Open Access Article
Open Access ArticleNano-evolution and protein-based enzymatic evolution predicts novel types of natural product nanozymes of traditional Chinese medicine: cases of herbzymes of Taishan-Huangjing (Rhizoma polygonati) and Goji (Lycium chinense)†
Guldan
Nazarbek
a,
Aidana
Kutzhanova
a,
Lazzat
Nurtay
a,
Chenglin
Mu
b,
Bexultan
Kazybay
 a,
Xugang
Li
b,
Cuiping
Ma
a,
Xugang
Li
b,
Cuiping
Ma
 c,
Amr
Amin
c,
Amr
Amin
 de and
Yingqiu
Xie
de and
Yingqiu
Xie
 *a
*a
aBiology Department, Nazarbayev University, 53 Kabanbay Batyr Ave, 010000, Nur-Sultan, Kazakhstan. E-mail: xieautumnus@yahoo.com; yingqiu.xie@nu.edu.kz; Tel: +7 7172694686
bSino-German Joint Research Center on Agricultural Biology, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, 271018, China. E-mail: Chenglinmusdau@163.com; xgli@sdau.edu.cn
cShandong Provincial Key Laboratory of Biochemical Engineering, Qingdao Nucleic Acid Rapid Detection Engineering Research Center, College of Marine Science and Biological Engineering, Qingdao University of Science and Technology, Qingdao, 266042, China. E-mail: mcp169@163.com
dBiology Department, UAE University, Al Ain 15551, UAE
eThe College, The University of Chicago, Chicago, IL 60637, USA. E-mail: amramin@uchicago.edu
First published on 12th August 2021
Abstract
Nanozymes and natural product-derived herbzymes have been identified in different types of enzymes simulating the natural protein-based enzyme function. How to explore and predict enzyme types of novel nanozymes when synthesized remains elusive. An informed analysis might be useful for the prediction. Here, we applied a protein-evolution analysis method to predict novel types of enzymes with experimental validation. First, reported nanozymes were analyzed by chemical classification and nano-evolution. We found that nanozymes are predominantly classified as protein-based EC1 oxidoreductase. In comparison, we analyzed the evolution of protein-based natural enzymes by a phylogenetic tree and the most conserved enzymes were found to be peroxidase and lyase. Therefore, the natural products of Rhizoma polygonati and Goji herbs were analyzed to explore and test the potent new types of natural nanozymes/herbzymes using the simplicity simulation of natural protein enzyme evolution as they contain these conserved enzyme types. The experimental validation showed that the natural products from the total extract of nanoscale traditional Chinese medicine Huangjing (RP, Rhizoma polygonati) from Mount-Tai (Taishan) exhibit fructose-bisphosphate aldolase of lyase while nanoscale Goji (Lycium chinense) extract exhibits peroxidase activities. Thus, the bioinformatics analysis would provide an additional tool for the virtual discovery of natural product nanozymes.
Introduction
Nanozymes are functional nanomaterials with enzyme-like activity and biocatalytic function that mimic natural enzymes (“learning from nature”) while retaining nanomaterial properties.1,2 Since the discovery of iron nanoparticles with intrinsic enzyme activity, more than 540 novel nanozymes have been reported with modifications since the first generation of classical Fe3O4 nanozyme with peroxidase-like activity.3 Recently, Meng et al. reported an improved peroxidase activity of a pyrite peroxidase nanozyme with super strong binding ability towards substrate hydrogen peroxide (H2O2) compared to the traditional nanozyme Fe3O4 and protein-based horseradish peroxidase, and additional glutathione oxidase-like activity, following the catalysis of H2O2 generation.4 The dual nanozyme finally targeted KRAS mutation with tumor catalytic therapy.4Currently, there are numerous types of nanozymes reported but most are oxidoreductase, hydrolases and hydrolases.1 In particular, peroxidase POD is widely studied and reported as a type of oxidoreductase.2 There are basically 6 or 7 large types of protein-based enzymes, namely natural enzymes, which are classified according to the Enzyme Commission (EC) classification as EC1, 2, 3, 4, 5, 6, 7 representing oxidoreductases, transferases, hydrolases, lyases, isomerases, ligases, and translocases, respectively.5 Frances H. Arnold proposed the concept of the directed evolution of enzymes by introducing random mutation of genes encoding target protein enzymes and specific chemical reaction-based screening of target mutants, with repeated cycles to obtain the final expected enzymes.6 The directed evolution of enzymes is artificially created to simulate natural evolution mechanisms with natural selection.7 For example, cytochrome P450 can be evolved by directed evolution with synthetic carbene and nitrene to improve the catalysis activity via genetic coding changes or mutation.7 Thus, engineered enzyme evolution can be realized by modifications.
Nanozymes can combine the advantages of natural enzymes and traditional artificial mimicking enzymes with high-efficiency due to adjustable catalytic activity and nanostructures.2 In addition, nanozymes can function under extreme pH and high temperature conditions.1 The catalytic function of a nanozyme is related to its own nano effects such as size, surface area, and structure.2 These effects also provide an operational way for the optimization of simulated enzymes by manually controlling the nano effects to regulate the catalytic activity and substrate selectivity, which are simulating the natural enzyme evolution and selection. For example, Gao et al. synthesized Fe3O4 nanoparticles of different sizes and the relative catalytic activity was enhanced 5 times using 30 nm rather than 300 nm.8
A type of natural product-based nanozyme and nanoassembly has been arising as a new star in nano-natural product research. These include the natural compound-based self-assembly and nanozyme, namely herbzymes.9,10 Recently, we reported natural product-based nanozymes of protease-like, phosphatase-like, and α-amylases (EC 3.2.1.1), which are mainly hydrolases.9 However, whether natural products nanoparticles exert other types of enzyme activity and the evolution of nanozymes remains elusive.
Although numerous nanozymes were discovered and applied in biomedicine, their evolution is still unclear. Here, this study aims to analyze the nano-evolution of nanozymes and natural enzymatic phylogenetic trees to predict and discover novel types of nanozymes. We compared and analyzed the association between nanozyme types using six types of natural enzymes, and then identified evolutionary links to predict new potential nanozyme types with experimental validations and found a novel type of natural product nanozyme.
Methods
Evolutionary analysis of the protein-based natural enzyme
Sequences collected for this study were aligned using MEGAX. Maximum likelihood (ML) trees were generated using the RAxML-HPC2 on XSEDE (8.2.10) in the CIPRES Science Gateway platform by the GTR model of evolution, and fast bootstrap analyses of 1000 pseudoreplicates were completed. The combined maximum likelihood analyses were run using amino acid sequences that were downloaded from Genbank.Chemical nanozyme evolution analysis
First, the collected data11 showing that the most current reported nanozymes were analyzed from the list of recent review and searched papers in Pubmed, and then, the evolution of the same type of element-based families were divided according to the enzyme types and the elements of nanozymes. In detail, before making the self-made chemical evolutionary tree, all the data were classified into six categories according to the types of catalytic reactions of traditional classifications of EC1,2,3,4,5,6 representing oxidoreductase, transferase, hydrolase, lyase, isomerase, and ligase, respectively and an alternative type EC7 for translocases. Then, the chemical element forms of each enzyme were found successively and the branches were classified. Homology was calculated according to the element composition percentage, and the semi-quantitative evolution maps were drawn according to the homology.In the process of making a chemical evolutionary tree, the data of elements used were sorted into the same group, and the element frequency was counted and plotted according to the number of occurrences. The frequency of seven types of enzymes was plotted and analyzed according to the number of elements involved.
Preparation of herbal nanoparticles, fluorescence measurement and scanning electron microscopy (SEM)
Herbal nanoparticles were prepared from Mount Tai area-derived traditional Chinese medicine Rhizoma polygonat (RP) also named as Huangjing, and confirmed as described previously.9 For the Goji (Lycium chinense) nanoparticles prepared by a baking process,9 extract fluorescence was measured by a fluorometer described previously.9 The SEM analysis of baked Goji nanoparticles was performed by dropping Goji extract on a carrier and air-drying it before putting it into a scanning electron microscope.9Prediction of novel enzyme types of herbzyme/nanozyme
Based on the above evolutionary analysis of protein-based maximum likelihood (ML) trees and nanozyme-based natural enzyme type analysis, the most conserved types of enzyme based on EC1,2,3,4,5,6 representing oxidoreductase, transferase, hydrolase, lyase, isomerase, and ligase, respectively, were searched and analyzed.11 The most predominant chemical-based nanozymes were classified as EC1 oxidoreductase, while that of the protein-based enzymes was found to be fructose-bisphosphate aldolase. Thus, the hypothesis is that the nanozyme might have evolved from similarity to simple conserved protein enzymes to a complicated enzyme type. The predicted enzyme types of oxidoreductase and fructose-bisphosphate aldolase were further used for prediction and validation by experiment using herbal extracts of RP and Goji.Experimental validation of the fructose-bisphosphate aldolase of the lyase and peroxidase assay
The fructose-bisphosphate aldolase assay was performed according to the protocol from the manufacture of a fructose-bisphosphate aldolase assay kit (BC2270, Solarbio; http://www.solarbio.com/goods-6266.html). Briefly, after thoroughly mixing with reagents from kits, reaction mixtures' absorbance was measured as value A1 at 340 nm at 10 s, followed by incubation at 37 °C in a water bath. Finally, the absorbance value A2 at 310 s was measured again, which was recorded as A2 measurement. Controls of the A1 blank and A2 blank were also measured for the final calculation by the liquid volume method as FBA enzyme activity (U mL−1) = ΔA ÷ (ε × d) × 109 × Vreaction total ÷ Vsample ÷ T = 321.54 × ΔA (ΔA measurement tube = A1 measurement − A2; ΔA blank tube = A1 blank − A2 blank; ΔA = ΔA measuring tube − ΔA blank tube; Vreaction total: the total volume of the reaction system; VSample: add sample volume, 0.1 mL; ε: NADH molar extinction coefficient, 6.22 × 103 L mol−1 cm−1; d: light path of the cuvette, 1 cm; Vsample total: add extract volume, 1 mL; T: reaction time, 5 min; 109, unit conversion factor, 1 mol = 109 nmol) (http://www.solarbio.com/pdf/2020-shsj/BC2270.pdfHYPERLINK "http://www.solarbio.com/pdf/2020-shsj/BC2270.pdf" \o "http://www.solarbio.com/pdf/2020-shsj/BC2270.pdf"http://www.solarbio.com/pdf/2020-shsj/BC2270.pdf).Peroxidase assay was analyzed by previously reported methods.8 In brief, the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate used for the assay was purchased from Thermo Scientific (TMB Substrate Solution). The Goji-mediated herbzyme assay was performed at different TMB concentrations of the substrate by adjusting the volume and reaction was incubated at room temperature for 30 min. The optical density (OD) was measured after the reaction. The pH effect on the enzymatic activity of peroxidase extracted from the Goji was also measured.
Results
Nano-evolution of chemical nanozymes
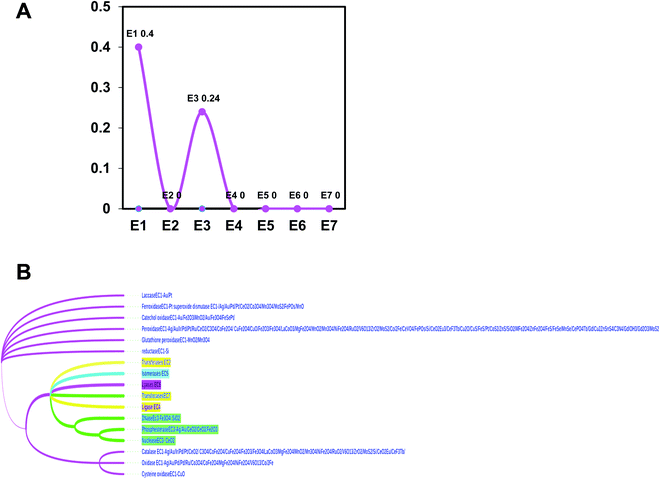
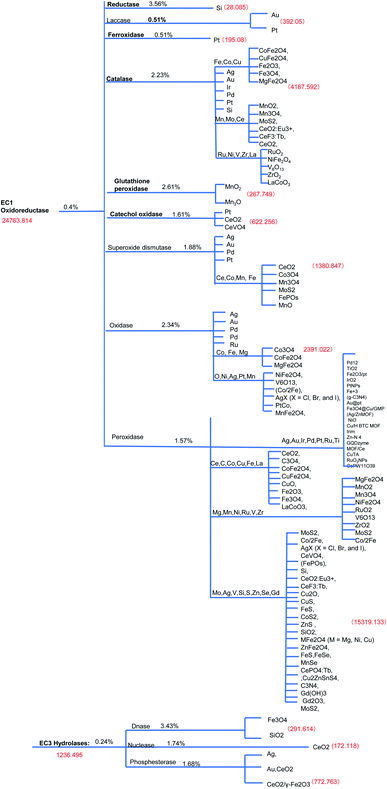
First, the most frequently used elements in the reported nanozymes are approximately 40 (ESI Fig. 1†). Frequency plots of elements showed that elements with the most number of occurrences are: O, V, S, Mn, Fe, Co, Ce, Cu, Au, Pt, etc. Among them, Fe appears the most, and the remaining elements appear less than 20 times. In addition, there are two major types of nanozymes as oxidoreductases and hydrolases in the chemical nanozyme list, which frequently appear with 40% and 24%, respectively (Fig. 1A).The most recent reviewed list of nanozymes with updated search showed that EC1 oxidoreductase (0.4%) group contains catalase (2.23%), oxidase (2.34%), peroxidase (1.57%), laccase (0.51%), glutathione peroxidase (2.61%), iron oxides (0.51%) reductase (3.56%), glutathione oxidase (1.61%), superoxide dismutase (1.88%) and catechol oxidase (1.61%) and EC2 hydrolase (0.24%) consists of nuclease (1.74%), DNase (3.43%), and phosphesterase (1.68%) (Figure 1B and Figure 2).11 The evolutionary tree of nanozyme elements also branched according to the distance of the element family, which collected most data of the latest literature from 2018 review to 2020 new publications reviewed (ESI references†).11 Our data suggest that peroxidase, catalase, and oxidase were the most conserved types among tested, while non-conservative types are reductases and ferroxidase.
Phylogenetic tree analysis of natural enzyme evolution to predict novel types of nanozyme
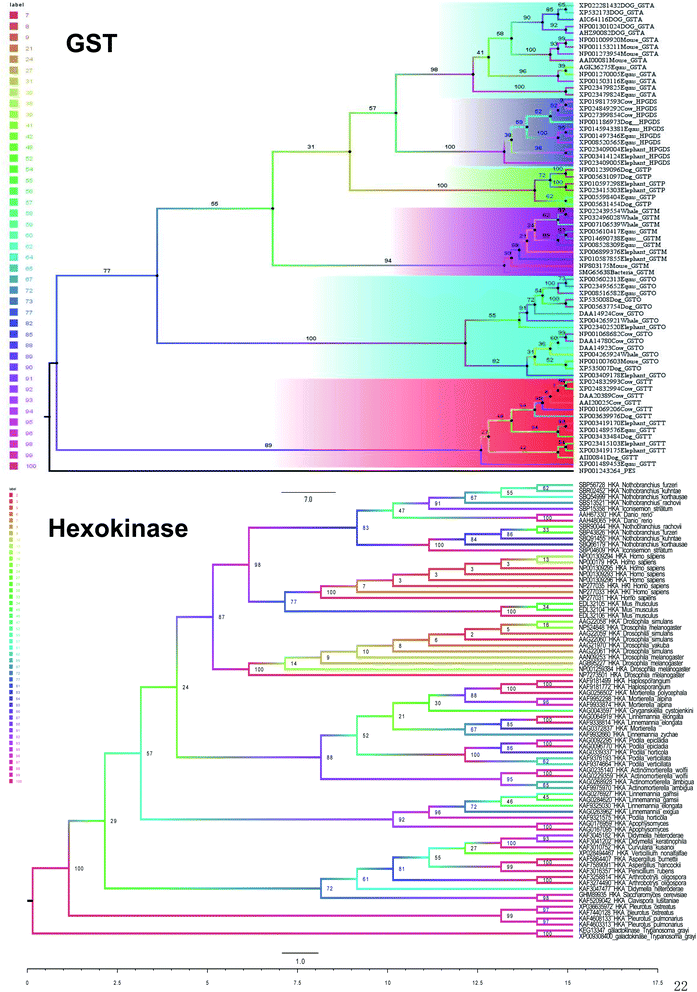
To better explore and understand the different types of nanozymes, we further tested and compared each type of natural enzyme evolutionary conservancy.Hexokinase (HK) (EC 2.7.1.1) is also a class of transferase, which can catalyze the phosphorylation of hexose in sugar catabolism and has a dual role of catalysis and regulation. In mammals, of which four subtypes of HK-I, -II, -III and -IV, the first 3 have higher affinity towards glucose than HK-IV.15 To find the evolutionary relationship among different species of hexokinase, we used the maximum likelihood method to create the relationship with galactokinase as the outgroup. The topology of the maximum likelihood tree is that all hexokinase species form large branches have a support rate of 100%. The first hexokinase branch is the animal branch, which includes humans, mice, and fish, with a bootstrap value of 87%, followed by the plant branch, which has a bootstrap value of 88% (Fig. 4). There is another bacterium branch with a self-expanding value of 72%. Animal branch and plant branch constitute a large branch with a low support rate of bootstrap support (BS) at 24%.
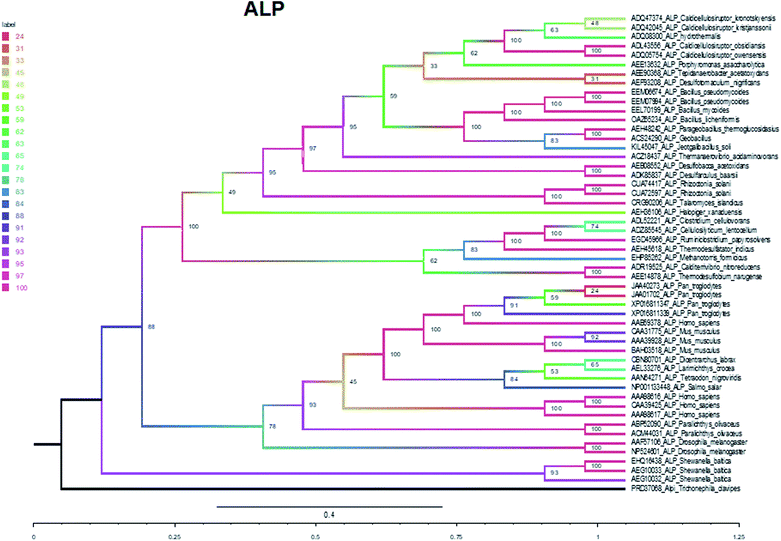
The phylogenetic tree of ALP was established with the maximum likelihood tree, in which ALPI (alkaline phosphatase intestinal) was selected as the outer group. Based on the topological structure of the maximum likelihood tree, the evolutionary tree is divided into two branches: bacteria and animal (Fig. 5). The support rate of the bacterium branch is 100%, and the support rate of the animal branch is 78%. The bacterium and animal branches form large branches with an approval rate of 88% (Fig. 5).
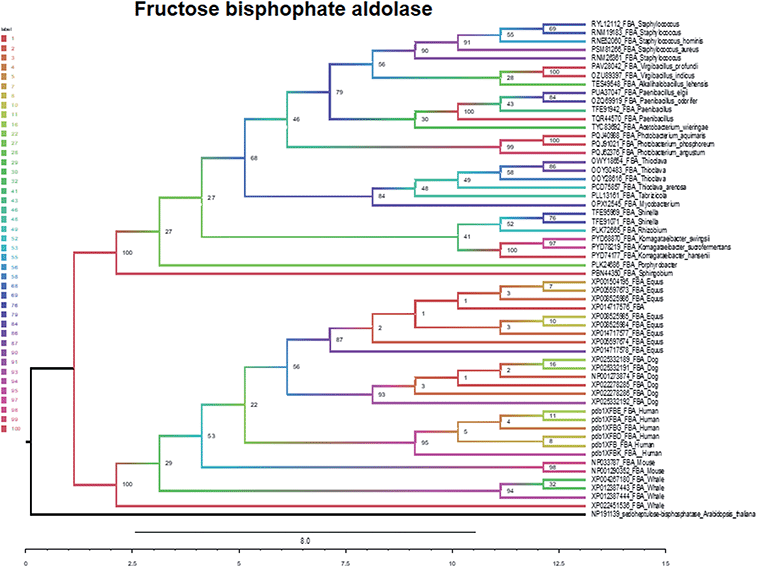
The evolutionary relations of fructose-bisphosphate aldolase were established using the maximum likelihood tree, and sedoheptulose-bisphosphatase was selected as the outer group. The maximum likelihood tree is divided into two major branches as animal (BS: 100%) and bacteria branches (BS: 100%) (Fig. 6).
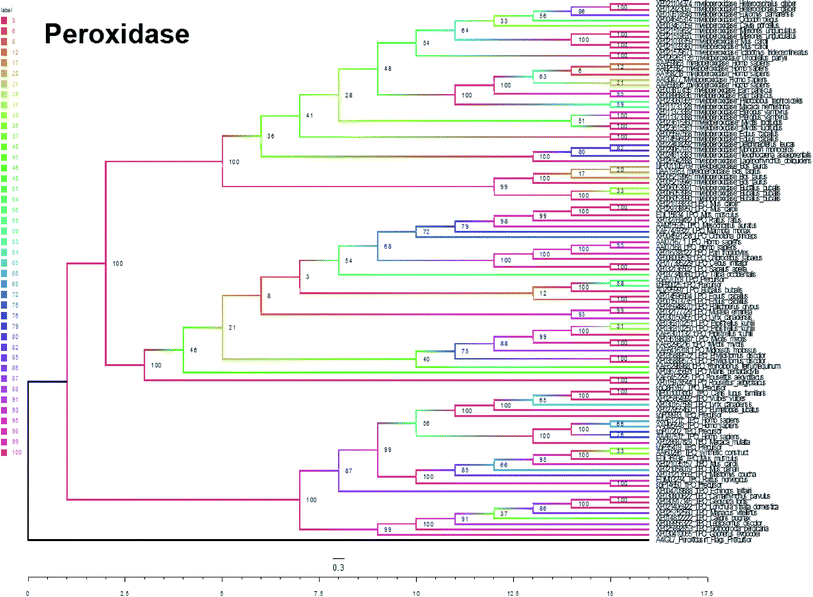
In summary, five evolutionary trees were established using the maximum likelihood tree. By observing the topological structure of the six phylogenetic trees here, it was found that the fructose-bisphosphate aldolase and peroxidase enzymes are the most conserved among the various species, which suggests that their potent bio-inspired nanoparticles would be prevalent to easily evolve and exert the functions of the two enzyme types.
Experimental validation of novel types of natural product nanoparticle-based-nanozymes: herbzymes
Given fructose-bisphosphate aldolase and peroxidase are the most conserved nanozyme types among those tested, we further explored the possibility of the prediction of natural product-nanoparticle based-nanozyme of herbzymes in these two new types as there is a report on the ALP activity of herbzyme. We first found that Rhizoma polygonati (RP) has the fructose-bisphosphate aldolase activity compared to the control (Fig. 7A). Moreover, the Goji extract of the nanoscale natural product showed quantum fluorescence at 420 nm and peroxidase activity when using TMB as the substrate with the highest activity at pH 10 and concentration-dependent kinetics when less than Vmax (Fig. 7B–E). Thus, in addition to the herbzyme with phosphatase activity of RP, as we reported before,9 fructose-bisphosphate aldolase of lyases by RP and peroxidase of Goji were discovered from prediction and validation.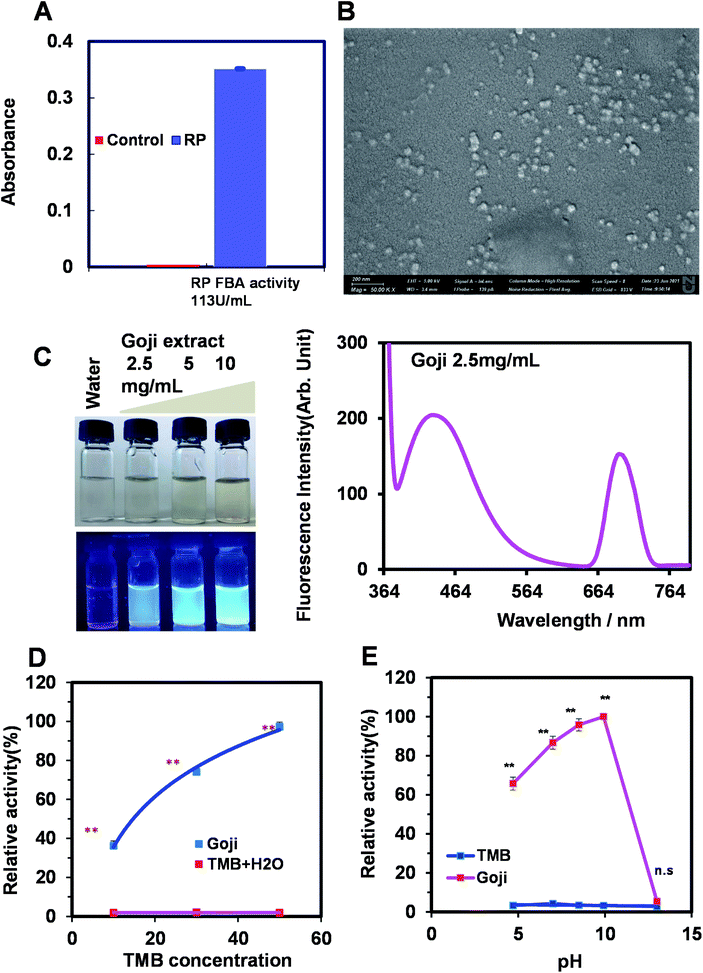 | ||
| Fig. 7 Experimental validation of predicted novel type of herbzymes. (A) Fructose bisphosphate aldolase (FBA) of RP nanoparticles using the RP extract with a FBA assay kit. Nanoscale characteristics of the RP extract were shown by us previously.9 (B) Nanoparticle characteristics of the Goji extract by fluorescence. (C) Nanoparticle characteristics of the Goji extract by size investigated by SEM. (D) Nanoscale Goji extract exhibits the nanozyme activity of peroxidase using TMB as the substrate showing the enzyme kinetics by different concentrations of the substrate. (E) pH effect on the nanoscale Goji extract on peroxidase activity. | ||
Discussion
Here, we report a bioinformatics-based analysis of protein enzyme evolution and comparisons among 6 types of classified enzymes with the nano-evolution of material-based nanozymes, and from the results, we further predicted the bio-inspired nanozyme of herbzyme's novel types corresponding to the protein enzyme types. Our results suggest that self-assembly of natural product of herbal nanoparticles may contribute to the fructose-bisphophate aldolase and peroxidase activities of the natural products which is consistent with our previous report.9From protein-based enzyme evolution, we found that fructose-bisphosphate aldolase is much conserved among different species. According to Say et al.,25 fructose-bisphosphate aldolase is a conserved bifunctional enzyme with aldolase catalytic activity in archaea and bacteria. Similarly, Shams et al.26 indicate that fructose-bisphosphate aldolase housekeeping has a metabolic role as it is a moonlighting protein meaning that it can perform several key functions. It is due to its involvement in the Calvin cycle and glycolysis in addition to gluconeogenesis, which are believed to be acquired additional functions as ancestral organisms evolved. Furthermore, the deletion experiment of genes responsible for fructose-bisphosphate aldolase in C. albicans27 suggests that fructose-bisphosphate aldolase also plays a critical role in the organism's growth. These studies clearly demonstrate the significance of fructose-bisphosphate aldolase for the survival of the organisms, hence indicating why it is highly conserved.
Peroxidase has been largely reported in metal material-based nanozymes. We here found the highly conserved possibility in protein-based enzymes, which correlated with the material-based nanozymes as the highly discovered enzyme of the EC family type. We then tested the herb natural product and we found some natural herbzymes have peroxidase activity, but not all herbs (data not shown) as we found RP did not have this type of enzyme function at this condition in vitro. The RP contains many polysaccharides, and the herbal processing induced chemical reaction results in the loss or generation of different sugars as reported from other herbs too.28,29 Whether it might be repressed or promoted by surface chemical groups, the novel types of nanoscale herbzymes may be predicted by the protein-based enzyme evolutionary conservation analysis.
Conclusion
Nanomaterial-evolution and protein-based enzyme evolutionary analysis showed relatively most conserved types of protein enzymes. The natural product-derived herbzyme was predicted to be a novel type of enzyme by analysis through bio-inspired simulation and experimental validation for the discovery of new types of nanozymes/herbzymes such as fructose-bisphosphate aldolase of lyases and peroxidase.Funding
Yingqiu Xie received the research grant from the Nazarbayev University Faculty-Development Competitive Research Grants Program (ID 16797152) with title of “Targeting cancer stem-like cells of castration-resistant prostate cancer through combinatorial inhibition of MET/nuclear MET and β-Catenin pathways: potential therapeutic intellectual property in prostate cancer treatment (110119FD4531)” to YX and (ID 16796808) with title of “Phosphatase-like nanozyme activity of carbon nanodots and its potential as supplement for kinase inhibitor drug treating prostate cancer: potential intellectual property discovered in food product (110119FD4542)” to HF and YX. YX thanks the United Arab Emirates University (UAEU) – 2019 Asian Universities Alliance (AUA) fellowship (YX and AA).Author contributions
Yingqiu Xie developed the novel concept, planned experiments, and wrote paper. Guldan Nazarbek, Lazzat Nurtay, performed enzyme and nanozyme evolutionary analysis and wrote paper. Aidana Kutzhanova, Chenglin Mu performed herbzyme experiments, analyzed data, and wrote some methods. Cuiping Ma and Amr Amin revised paper, and provided consulting. Xugang Li, and Bexultan Kazybay analyzed the results, discussed the data, and wrote part of the paper.Conflicts of interest
Y. X. received financial support for research, collaboration and consulting.Acknowledgements
We would like to thank the undergraduate students such as Assel Ibadulla who contributed to their support, assistance and valuable comments. The authors thank the Nazarbayev University Faculty-Development Competitive Research Grants Program (ID of 16797152 and ID 16796808), Shandong Taishanghuangjing Biotechnology Co. Ltd., Taian, and UAEU 2019 AUA fellowship.References
- M. Liang and X. Yan, Nanozymes: From New Concepts, Mechanisms, and Standards to Applications, Acc. Chem. Res., 2019, 52(8), 2190–2200, DOI:10.1021/acs.accounts.9b00140.
- R. Zhang, K. Fan and X. Yan, Nanozymes: created by learning from nature, Sci. China Life Sci., 2020, 63(8), 1183–1200, DOI:10.1007/s11427-019-1570-7.
- B. Jiang, L. Fang, K. Wu, X. Yan and K. Fan, Ferritins as natural and artificial nanozymes for theranostics, Theranostics, 2020, 10(2), 687–706, DOI:10.7150/thno.39827.
- X. Meng, D. Li, L. Chen, H. He, Q. Wang, C. Hong, J. He, X. Gao, Y. Yang, B. Jiang, G. Nie, X. Yan, L. Gao and K. Fan, High-Performance Self-Cascade Pyrite Nanozymes for Apoptosis-Ferroptosis Synergistic Tumor Therapy, ACS Nano, 2021, 15(3), 5735–5751, DOI:10.1021/acsnano.1c01248.
- P. K. Robinson, Enzymes: principles and biotechnological applications, Essays Biochem., 2015, 59, 1–41, DOI:10.1042/bse0590001 . Erratum in: Essays Biochem., 2015, 59, 75..
- F. H. Arnold, Innovation by Evolution: Bringing New Chemistry to Life (Nobel Lecture), Angew. Chem., Int. Ed., 2019, 58(41), 14420–14426, DOI:10.1002/anie.201907729.
- Y. Yang and F. H. Arnold, Navigating the Unnatural Reaction Space: Directed Evolution of Heme Proteins for Selective Carbene and Nitrene Transfer, Acc. Chem. Res., 2021, 54(5), 1209–1225, DOI:10.1021/acs.accounts.0c00591.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Intrinsic peroxidase-like activity of ferromagnetic nanoparticles, Nat. Nanotechnol., 2007, 2(9), 577–583, DOI:10.1038/nnano.2007.260.
- E. Benassi, H. Fan, Q. Sun, K. Dukenbayev, Q. Wang, A. Shaimoldina, A. Tassanbiyeva, L. Nurtay, A. Nurkesh, A. Kutzhanova, C. Mu, A. Dautov, M. Razbekova, A. Kabylda, Q. Yang, Z. Li, A. Amin, X. Li and Y. Xie, Generation of particle assemblies mimicking enzymatic activity by processing of herbal food: the case of rhizoma polygonati and other natural ingredients in traditional Chinese medicine, Nanoscale Adv., 2021, 3, 2222–2235, 10.1039/D0NA00958J.
- T. Li, P. Wang, W. Guo, X. Huang, X. Tian, G. Wu, B. Xu, F. Li, C. Yan, X. J. Liang and H. Lei, Natural Berberine-Based Chinese Herb Medicine Assembled Nanostructures with Modified Antibacterial Application, ACS Nano, 2019, 13(6), 6770–6781, DOI:10.1021/acsnano.9b01346.
- J. Wu, X. Wang, Q. Wang, Z. Lou, S. Li, Y. Zhu, L. Qin and H. Wei, Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes (II), Chem. Soc. Rev., 2019, 48(4), 1004–1076, 10.1039/C8CS00457A.
- A. A. Khan, A. H. Rahmani, Y. H. Aldebasi and S. M. Aly, Biochemical and pathological studies on peroxidases-an updated review, Glob. J. Health Sci., 2014, 6(5), 87–98, DOI:10.5539/gjhs.v6n5p87.
- Y. Sugano, DyP-type peroxidases comprise a novel heme peroxidase family, Cell. Mol. Life Sci., 2009, 66(8), 1387–1403, DOI:10.1007/s00018-008-8651-8.
- R. R. Singh and K. M. Reindl, Glutathione S-Transferases in Cancer, Antioxidants, 2021, 10(5), 701, DOI:10.3390/antiox10050701.
- F. Ciscato, L. Ferrone, I. Masgras, C. Laquatra and A. Rasola, Hexokinase 2 in Cancer: A Prima Donna Playing Multiple Characters, Int. J. Mol. Sci., 2021, 22(9), 4716, DOI:10.3390/ijms22094716.
- X. Xie, Y. Wang, X. Zhou, J. Chen, M. Wang and X. Su, Fe-N-C single-atom nanozymes with peroxidase-like activity for the detection of alkaline phosphatase, Analyst, 2021, 146(3), 896–903, 10.1039/D0AN01846E.
- Y. Xie, H. Fan, W. Lu, Q. Yang, A. Nurkesh, T. Yeleussizov, A. Maipas, J. Lu, L. Manarbek, Z. Chen and E. Benassi, Nuclear MET requires ARF and is inhibited by carbon nanodots through binding to phospho-tyrosine in prostate cancer, Oncogene, 2019, 38(16), 2967–2983, DOI:10.1038/s41388-018-0608-2.
- E. Lorentzen, B. Siebers, R. Hensel and E. Pohl, Structure, function and evolution of the Archaeal class I fructose-1,6-bisphosphate aldolase, Biochem. Soc. Trans., 2004, 32(Pt 2), 259–263, DOI:10.1042/bst0320259.
- G. J. Thomson, G. J. Howlett, A. E. Ashcroft and A. Berry, The dhnA gene of Escherichia coli encodes a Class I fructose bisphosphate aldolase, Biochem. J., 1998, 331(2), 437–445, DOI:10.1042/bj3310437.
- S. Banerjee, F. Anderson and G. K. Farber, The evolution of sugar isomerases, Protein Eng., 1995, 8(12), 1189–1195, DOI:10.1093/protein/8.12.1189.
- S. Martinez Cuesta, N. Furnham, S. A. Rahman, I. Sillitoe and J. M. Thornton, The evolution of enzyme function in the isomerases, Curr. Opin. Struct. Biol., 2014, 26(100), 121–130, DOI:10.1016/j.sbi.2014.06.002.
- S. Shuman, DNA ligases: progress and prospects, J. Biol. Chem., 2009, 284(26), 17365–17369, DOI:10.1074/jbc.R900017200.
- A. Sallmyr, I. Rashid, S. K. Bhandari, T. Naila and A. E. Tomkinson, Human DNA ligases in replication and repair, DNA Repair, 2020, 93, 102908, DOI:10.1016/j.dnarep.2020.102908.
- N. Yutin and E. V. Koonin, Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: a case of hidden complexity, Biol. Direct, 2009, 4, 51, DOI:10.1186/1745-6150-4-51.
- R. F. Say and G. Fuchs, Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme, Nature, 2010, 464(7291), 1077–1081, DOI:10.1038/nature08884.
- F. Shams, N. J. Oldfield, K. G. Wooldridge and D. P. Turner, Fructose-1,6-bisphosphate aldolase (FBA)-a conserved glycolytic enzyme with virulence functions in bacteria: ‘ill met by moonlight’, Biochem. Soc. Trans., 2014, 42(6), 1792–1795, DOI:10.1042/BST20140203.
- A. Rodaki, T. Young and A. J. Brown, Effects of depleting the essential central metabolic enzyme fructose-1,6-bisphosphate aldolase on the growth and viability of Candida albicans: implications for antifungal drug target discovery, Eukaryot. Cell, 2006, 5(8), 1371–1377, DOI:10.1128/EC.00115-06.
- I. Pawlaczyk-Graja, S. Balicki and K. A. Wilk, Effect of various extraction methods on the structure of polyphenolic-polysaccharide conjugates from Fragaria vesca L. leaf, Int. J. Biol. Macromol., 2019, 130, 664–674, DOI:10.1016/j.ijbiomac.2019.03.013.
- Y. Xie, L. Nurtay, Q. Sun, Q. Wang, C. Mu, Z. Liang, A. Amin and X. Li, Rhizoma polygonati from Mount Tai: nutritional value and usefulness as a traditional Chinese medicine, source of herbzyme, and potential remediating agent for COVID-19 and food shortage, Acupuncture and Herbal Medicine, 2021, 1, 36–44 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1na00475a |
| This journal is © The Royal Society of Chemistry 2021 |