Targeted metabolomics reveals dynamic portrayal of amino acids and derivatives in triple-negative breast cancer cells and culture media†
Fang
Kou‡
a,
Bangjie
Zhu‡
ab,
Wenbin
Zhou
c,
Chunming
Lv
c,
Yu
Cheng
 *ab and
Hai
Wei
*a
*ab and
Hai
Wei
*a
aInstitute of Interdisciplinary Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 201203, China
bSchool of Pharmacy, Shanghai Jiao Tong University, Shanghai 200240, China
cShanghai Zhulian Intelligent Technology Ltd. Co., Shanghai 201323, China
First published on 23rd November 2020
Abstract
Triple-negative breast cancer (TNBC) is well-known for its metastatic aggressiveness and poor survival prognosis, accounting for nearly a quarter of cases in breast cancer. We performed intra- and extra-cellular profiling of 40 amino acids and derivatives on three cell lines and their culture media, including TNBC, non-TNBC and normal breast epithelial cells, using HILIC-MS/MS. Characteristic metabolic alteration of amino acids and derivatives was observed in TNBC cells, compared to non-TNBC cells, especially in correlated intra- and extra-cellular metabolic pathways. Intra-cellularly, quantified glutamic acid, β-alanine, aspartic acid, glutathione, N-acetyl-serine and N-acetyl-methionine were most significantly increased (>2-fold, p < 0.01 and VIP > 1) in TNBC cells. Extra-cellularly, significantly increased uptake of glutamine, serine, β-alanine, and lysine and elevated excretion of glutamic acid and L-cysteine-glutathione (p < 0.01 and VIP > 1) were observed by TNBC cells from or to their cell culture media. This study depicted a novel dynamic portrayal of metabolic dysregulation between TNBC and non-TNBC cells, correlated in both intra- and extra-cellular amino acid profiles. Quantification of these distinctive metabolites of TNBC cells might offer advanced understanding and new treatment targets for TNBC.
1. Introduction
Breast cancer is one of the top three most common cancers emerging in women in the world.1 Triple-negative breast cancer (TNBC) is named for the short amplification of three hormone receptors of estrogen, progesterone and human epidermal growth factor receptor 2 (ER, PR and HER2), accounting for nearly a quarter of cases of breast cancer.2 TNBCs tend to be higher in grade, harder to diagnose and more invasive. The receptor-targeting drugs for breast cancer, tamoxifen and trastuzumab, are not effective for TNBC patients because of mainly targeting the three hormone receptors not expressed in TNBC.3 Clinically, more frequent recurrence and distant metastasis, poor prognosis and lower survival rates are more commonly seen in TNBC patients.4TNBC cancers usually lack TP53 or express mutated TP53. In the Cancer Genome Atlas (TGCA), 68% of 102 TNBC cases were reported with mutations of TP53, in addition to 3% homozygous absence or 7% amplifications of MDM2/4.5 Consistent with the TGCA data, TP53 was mutated in 53.8 percent of the 104 TNBC tumors sequenced in Shah's study.6 It is reported that deletion of methionine could powerfully inhibit metastasis in two human TNBC cell lines, MDA-MB-231 and Hs 578T.7
Metabolic hallmarks are frequently reported to emerge in cancer cells, as they take more nutrients of glucose and amino acids compared to normal cells, because of their characteristically quick uncontrolled growth and aggressiveness.8–10 The metabolic profile of TNBC in African American women reports that specific metabolic pathways affect the up-regulation in TNBC tumors of energy metabolism and transmethylation, compared to non-TNBC of ER+ tumors.11
Of the endogenous metabolites, amino acids and their derivatives are involved in critical biological processes including energy metabolism, protein synthesis, transport of lipids, and neurotransmission.12 Elevated requirements for amino acids and derivatives are exhibited in many cancers, supplied by an extracellular source or increased endogenous synthesis de novo.13 Previous literature shows that glutamine is the common nutrient consumed most in many studies of cultured cancer cells.14,15 In distant migrant models of breast cancer, catabolism of proline was reported to support proliferation of breast cancer cells through proline dehydrogenase.16
Despite these molecular studies, quantitative profiling of amino acids and derivatives has not been reported to explore the characteristic signature in TNBC cancer cells, from the intra- and extra-cellular perspectives. We have established a quantitative profiling method of 40 endogenous amino acids and derivatives in cell lines.12 In this study, we aim to quantify 40 amino acids and derivatives on cell lysates and cell culture medium samples from the TNBC cell line of HCC1143, non-triple-negative breast cancer cell line (non-TNBC) of HCC202, and normal breast epithelial cell line of MCF10A, using hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry (HILIC-MS/MS). HCC1143 is a typical TNBC cell line with a positive p53 mutation and triple negative hormone receptors of ER, PR and HER2. HCC202 is a typical non-TNBC cell line with HER2+. In this study, we explored the characteristic metabolic hallmarks of amino acid and derivative profiling on TNBC and non-TNBC cell lines, from both the intra- and extra-cellular amino acid and derivative perspectives. The protocol for this study was approved by the Institutional Review Boards of Shanghai University of Traditional Chinese Medicine.
2. Materials and methods
2.1 Chemicals and reagents
Reagent grade chemicals and reagents were obtained from commercial suppliers and used without additional work-up, as described in detail in our previous paper.122.2 Preparation of the stock solutions, calibration solution and quality control samples
Preparation of the stock solutions, calibration solution, quality control samples and internal standard of amino acids and their derivatives was described in detail in our previous paper.12 A list of the amino acids and their derivatives studied is presented in Table 1.| Analyte | Concentration of cell lysates (mean ± SD) (ng per 106 cells) (n = 6) | Concentration of cell culture medium (mean ± SD) (ng per 106 cells) (n = 6) | Dynamic change of medium from HCC202 (mean ± SD, n = 6, ng per 106 cells per 24 hours) | Dynamic change of medium from HCC1143 (mean ± SD, n = 6, ng per 106 cells per 24 hours) | ||||
|---|---|---|---|---|---|---|---|---|
| HCC202 (non-TNBC) | HCC1143 (TNBC) | HCC202 (non-TNBC) | HCC1143 (TNBC) | Uptake | Excretion | Uptake | Excretion | |
| N.D., not detected. | ||||||||
| N-Acetylmethionine | N.D. | 2.27 ± 0.263 | N.D. | N.D. | — | — | — | |
| N-Acetyltryptophan | N.D. | N.D. | 26.7 ± 3.09 | 23.7 ± 2.17 | 0.589 | 3.56 | ||
| β-Alanine | 23.3 ± 11.1 | 423 ± 8.33 | 2104 ± 97.3 | 1707 ± 132 | 64.2 | 461 | ||
| Alanine | 293 ± 27.8 | 298 ± 22.7 | 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 793 ± 2033 793 ± 2033 |
38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 270 ± 2896 270 ± 2896 |
−4528 | −5004 | ||
| Arginine | 1955 ± 315 | 1680 ± 280 | 682![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860 ± 9721 860 ± 9721 |
668![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 507 ± 12 507 ± 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 316 316 |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 607 607 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 960 960 |
||
| Asparagine | 1262 ± 94.3 | 1150 ± 87.2 | 204![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 086 ± 5709 086 ± 5709 |
193![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 ± 5683 027 ± 5683 |
2010 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 069 069 |
||
| Aspartic aid | 323 ± 46.5 | 1413 ± 53.5 | 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 276 ± 2728 276 ± 2728 |
94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 846 ± 3255 846 ± 3255 |
−4108 | −3678 | ||
| Cycloleucine | N.D. | N.D. | N.D. | N.D. | — | — | — | |
| Cysteine | 45.2 ± 5.88 | 30.1 ± 5.71 | 1102 ± 77.7 | 1273 ± 59.8 | −154 | −325 | ||
| Glycine | 1610 ± 55.1 | 1621 ± 146 | 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 ± 6663 800 ± 6663 |
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 904 ± 7869 904 ± 7869 |
6307 | 2203 | ||
| Glycylglycine | 2.13 ± 0.502 | 2.40 ± 0.401 | 80.6 ± 7.37 | 65.7 ± 4.40 | −18.1 | −3.22 | ||
| Glycylproline | 0.855 ± 0 | 0.701 ± 0.043 | N.D. | N.D. | — | — | — | |
| Glycylvaline | N.D. | N.D. | 43.2 ± 15.4 | 40.8 ± 10.3 | 48.5 | 50.9 | ||
| Histidine | 1823 ± 97.7 | 1562 ± 84.0 | 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 431 ± 1803 431 ± 1803 |
64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 195 ± 1331 195 ± 1331 |
1616 | 4851 | ||
| Isoleucine | 683 ± 60.5 | 646 ± 61.7 | 211![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 744 ± 6528 744 ± 6528 |
205![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 048 ± 3370 048 ± 3370 |
6228 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 924 924 |
||
| Lysine | 671 ± 98.6 | 522.5 ± 60.7 | 154![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 593 ± 3677 593 ± 3677 |
143![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 841 ± 2019 841 ± 2019 |
2579 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 331 331 |
||
| Phenylalanine | 490 ± 42.2 | 395 ± 39.2 | 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 520 ± 2976 520 ± 2976 |
73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 085 ± 1851 085 ± 1851 |
3135 | 7569 | ||
| Proline | 234 ± 13.8 | 304 ± 18.4 | 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 743 ± 2175 743 ± 2175 |
83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 291 ± 2190 291 ± 2190 |
209 | 2661 | ||
| Pyroglutamic acid | 881 ± 96.6 | 486 ± 93.2 | 233![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 499 ± 4610 499 ± 4610 |
215![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 367 ± 7342 367 ± 7342 |
−65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 707 707 |
−47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 575 575 |
||
| Sarcosine | N.D. | N.D. | N.D. | N.D. | — | — | — | |
| Serine | 822 ± 68.5 | 635 ± 45.3 | 127![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 236 ± 4394 236 ± 4394 |
109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 563 ± 2930 563 ± 2930 |
−457 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 215 215 |
||
| Threonine | 532 ± 58.3 | 403 ± 46.6 | 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 785 ± 5865 785 ± 5865 |
82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 695 ± 3848 695 ± 3848 |
2103 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 193 193 |
||
| Tryptophan | 148 ± 14.9 | 111 ± 9.73 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 629 ± 615 629 ± 615 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 617 ± 558 617 ± 558 |
779 | 1791 | ||
| trans-4-Hydroxyproline | 552 ± 39.6 | 507 ± 33.8 | 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 685 ± 2950 685 ± 2950 |
89![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861 ± 1552 861 ± 1552 |
694 | 1518 | ||
| Tyrosine | 131 ± 11.2 | 132 ± 11.5 | 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ± 3244 100 ± 3244 |
72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 866 ± 1817 866 ± 1817 |
— | 5214 | ||
| Valine | 152 ± 11.4 | 142 ± 17.4 | 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 044 ± 1649 044 ± 1649 |
86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 983 ± 1807 983 ± 1807 |
3361 | 8423 | ||
| Citrulline | 23.0 ± 3.14 | 23.4 ± 2.83 | 7048 ± 316 | 6981 ± 484 | 407 | 474 | ||
| Glutathione | 4533 ± 89.1 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 133 ± 266 133 ± 266 |
N.D. | N.D. | — | — | — | |
| Glycylleucine | 0.768 ± 0 | N.D. | 15.0 ± 2.78 | 15.0 ± 1.47 | 2.64 | 2.63 | ||
| N-Phenylacetylglycine | 30.5 ± 4.04 | 23.5 ± 3.47 | 6464 ± 374 | 6551 ± 276 | −297 | −383 | ||
| N-Acetyl-Asp-Glu | N.D. | 16.9 ± 3.21 | N.D. | N.D. | — | — | — | |
| N-Acetylserine | 12.9 ± 1.81 | 31.4 ± 3.73 | 229 ± 42.6 | 218 ± 22.0 | 13.1 | 23.8 | ||
| N-Acetyllysine | 0.818 ± 0.195 | 0.838 ± 0.228 | 99.2 ± 12.0 | 109 ± 15.0 | 1.12 | −8.53 | ||
| L-Cysteine-glutathione | N.D. | 94.1 ± 14.7 | 3006 ± 538 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 460 ± 1180 460 ± 1180 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 556 556 |
−12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 898 898 |
||
| Homocysteine | N.D. | 1.023 ± 0.234 | 14.2 ± 4.36 | 9.75 ± 6.22 | 2.56 | 6.98 | ||
| S-Adenosylhomocysteine | 3.02 ± 0.779 | 6.56 ± 1.86 | 12.7 ± 5.32 | 28.0 ± 8.74 | — | — | ||
| Methionine | 122 ± 10.5 | 109 ± 9.14 | 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 580 ± 1209 580 ± 1209 |
44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 465 ± 1300 465 ± 1300 |
1576 | 3691 | ||
| Glutamine | 6387 ± 557 | 4755 ± 431 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 102![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 460 ± 25 460 ± 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 144 144 |
948![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 613 ± 22 613 ± 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 094 094 |
83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
237![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 747 747 |
||
| Glutamic acid | 1106 ± 104 | 9323 ± 419 | 149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 189 ± 4373 189 ± 4373 |
195![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 791 ± 3279 791 ± 3279 |
−7484 | −54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 085 085 |
||
| Leucine | 625 ± 28.8 | 561 ± 61.5 | 222![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 973 ± 3794 973 ± 3794 |
217![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 745 ± 3924 745 ± 3924 |
5315 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 543 543 |
||
2.3 Cell culture, sample preparation and LC-MS/MS conditions
The TNBC cell line of HCC1143, non-TNBC cell line HCC202, and normal breast epithelial cell line MCF10A were purchased from the American Type Culture Collection (Manassas, VA, USA). The culture of the cell lines, cell sample collection, sample preparation, LC-MS/MS conditions and Analyst 1.6.2 software for data acquisition and processing were described in detail in our previous paper.12 Triplicate samples of blank culture medium were collected, immediately frozen and stored at −80 °C until analysis. Six-well plates were cultured for each cell line. At harvest time, each cell incubation medium of 200 μL was collected after cell counting. The cellular pellets were collected after the remaining medium was removed.12 Both the culture medium and cell samples were immediately frozen in liquid nitrogen and then stored at −80 °C until analysis.2.4 Statistical analysis
In the analysis of the targeted profiles among the TNBC and non-TNBC cell lines, and the breast epithelial cell line, univariate and multi-variate statistical analysis were conducted using SPSS v.22.0 software (SPSS Inc., Chicago, IL, USA) and SIMCA-P 11.0 (Umetrics, Umeå, Sweden), as described in our previous paper.12 Volcano analysis and Z-score heatmaps of clustering analysis were performed based on statistical analysis software packages in R studio that were widely used in omics approaches (http://cran.r-project.org/).173. Results
Of the 40 targeted chemicals of amino acids, 36 intra-cellular and 34 extra-cellular analytes were detected and quantified respectively in the three cell lines including TNBC, non-TNBC, and breast epithelial cells.The intra-cellular concentrations on cell lysates were expressed as the mean concentration (given in ng per 106 cells) and standard deviation (Table 1), as described in our previous literature.12 The extra-cellular concentrations on cell culture media were expressed as the mean concentration (given in ng per 106 cells) and standard deviation, and the uptake or excretion by the cells from or to the media (given in ng per 106 cells per 24 h) was calculated from the mean value of blank medium samples minus individual culture medium samples (Table 1). The concentrations of cell lysates and medium samples from normal breast epithelial cells of MCF10A, and blank cell culture medium samples are shown in Table S1 (ESI†).
The differential metabolites (p < 0.05 and VIP > 1) of amino acids and derivatives in intra- and extra-cellular profiling are shown in Table 2 on cell lysates and cell culture media from the cell lines of non-TNBC and TNBC, with univariate and multivariate statistical analysis. The statistical intra- and extra-cellular comparisons among the three cell lines of normal breast cells, non-TNBC and TNBC are shown in Table S2 (ESI†). The statistical results of intra- and extra-cellular comparison of non-TNBC versus normal breast cells, and TNBC versus normal breast cells are shown in Tables S3 and S4 (ESI†), respectively. A p-value <0.05 was considered to be a statistically significant difference throughout the study, using univariate statistical analysis of the Mann–Whitney U test or Kruskal–Wallis test.
| Analyte | Cell lysates of HCC 1143 cells (TNBC) vs. HCC 202 cells (non-TNBC) | Cell culture medium from HCC 1143 cells (TNBC) vs. HCC 202 cells (non-TNBC) | ||||
|---|---|---|---|---|---|---|
| p | VIP valueb | Fold change | p | VIP valueb | Fold changec | |
| a p obtained by the Mann–Whitney U test. b VIP obtained by OPLS-DA analysis. c The fold change of the cell culture medium was calculated by the ratio of uptake or excretion of the metabolites between the two cell lines. A negative value represents the opposite direction of uptake or excretion of the metabolite in the two cell lines. d Some concentration of the metabolite was below the LLOQ in the data. The LLOQ values of the metabolites were used when fold change was calculated. | ||||||
| N-Acetylmethionine | 0.003 | 1.4 | 3.79d | — | — | — |
| N-Acetylserine | 0.002 | 1.3 | 2.44 | — | — | — |
| Aspartic aid | 0.002 | 1.4 | 4.38 | — | — | — |
| Glutathione | 0.005 | 1.4 | 3.56 | — | — | — |
| β-Alanine | 0.002 | 1.4 | 18.17 | 0.002 | 1.4 | 7.18 |
| Serine | 0.002 | 1.2 | 0.77 | 0.002 | 1.4 | −37.64 |
| Glutamine | 0.002 | 1.2 | 0.74 | 0.002 | 1.5 | 2.83 |
| Glutamic acid | 0.002 | 1.4 | 8.43 | 0.002 | 1.5 | 7.23 |
| L-Cysteine-glutathione | 0.003 | 1.4 | 9.41d | 0.002 | 1.5 | −1.03 |
| Lysine | — | — | — | 0.002 | 1.4 | 5.17 |
| Pyroglutamic acid | 0.002 | 1.3 | 0.55 | 0.002 | 1.3 | 0.72 |
| Cysteine | 0.004 | 1.2 | 0.67 | 0.004 | 1.3 | 2.10 |
| Histidine | 0.002 | 1.2 | 0.86 | 0.015 | 1.2 | 3.00 |
| Phenylalanine | 0.005 | 1.1 | 0.81 | 0.009 | 1.1 | 2.41 |
| Proline | 0.002 | 1.3 | 1.30 | — | — | — |
| Threonine | 0.002 | 1.1 | 0.76 | 0.015 | 1.1 | 5.32 |
| Tryptophan | 0.002 | 1.2 | 0.75 | 0.015 | — | 2.30 |
| N-Acetyl-Asp-Glu | 0.003 | 1.3 | 2.81d | — | — | — |
| S-Adenosylhomocysteine | 0.005 | 1.1 | 2.17 | — | — | — |
| Tyrosine | — | — | — | 0.026 | 1.1 | — |
| Valine | — | — | — | 0.002 | 1.3 | 2.51 |
| Asparagine | — | — | — | 0.015 | 1.1 | 6.50 |
| Glycylglycine | — | — | — | 0.015 | 1.2 | 0.18 |
Unsupervised principal component analysis (PCA) and supervised orthogonal partial least squares-discriminant (OPLS-DA) in multivariate statistical analysis were used to explore the classification of the different cell lines based on the similarity and difference in the score plots. The quantified concentrations of metabolites were unit variate-scaled (scaled to unit variance) prior to mathematical modeling. The R2Y and Q2Y values from OPLS-DA modeling with typical seven-fold cross-validation were used for assessment of the goodness of fit and model predictability. A 1000-iteration permutation test was performed to evaluate the quality of the computed OPLS-DA model and a negative intercept of Q2Y suggested a truly reliable model. The parameters of the PCA and OPLS-DA models among the three cell lines are shown in Table S5 (ESI†). VIP > 1 was set as the threshold of a significant difference in PCA and OPLS-DA of the multi-variate statistical analysis.
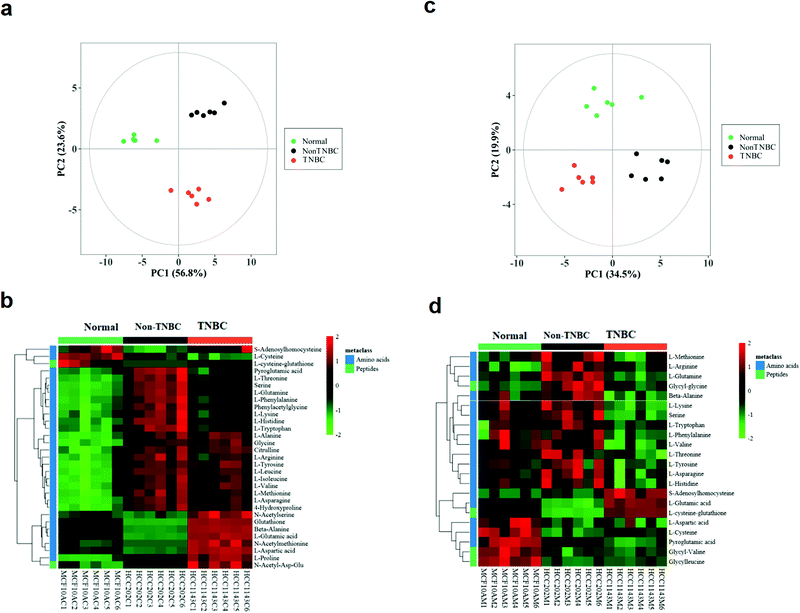
The intra-cellular data from the three groups of normal, non-TNBC and TNBC cells were clearly separated in the PCA score plot (Fig. 1a). The intra-cellular Z-score heatmap of clustering analysis of the three groups exhibited totally different metabolic characteristics in the amino acid profile based on the univariate and multivariate statistical analysis (Fig. 1b). Each red or green square in the heatmap stands for the clustering value of a metabolite in a sample within the cohort, suggesting relative up- or down-regulation of a metabolite among the three groups on the horizontal axis. The vertical axis shows the hierarchical classification of metabolites. The extra-cellular data of the three groups also shared the well separated tendency in the PCA score plot and Z-score heatmap of clustering analysis (Fig. 1c and d). In the extra-cellular Z-score heatmap, the dataset was recalculated (individual value/mean value of blank medium samples), with >1 being excreted and <1 consumed.
The intra- and extra-cellular PCA score plot and Z-score heatmap of clustering analysis between the normal breast cells and non-TNBC are shown in Fig. S1 (ESI†). The intra- and extra-cellular PCA score plot and Z-score heatmap of clustering analysis between the normal breast cells and TNBC are shown in Fig. S2 (ESI†). These results showed the remarkable differentiation between the normal and non-TNBC, and between the normal and TNBC, both intra-cellularly and extra-cellularly.
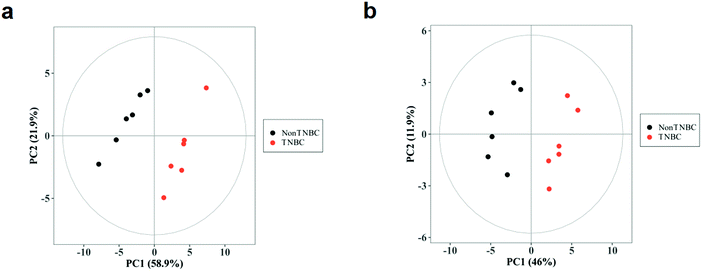
Furthermore, the significant difference between the intra-cellular non-TNBC and TNBC cells was analyzed using PCA and OPLS-DA analysis. The PCA score plots of the cell lysates and cell culture media with a clear separation between the non-TNBC and TNBC are shown in Fig. 2a and b. One predictive component and one orthognal component were used for the OPLS-DA models. The differentiation between the intra- and extra-cellular non-TNBC and TNBC cells was perfect, leading to good fitness and prediction of the models. The parameters of all these PCA and OPLS-DA models among the three cell lines are shown in Table S5 (ESI†).
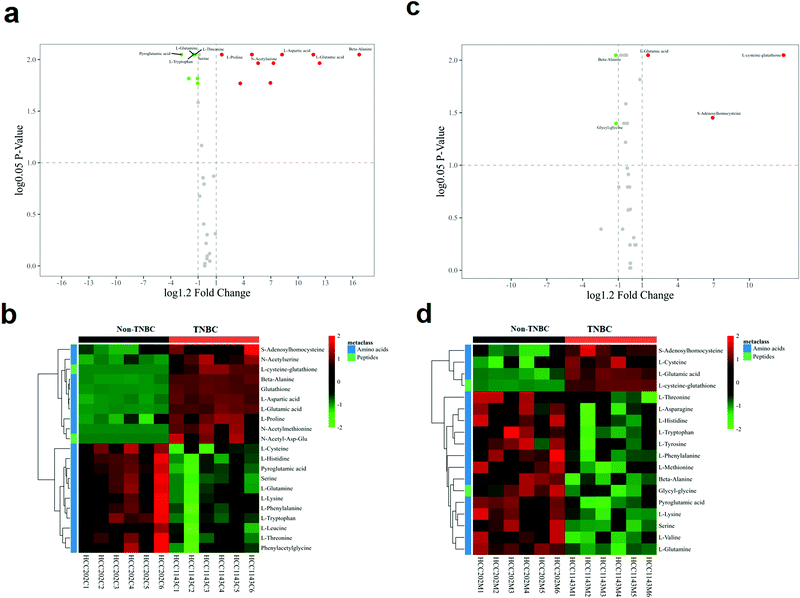
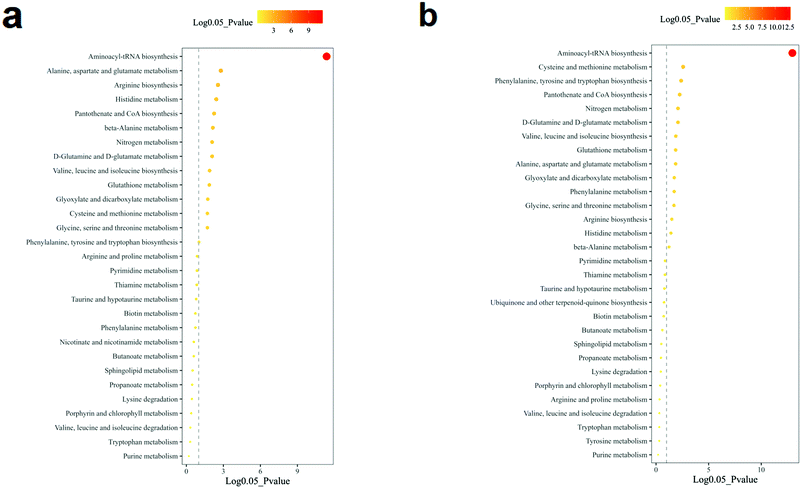
The volcano analysis between the two groups of non-TNBC and TNBC cells is illustrated in Fig. 3a. The Z-score heatmap of clustering analysis between the intra-cellular non-TNBC and TNBC cells is illustrated in Fig. 3b, showing up- or down-regulation of metabolites between non-TNBC and TNBC cells horizontally and a hierarchical classification vertically. The extra-cellular data on the culture medium samples from the non-TNBC and TNBC cells is exhibited in the volcano analysis and Z-score heatmap of clustering analysis in Fig. 3c and d. The intra- and extra-cellular Z-score heatmap together provided a whole dynamic portrayal of the significant difference in metabolic alteration between the non-TNBC and TNBC cells. Encyclopedia of Genes and Genomes (KEGG) metabolic pathway enrichment analyses were performed in intra- and extra-cellular non-TNBC and TNBC, as shown in Fig. 4a and b.
The boxplots of the most significantly different metabolites of amino acids and derivatives are depicted on the non-TNBC and TNBC cells, intra- and extra-cellularly, respectively (Fig. 5a and b). Intra-cellularly, quantified N-acetyl-methionine, N-acetyl-serine, β-alanine, aspartic acid, glutathione and glutamic acid were most significantly increased in TNBC cells, compared to non-TNBC cells (given in ng per 106 cells). Extra-cellularly, significantly increased uptake of glutamine, serine, β-alanine, and lysine and elevated excretion of glutamic acid and L-cysteine-glutathione were observed in the cell culture medium samples by TNBC cells from or to their cell culture media, compared to those by non-TNBC cells. The extra-cellular data was not shown in quantified values, but recalculated by a ratio of the individual value/mean value of blank medium samples, with >1 being dysregulation of excretion and <1 of uptake.
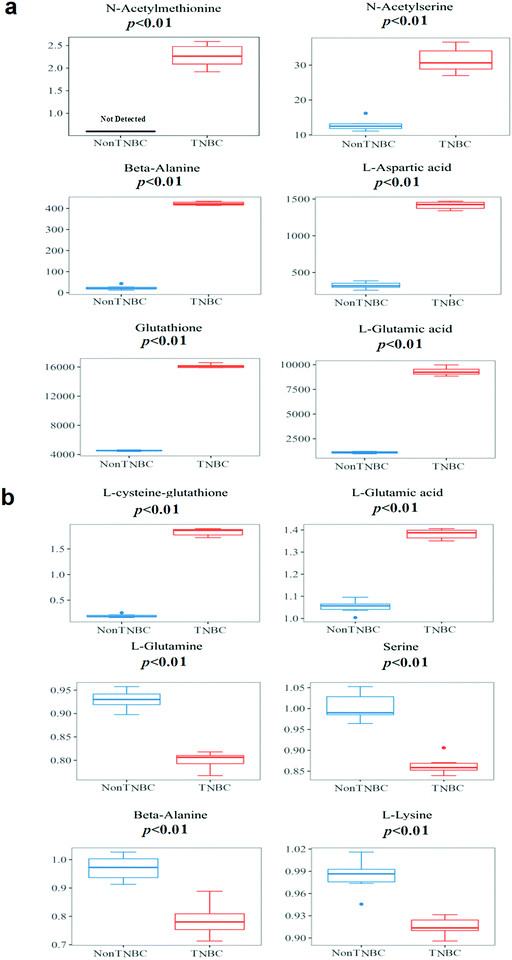 | ||
| Fig. 5 The boxplots of the most significantly different metabolites of intra- and extra-cellular profiling of amino acids and derivatives on cell lysates and cell culture media from the cell lines of non-TNBC and TNBC. (a) Quantified values of cell lysates (given in ng per 106 cells); and (b) cell culture media. The extra-cellular data is not shown in quantified values, but recalculated by a ratio of the individual value/mean value of blank medium samples, with >1 being dysregulation of excretion and <1 of uptake. The quantified extra-cellular data is shown in Table 1. The p-value for each metabolite was obtained using the Mann–Whitney U test. | ||
4. Discussion
In this study, increased amino acid pools of most analytes were observed in both breast cells of TNBC and non-TNBC, compared to normal breast epithelial cells (Tables S3 and S4, ESI†). Furthermore, characteristic metabolic dysregulation of amino acids and derivatives between non-TNBC and TNBC cells was observed, especially in the metabolic pathways involving glutamine and glutamic acid, with both intra- and extra-cellular alterations correlated.The dysregulation of N-acetyl-L-methionine was quantified and found for the first time with a significant increase (>2-fold, VIP > 1 and p < 0.01) in TNBC cells, compared to non-TNBC (Fig. 5a). Methionine is the only sulfur containing amino acid of the eight essential amino acids in the human diet, which plays a critical role in sustaining methylation and the redox state in cells. H. Jeon et al. reported that the deprivation of methionine suppressed distant metastasis of TNBC, reducing the phosphorylation, activity and mRNA expression of MMP-2 and MMP-9 in cells of MDA-MB-231 while raising metalloproteinase 1 in mRNA expression.7N-Acetyl-L-methionine has been long reported to be accessible and a replacement in the human diet for the methionine requirement.18,19 Smith et al. demonstrated the detected cellular endogenous presence of N-acetyl-methionine in brain derived cell lines, as well as mouse and human brain tissue.20
N-Acetyl-serine was the other acetyl-amino acid that was most significantly increased (>2-fold, VIP > 1 and p < 0.01) in TNBC cells, compared to the non-TNBC in our study (Fig. 5a). N-Acetyl-serine was previously reported to be differentially regulated in global metabonomic studies of cancers,21 but was never quantified. In our study, we first detected the concentration of N-acetyl-serine, at the level of 12.9 ± 1.81 ng per 106 cells in the non-TNBC cells and 32.3 ± 3.40 ng per 106 cells in the TNBC cells, being significantly different between the TNBC and non-TNBC cells (Table 2). The mechanism underlying the dysregulation of N-acetyl-serine in TNBC cells is still unknown and needs more further exploration.
Besides the two acetyl-amino acids, β-alanine, aspartic acid, and glutamic acid were also intra-cellularly quantified and found to be significantly increased (>2-fold, VIP > 1 and p < 0.01) in TNBC cells, compared to the non-TNBC (Fig. 5a). Meanwhile, β-alanine, glutamine, serine, glutamic acid and L-cysteine-glutathione were also extra-cellularly quantified and dysregulated in the cell culture medium samples from TNBC cells, compared to those from the non-TNBC.
β-Alanine was reported to be elevated in global metabolomics of breast cancer tissue, and also more increased in the subtype of ER-breast cancer than in that of ER+.22 4-Aminobutyrate aminotransferase (ABAT) depended on β-alanine as its substrate. Transcript expression of ABAT was co-relative to the ER status, with lower transcript expression of ABAT being related to lower expression of the ABAT protein and a higher concentration of beta-alanine.22 Consistent with the global metabolomics of clinical breast cancer tissue study, we found that the quantified β-alanine was significantly intra-cellularly increased (>2-fold, VIP > 1 and p < 0.01) in TNBC cells, compared to the non-TNBC cells. Extra-cellularly, β-alanine had an elevated uptake (VIP > 1 and p < 0.01) by the TNBC cells from the cell culture medium extra-cellularly, compared to that by the non-TNBC cells (Fig. 5a and b).
Aspartic acid, glutathione, L-cysteine-glutathione, glutamine and glutamic acid are all involved in glutamine metabolism. Glutamine and glucose were the nutrients consumed most rapidly in many different cancer cell lines.23,24 Glutamine was also the one amino acid that is deleted most quickly in blood serum of many cancer patients.23–25 Glutamine supplied nitrogen for both nucleotide synthesis and synthesis of non-essential amino acids, with aspartic acid, glutamic acid and so on forming the downstream metabolites.26 Meanwhile, glutamine was related to the synthesis of glutathione, sustaining the redox balance.27 The significantly decreased glutamine in TNBC cells was observed in previous literature because it usually had a large uptake and was deleted quickly for consumption in the TNBC cell line.28 As the common substrate of the most increased amino acids of aspartic acid, glutathione and glutamic acid in TNBCs, glutamine in our study also exhibited decreased intra-cellular regulation in the TNBC, compared to the non-TNBC cells. Extra-cellularly, glutamine showed a significantly increased uptake by the TNBC cells from the cell culture medium. The intra- and extra-cellular dysregulations of glutamine provide a novel dynamic portrayal of the characteristic requirements of TNBC, with more supply of glutamine and more active metabolic recycling than that in non-TNBC (Fig. 5b). Similar to glutamine, serine was also significantly increased in the uptake by TNBC from the cell culture medium. Serine was also important for the proliferation of cancer cells in previous literature. Precursors for nucleotide synthesis depend on the one-carbon units produced by serine.29 Many tumors were reported to sustain proliferation, relying on a stable extracellular supply of serine.30 The elevated uptake of serine by the TNBC cells was correlated to the intra-cellular up-regulation of N-acetyl-serine in the TNBC (Fig. 5a and b). Lysine, one of the essential amino acids and usually supplied by exogenous sources, was increased in the uptake by the TNBC cells from the culture media (Fig. 5b).
Glutamic acid, the main downstream metabolite of glutamine, plays many important roles in biological processes. Glutamic acid was converted to α-KG without the nitrogen, either released as ammonium (deamination) or transferred to an α-ketoacid (transamination), which generated the corresponding amino acid. Glutamic acid also underwent ligation to cysteine, forming γ-glutamylcysteine. And then glycine would be condensed with γ-glutamylcysteine, yielding the antioxidant glutathione.13 Glutathione and L-cysteine-glutathione were critical in the role of the antioxidant system.31 Aspartic acid and glutamine provided the substrate source for synthesis of asparagine through the asparagine synthetase (ASNS) enzyme. Yet, some types of cancer cells depend on asparagine supply from blood serum, having no ASNS expression, such as leukemic lymphoblasts.13 Aspartic acid is the substrate for synthesis of asparagine through ASNS. Intra-cellularly, largely increased aspartic acid, glutathione and glutamic acid (>2-fold, VIP > 1 and p < 0.01) were observed in the TNBC in our study. Extra-cellularly, glutamic acid and L-cysteine-glutathione were increased in the excretion (VIP > 1 and p < 0.01) by the TNBC cells to their cell culture media (Fig. 5a and b).
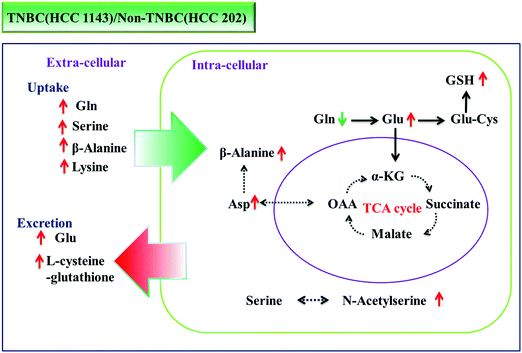
Using large-scale targeted proteomics, Kodama demonstrated that glutamine metabolism promotes malignant progression of cancer by a shift of nitrogen from the anaplerotic pathway into the TCA cycle to nucleotide biosynthesis through glutaminase (GLS1) and phosphoribosyl pyrophosphate amidotransferase (PPAT).32 In the current study, metabolic pathways involving glutamine and glutamic acid are also found to be clearly increased in the TNBC cells (the subtype of breast cancer with more metastatic aggressiveness), compared to the non-TNBC cells, with both intra- and extra-cellularly correlated. Glutamine is more consumed (0.74-fold intra-cellularly; 2.83-fold in uptake; both p < 0.002), and glutamic acid is more produced (8.43-fold, p < 0.002) and excreted more (7.23-fold in excretion, p < 0.002) to the culture medium by TNBC cells. Fig. 6 illustrates the dynamic portrayal of disturbed metabolic pathways involving glutamine, glutamic acid and other key amino acids in TNBC cells and the interactions with their micro-environment, which were different from those of non-TNBC cells, both intra- and extra-cellularly.
Some certain amino acids have been considered as potential targets in previous cancer studies. Glutamine, serine and glycine are observed to be the amino acids that are most quickly consumed in many cultured cancer cells.14,15 Deprivation of methionine was demonstrated to inhibit the invasion of triple-negative breast cancer.33 The findings in this study confirm the increased amino acid pool in cancer cells, and further illustrate the dynamic uptake or excretion of a large amount of amino acids in the quantitative profile. These dysregulated amino acids might be transferred to the target in the studies of human TNBC, compared to non-TNBC. For example, the most consumed amino acids in the present study, such as glutamine, serine, β-alanine and lysine, seem critical in the growth of TNBC, which might lead to further studies of deprivation or reduced supply in the diet to inhibit the rapid proliferation of TNBC. The glutamine metabolic pathway is more enhanced in TNBC than that in non-TNBC, with more consumed glutamine and more excreted glutamic acid, which might provide new insight or novel targets for the treatment of TNBC. Based on the quantitative profiling results, specific functional studies on dysregulated amino acids could be further investigated.
5. Conclusion
In conclusion, characteristic metabolic alteration of amino acids and derivatives was observed in both intra- and extra-cellular metabolomic profiles between TNBC and non-TNBC cells, especially in up-regulated pathways involving glutamine and glutamic acid metabolism. Intra-cellularly, quantified N-acetyl-methionine, N-acetyl-serine, β-alanine, aspartic acid, glutathione and glutamic acid were most significantly increased (>2-fold, p < 0.01 and VIP > 1) in TNBC cells, compared to non-TNBC cells. Extra-cellularly, significantly increased uptake of glutamine, serine, β-alanine, and lysine and elevated excretion of glutamic acid and L-cysteine-glutathione (p < 0.01 and VIP > 1) were observed in the cell culture medium samples by TNBC cells from or to their cell culture media, compared to those by non-TNBC cells. The different interaction of the non-TNBC and TNBC cells with their micro-environments was correlated with their different intra-cellular metabolic disturbance. Quantification of these distinctive metabolites of TNBC cells might provide novel insights and new targets for treatment.Author contribution statement
YC and HW conceived and supervised the study; FK, BZ, YC and HW designed the experiments; BZ performed the experiments; BZ, WZ, CL, FK and YC analyzed the data; FK, BZ, YC and HW wrote and revised the manuscript.Abbreviations
| TNBC | Triple-negative breast cancer |
| HILIC-MS/MS | Hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry |
| non-TNBC | Non-triple-negative breast cancer cell line |
| ER | Estrogen |
| PR | Progesterone |
| HER2 | Human epidermal growth factor receptor 2 |
| TGCA | The Cancer Genome Atlas |
| PCA | Principal component analysis |
| OPLS-DA | Orthogonal partial least squares-discriminant |
| ABAT | 4-Aminobutyrate aminotransferase |
| ASNS | Asparagine synthetase |
| GLS1 | Glutaminase |
| PPAT | Phosphoribosyl pyrophosphate amidotransferase. |
Conflicts of interest
The authors declare no conflicts of interest.References
- L. A. Torre, F. Bray, R. L. Siegel, J. Ferlay, J. Lortet-Tieulent and A. Jemal, Global cancer statistics, 2012, Ca-Cancer J. Clin., 2015, 65, 87–108 CrossRef.
- B. D. Lehmann and J. A. Pietenpol, Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes, J. Pathol., 2014, 232, 142–150 CrossRef.
- Q. Jiao, A. Wu, G. Shao, H. Peng, M. Wang, S. Ji, P. Liu and J. Zhang, The latest progress in research on triple negative breast cancer (TNBC): risk factors, possible therapeutic targets and prognostic markers, J. Thorac. Dis., 2014, 6, 1329–1335 Search PubMed.
- A. D. Elias, Triple-negative breast cancer: a short review, Am. J. Clin. Oncol., 2010, 33, 637–645 CrossRef.
- E. Cerami, J. Gao, U. Dogrusoz, B. E. Gross, S. O. Sumer, B. A. Aksoy, A. Jacobsen, C. J. Byrne, M. L. Heuer, E. Larsson, Y. Antipin, B. Reva, A. P. Goldberg, C. Sander and N. Schultz, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Cancer Discovery, 2012, 2, 401–404 CrossRef.
- S. P. Shah, A. Roth, R. Goya, A. Oloumi, G. Ha, Y. Zhao, G. Turashvili, J. Ding, K. Tse, G. Haffari, A. Bashashati, L. M. Prentice, J. Khattra, A. Burleigh, D. Yap, V. Bernard, A. McPherson, K. Shumansky, A. Crisan, R. Giuliany, A. Heravi-Moussavi, J. Rosner, D. Lai, I. Birol, R. Varhol, A. Tam, N. Dhalla, T. Zeng, K. Ma, S. K. Chan, M. Griffith, A. Moradian, S. W. Cheng, G. B. Morin, P. Watson, K. Gelmon, S. Chia, S. F. Chin, C. Curtis, O. M. Rueda, P. D. Pharoah, S. Damaraju, J. Mackey, K. Hoon, T. Harkins, V. Tadigotla, M. Sigaroudinia, P. Gascard, T. Tlsty, J. F. Costello, I. M. Meyer, C. J. Eaves, W. W. Wasserman, S. Jones, D. Huntsman, M. Hirst, C. Caldas, M. A. Marra and S. Aparicio, The clonal and mutational evolution spectrum of primary triple-negative breast cancers, Nature, 2012, 486, 395–399 CrossRef CAS.
- J. H. K. Hyein Jeon, E. Lee, Y. Jin Jang, J. Eun Son, J. Yeon Kwon, T.-g. Lim, S. Kim, J. Han Yoon Park, J.-E. Kim and K. Won Lee, Methionine deprivation suppresses triple-negative breast cancer metastasis in vitro and in vivo, Oncotarget, 2016, 7, 67223–67234 CrossRef.
- D. R. Wise and C. B. Thompson, Glutamine addiction: a new therapeutic target in cancer, Trends Biochem. Sci., 2010, 35, 427–433 CrossRef CAS.
- Y. D. Bhutia, E. Babu, S. Ramachandran and V. Ganapathy, Amino Acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs, Cancer Res., 2015, 75, 1782–1788 CrossRef CAS.
- C. V. Dang, Rethinking the Warburg effect with Myc micromanaging glutamine metabolism, Cancer Res., 2010, 70, 859–862 CrossRef CAS.
- Y. M. Kanaan, B. P. Sampey, D. Beyene, A. K. Esnakula, T. J. Naab, L. J. Ricks-Santi, S. Dasi, A. Day, K. W. Blackman, W. Frederick, R. L. Copeland, Sr. and E. Gabrielson, R.L. Dewitty, Jr., Metabolic profile of triple-negative breast cancer in African-American women reveals potential biomarkers of aggressive disease, Cancer genomics &, Proteomics, 2014, 11, 279–294 Search PubMed.
- B. Zhu, L. Li, H. Wei, W. Zhou, W. Zhou, F. Li, P. Lin, J. Sheng, Q. Wang, C. Yan and Y. Cheng, A simultaneously quantitative profiling method for 40 endogenous amino acids and derivatives in cell lines using hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry, Talanta, 2020, 207, 120256 CrossRef CAS.
- M. J. Lukey, W. P. Katt and R. A. Cerione, Targeting amino acid metabolism for cancer therapy, Drug Discovery Today, 2017, 22, 796–804 CrossRef CAS.
- M. Jain, R. Nilsson, S. Sharma, N. Madhusudhan, T. Kitami, A. L. Souza, R. Kafri, M. W. Kirschner, C. B. Clish and V. K. Mootha, Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation, Science, 2012, 336, 1040–1044 CrossRef CAS.
- A. M. Hosios, V. C. Hecht, L. V. Danai, M. O. Johnson, J. C. Rathmell, M. L. Steinhauser, S. R. Manalis and M. G. Vander Heiden, Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells, Dev. Cell, 2016, 36, 540–549 CrossRef CAS.
- I. Elia, D. Broekaert, S. Christen, R. Boon, E. Radaelli, M. F. Orth, C. Verfaillie, T. G. P. Grunewald and S. M. Fendt, Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells, Nat. Commun., 2017, 8, 15267 CrossRef.
- V. Cipriani, L. Lores-Motta, F. He, D. Fathalla, V. Tilakaratna, S. McHarg, N. Bayatti, I. E. Acar, C. B. Hoyng, S. Fauser, A. T. Moore, J. R. W. Yates, E. K. de Jong, B. P. Morgan, A. I. den Hollander, P. N. Bishop and S. J. Clark, Increased circulating levels of Factor H-Related Protein 4 are strongly associated with age-related macular degeneration, Nat. Commun., 2020, 11, 778 CrossRef CAS.
- R. W. Boggs, J. T. Rotruck and R. A. Damico, Acetylmethionine as a source of methionine for the rat, The, J. Nutr., 1975, 105, 326–330 CrossRef CAS.
- J. T. Rotruck and R. W. Boggs, Comparative metabolism of L-methionine and N-acetylated derivatives of methionine, J. Nutr., 1975, 105, 331–337 CrossRef CAS.
- T. Smith, M. S. Ghandour and P. L. Wood, Detection of N-acetyl methionine in human and murine brain and neuronal and glial derived cell lines, J. Neurochem., 2011, 118, 187–194 CrossRef CAS.
- I. Zarei, R. C. Oppel, E. C. Borresen, R. J. Brown and E. P. Ryan, Modulation of plasma and urine metabolome in colorectal cancer survivors consuming rice bran, Integr. Food, Nutr. Metab., 2019, 6, 1–14 Search PubMed.
- J. Budczies, S. F. Brockmoller, B. M. Muller, D. K. Barupal, C. Richter-Ehrenstein, A. Kleine-Tebbe, J. L. Griffin, M. Oresic, M. Dietel, C. Denkert and O. Fiehn, Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: alterations in glutamine and beta-alanine metabolism, J. Proteomics, 2013, 94, 279–288 CrossRef CAS.
- J. L. Coloff, J. P. Murphy, C. R. Braun, I. S. Harris, L. M. Shelton, K. Kami, S. P. Gygi, L. M. Selfors and J. S. Brugge, Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells, Cell Metab., 2016, 23, 867–880 CrossRef CAS.
- R. J. DeBerardinis, A. Mancuso, E. Daikhin, I. Nissim, M. Yudkoff, S. Wehrli and C. B. Thompson, Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 19345–19350 CrossRef CAS.
- Y. Cheng, G. Xie, T. Chen, Y. Qiu, X. Zou, M. Zheng, B. Tan, B. Feng, T. Dong, P. He, L. Zhao, A. Zhao, L. X. Xu, Y. Zhang and W. Jia, Distinct urinary metabolic profile of human colorectal cancer, J. Proteome Res., 2012, 11, 1354–1363 CrossRef CAS.
- Y. J. Cha, E. S. Kim and J. S. Koo, Amino Acid Transporters and Glutamine Metabolism in Breast Cancer, Int. J. Mol. Sci., 2018, 19(3), 907 CrossRef.
- M. Yudkoff, D. Pleasure, L. Cregar, Z. P. Lin, I. Nissim, J. Stern and I. Nissim, Glutathione turnover in cultured astrocytes: studies with [15N]glutamate, J. Neurochem., 1990, 55, 137–145 CrossRef CAS.
- G. D. Chen Xiaowu, Sun Qinsheng, Chen Yuzong and Liu Hongxia, & Jiang Yuyang Metabolic Profiling of Amino Acids by Liquid Chromatography–Tandem Mass Spectrometry (LC–MS) to Characterize the Significance of Glutamine in Triple-Negative Breast Cancer (TNBC), Anal. Lett., 2019, 52, 1068–1082 CrossRef.
- K. R. Mattaini, M. R. Sullivan and M. G. Vander Heiden, The importance of serine metabolism in cancer, J. Cell Biol., 2016, 214, 249–257 CrossRef CAS.
- O. D. Maddocks, C. R. Berkers, S. M. Mason, L. Zheng, K. Blyth, E. Gottlieb and K. H. Vousden, Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells, Nature, 2013, 493, 542–546 CrossRef CAS.
- S. Ashfaq, J. L. Abramson, D. P. Jones, S. D. Rhodes, W. S. Weintraub, W. C. Hooper, V. Vaccarino, R. W. Alexander, D. G. Harrison and A. A. Quyyumi, Endothelial function and aminothiol biomarkers of oxidative stress in healthy adults, Hypertension, 2008, 52, 80–85 CrossRef CAS.
- M. Kodama, K. Oshikawa, H. Shimizu, S. Yoshioka, M. Takahashi, Y. Izumi, T. Bamba, C. Tateishi, T. Tomonaga, M. Matsumoto and K. I. Nakayama, A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer, Nat. Commun., 2020, 11, 1320 CrossRef CAS.
- H. Jeon, J. H. Kim, E. Lee, Y. J. Jang, J. E. Son, J. Y. Kwon, T. G. Lim, S. Kim, J. H. Park, J. E. Kim and K. W. Lee, Methionine deprivation suppresses triple-negative breast cancer metastasis in vitro and in vivo, Oncotarget, 2016, 7, 67223–67234 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0mo00126k |
| ‡ These authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2021 |