C. Rinaldi, Biochem. J., 2004, 380, 859–865 CrossRef CAS.
- Z. Zhou, D. Wei, Y. Guan, A. Zheng and J. Zhong, J. Appl. Microbiol., 2010, 108, 898–907 CrossRef CAS.
- J. Zhao, T. Peng, S. Liang, M. Ma, Z. Zeng, P. Yu, D. Gong and S. Deng, Food Control, 2020, 109, 106953 CrossRef CAS.
- S. M. Navarro Gallón, E. Alpaslan, M. Wang, P. Larese-Casanova, M. E. Londoño, L. Atehortúa, J. J. Pavón and T. J. Webster, Mater. Sci. Eng., C, 2019, 99, 685–695 CrossRef.
- M. Felder, B. S. Trüeb, A. O. Stucki, S. Borcard, J. D. Stucki, B. Schnyder, T. Geiser and O. Guenat, Front. Bioeng. Biotechnol., 2019, 7, 3 CrossRef.
- K.-S. Lin, A. K. Adhikari, Z.-Y. Tsai, Y.-P. Chen, T.-T. Chien and H.-B. Tsai, Catal. Today, 2011, 174, 88–96 CrossRef CAS.
- S. Sagadevan, Z. Z. Chowdhury and R. F. Rafique, Mater. Res., 2018, 21, e20160533 CAS.
- A. Costa, E. Tortella, M. Morelli and R. Kiminami, J. Magn. Magn. Mater., 2003, 256, 174–182 CrossRef CAS.
- S. Bhosale, P. Ekambe, S. Bhoraskar and V. Mathe, Appl. Surf. Sci., 2018, 441, 724–733 CrossRef CAS.
- N. Sattarahmady, M. Heidari, T. Zare, M. Lotfi and H. Heli, Appl. Magn. Reson., 2016, 47, 925–935 CrossRef CAS.
- I. Khan, K. Saeed and I. Khan, Arabian J. Chem., 2019, 12, 908–931 CrossRef CAS.
- S. V. K. Rompicharla, P. Kumari, H. Bhatt, B. Ghosh and S. Biswas, Int. J. Pharm., 2019, 557, 329–341 CrossRef CAS.
- X. Lasheras, M. Insausti, I. Gil de Muro, E. Garaio, F. Plazaola, M. Moros, L. De Matteis, J. s. M. de la Fuente and L. Lezama, J. Phys. Chem. C, 2016, 120, 3492–3500 CrossRef CAS.
- K. M. de la Harpe, P. P. Kondiah, Y. E. Choonara, T. Marimuthu, L. C. du Toit and V. Pillay, Cells, 2019, 8, 1209 CrossRef CAS.
- R. Irshad, K. Tahir, B. Li, A. Ahmad, A. R. Siddiqui and S. Nazir, J. Photochem. Photobiol., B, 2017, 170, 241–246 CrossRef CAS.
- Q. Ran, Y. Xiang, Y. Liu, L. Xiang, F. Li, X. Deng, Y. Xiao, L. Chen, L. Chen and Z. Li, Sci. Rep., 2015, 5, 16209 CrossRef CAS.
- P. Koli and K. Kapadnis, Int. J. Chem. Phys. Sci., 2014, 4, 357–360 Search PubMed.
- P. C. Lee, C. C. Chu, Y. J. Tsai, Y. C. Chuang and F. D. Lung, Chem. Biol. Drug Des., 2019, 94(2), 1537–1544 CAS.
- N. Babitha, L. S. Priya, S. R. Christy, A. Manikandan, A. Dinesh, M. Durka and S. Arunadevi, J. Nanosci. Nanotechnol., 2019, 19, 2888–2894 CrossRef CAS.
- V. Sumithra, A. Manikandan, M. Durka, S. K. Jaganathan, A. Dinesh, N. Ramalakshmi and S. A. Antony, Adv. Sci., Eng. Med., 2017, 9, 483–488 CrossRef CAS.
- K. Elayakumar, A. Manikandan, A. Dinesh, K. Thanrasu, K. Kanmani Raja, R. Thilak Kumar, Y. Slimani, S. K. Jaganathan and A. Baykal, J. Magn. Magn. Mater., 2019, 478, 140–147 CrossRef CAS.
- S. Kang, M. Pinault, L. D. Pfefferle and M. Elimelech, Langmuir, 2007, 23, 8670–8673 CrossRef CAS.
- R. P. Sharma, S. D. Raut, R. M. Mulani, A. S. Kadam and R. S. Mane, Int. Nano Lett., 2019, 9, 141–147 CrossRef CAS.
- C. Ehi-Eromosele, J. Olugbuyirozz, O. Taiwo, O. Bamgboye and C. Ango, Bull. Chem. Soc. Ethiop., 2018, 32, 451–458 CrossRef CAS.
- D. Thakur, S. Govindaraju, K. Yun and J.-S. Noh, Nanomaterials, 2019, 9, 1431 CrossRef CAS.
- B. G. Manju and P. Raji, Appl. Phys. A: Mater. Sci. Process., 2019, 125, 313 CrossRef.
- B. G. Manju and P. Raji, J. Electron. Mater., 2019, 48, 7710–7720 CrossRef.
- B. Ankamwar, Biomedical Engineering-Technical Applications in Medicine, 2012, pp. 93–114 Search PubMed.
- M. Elsabahy and K. L. Wooley, Chem. Soc. Rev., 2012, 41, 2545–2561 RSC.
- S. Rehman, B. R. Jermy, S. Akhtar, J. F. Borgio, S. Abdul Azeez, V. Ravinayagam, R. Al Jindan, Z. H. Alsalem, A. Buhameid and A. Gani, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 2072–2082 CrossRef CAS.
- A. Iswarya, B. Vaseeharan, M. Anjugam, B. Ashokkumar, M. Govindarajan, N. S. Alharbi, S. Kadaikunnan, J. M. Khaled and G. Benelli, Colloids Surf., B, 2017, 158, 257–269 CrossRef CAS.
- O. Bondarenko, K. Juganson, A. Ivask, K. Kasemets, M. Mortimer and A. Kahru, Arch. Toxicol., 2013, 87, 1181–1200 CrossRef CAS.
- A. Sukhanova, S. Bozrova, P. Sokolov, M. Berestovoy, A. Karaulov and I. Nabiev, Nanoscale Res. Lett., 2018, 13, 44 CrossRef.
- F. Huang, J. Kong, J. Ju, Y. Zhang, Y. Guo, Y. Cheng, H. Qian, Y. Xie and W. Yao, Sci. Rep., 2019, 9, 490 Search PubMed.
- S. A. Anuj, H. P. Gajera, D. G. Hirpara and B. A. Golakiya, Eur. J. Pharm. Sci., 2019, 127, 208–216 CrossRef CAS.
- M. Arakha, S. Pal, D. Samantarrai, T. K. Panigrahi, B. C. Mallick, K. Pramanik, B. Mallick and S. Jha, Sci. Rep., 2015, 5, 14813 CrossRef CAS.
- M. Arakha, S. Pal, D. Samantarrai, T. K. Panigrahi, B. C. Mallick, K. Pramanik, B. Mallick and S. Jha, Sci. Rep., 2015, 5, 14813 CrossRef CAS.
- T. Wang, Z. Jiang, T. An, G. Li, H. Zhao and P. K. Wong, Environ. Sci. Technol., 2018, 52, 4774–4784 CAS.
- K. Muthukumar, D. S. Lakshmi, S. D. Acharya, S. Natarajan, A. Mukherjee and H. C. Bajaj, Mater. Chem. Phys., 2018, 209, 172–179 CrossRef CAS.
- O. M. Bondarenko, M. Sihtmäe, J. Kuzmičiova, L. Ragelienė, A. Kahru and R. Daugelavičius, Int. J. Nanomed., 2018, 13, 6779 CrossRef CAS.
- A. Parashar, S. Sikarwar and R. Jain, J. Dispersion Sci. Technol., 2019, 884–894 Search PubMed.
- S. J. Kelly, X. Wen, D. P. Arnold and J. S. Andrew, AIP Adv., 2016, 6, 056105 CrossRef.
- W. R. Li, X. B. Xie, Q. S. Shi, S. S. Duan, Y. S. Ouyang and Y. B. Chen, BioMetals, 2011, 24, 135–141 CAS.
- J. Yang, J. Y. Lee and H. P. Too, Anal. Chim. Acta, 2007, 588, 34–41 CAS.
- L. Salvioni, E. Galbiati, V. Collico, G. Alessio, S. Avvakumova, F. Corsi, P. Tortora, D. Prosperi and M. Colombo, Int. J. Nanomed., 2017, 12, 2517 CrossRef CAS.
- R. P. Sharma, S. D. Raut, R. M. Mulani, A. S. Kadam and R. S. Mane, Int. Nano Lett., 2019, 9, 141–147 CrossRef CAS.
- Y. Gao, M. Arokia Vijaya Anand, V. Ramachandran, V. Karthikkumar, V. Shalini, S. Vijayalakshmi and D. Ernest, J. Cluster Sci., 2019, 30, 937–946 CrossRef CAS.
- K. Steffy, G. Shanthi, A. S. Maroky and S. Selvakumar, J. Infect. Public Health, 2018, 11, 463–471 CrossRef.
- L. Zhang, L. Wu, Y. Mi and Y. Si, Bull. Environ. Contam. Toxicol., 2019, 1–6 Search PubMed.
- X. Lin, J. Li, S. Ma, G. Liu, K. Yang, M. Tong and D. Lin, PLoS One, 2014, 9, e110247 CrossRef.
- M. M. Naik, H. S. B. Naik, N. Kottam, M. Vinuth, G. Nagaraju and M. C. Prabhakara, J. Sol-Gel Sci. Technol., 2019, 91, 578–595 CrossRef CAS.
- S. Dong, J. Xu, T. Jia, M. Xu, C. Zhong, G. Yang, J. Li, D. Yang, F. He and S. Gai, Chem. Sci., 2019, 10, 4259–4271 RSC.
- V. P. Giri, S. Pandey, M. Kumari, S. K. Paswan, A. Tripathi, M. Srivastava, C. V. Rao, R. Katiyar, C. S. Nautiyal and A. Mishra, FEMS Microbiol. Lett., 2019, 366(16), fnz201 CrossRef CAS.
- M. Moniri, A. B. Moghaddam, S. Azizi, R. A. Rahim, S. W. Zuhainis, M. Navaderi and R. Mohamad, Int. J. Nanomed., 2018, 13, 5097 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |