Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effect-directed analysis and chemical identification of agonists of peroxisome proliferator-activated receptors in white button mushroom†
Long
Pham Ngoc
 *abc,
Rick
Helmus
*abc,
Rick
Helmus
 d,
Andreas W.
Ehlers
ef,
Kees
Swart
a,
Harry
Besselink
a,
Eva
de Rijke
d,
Andreas W.
Ehlers
ef,
Kees
Swart
a,
Harry
Besselink
a,
Eva
de Rijke
 d,
Ha
Dang Thi Cam
b,
Abraham
Brouwer
ac and
Bart
van der Burg
d,
Ha
Dang Thi Cam
b,
Abraham
Brouwer
ac and
Bart
van der Burg
 a
a
aBioDetection Systems, Science Park 406, 1098 XH Amsterdam, The Netherlands. E-mail: n.l.pham@vu.nl; Tel: +31204350750
bInstitute of Biotechnology, Vietnam Academy of Science and Technology (VAST), 18 Hoang Quoc Viet, Cau Giay, Hanoi, Vietnam
cDepartment of Ecological Science, Vrije Universitei, De Boelelaan 1085, 1081 HV, Amsterdam, The Netherlands
dInstitute of Biodiversity & Ecosystem Dynamics, University of Amsterdam, Science Park 904, 1090 GE, Amsterdam, The Netherlands
eVan't Hoff Institute for Molecular Sciences, University of Amsterdam, Science Park 904, 1090 GD Amsterdam, The Netherlands
fDepartment of Chemistry, Science Faculty, University of Johannesburg, PO Box 254, Auckland Park, Johannesburg, South Africa
First published on 7th December 2020
Abstract
Obesity has a serious effect on human health. It relates to metabolic syndrome, including the associated disorders such as type 2 diabetes, heart disease, stroke and hyperemia. The peroxisome proliferator-activated receptors (PPARs) are important receptors to control fat metabolism in the human body. Because of the safety concerns of synthetic drugs targeting PPARs, ligands from natural sources have drawn interest. Earlier, we have found high PPAR activities in extracts from Agaricus bisporus (white button mushroom, WBM). WBM contains a wide range of candidate compounds which could be agonists of PPARs. To identify which compounds are responsible for PPAR activation by WBM extracts, we used fractionation coupled to effect-directed analysis with reporter gene assays specific for all three PPARs for purification and LC/MS-TOF and NMR for compound identification in purified active fractions. Surprisingly, we identified the relatively common dietary fatty acid, linoleic acid, as the main ligand of PPARs in WBM. Possibly, the relatively low levels of linoleic acid in WBM are sufficient and instrumental in inducing its anti-obesogenic effects, avoiding high energy intake and negative health effects associated with high levels of linoleic acid consumption. However, it could not be excluded that a minor relatively potent compound contributes towards PPAR activation, while the anti-obesity effects of WBM may be further enhanced by receptor expression modulating compounds or compounds with completely PPAR unrelated modes of action.
Introduction
Obesity has a significant impact on human health, leading to an increased incidence of metabolic syndrome, which is characterized by central obesity, high blood pressure, high blood sugar, high level of triglycerides, and low serum high-density lipoprotein (HDL). This can lead to type 2 diabetes, heart disease and hyperemia.1–3 Thus, there is a great demand for safe treatments of these disorders. Several drugs are used to treat obesity-related disorders. These include fibrates to treat hyperlipidemia and thiazolidinediones (TZD) to treat type 2 diabetes. Remarkably, these drugs were found to be ligands for peroxisome proliferator-activated receptors (PPARs). PPARs are a subgroup of nuclear hormone receptors and are essential in the regulation of fat storage and metabolism.4,5 There are three isoforms of PPARs, i.e., PPARα, PPARγ, and PPARδ, with partly overlapping and specific functions, and different tissue distribution. PPARα is a key player in lipid metabolism, while PPARγ has a major role in stimulating adipogenesis and improving insulin sensitivity. Fibrates are ligands for PPARα, while TZD targets PPARγ.5 PPARδ also is important in fat metabolism, generating heat when uncoupled to energy production, playing a role in the switch from glucose to fat burning.4,5 Unfortunately there are serious side effects related to the use of TZDs such as rosiglitazone, while so far attempts have failed to generate safe drugs for the most interesting receptor in this context, PPARδ.6,7 There is a great need to identify alternative PPAR ligands, which could include non-drug related approaches. It should be noted that there is a possibility that bioactive compounds activating the PPARs would give the same side effects as the current pharmaceuticals, if these side effects were inherent to PPAR activation. This indeed could be the case, and there are concerns and controversies regarding the effects of PPARδ activation on cancer progression.8 This, however, may be caused by the complex nature of cancer progression and the many possible points of interaction, together with the differences in the animal and cellular models used.8 To avoid misinterpretation of these data, the best possible test should be carried out in humans. Since this is not feasible, an alternative is the identification of active molecules in a food item with a long history of safe use, as has been done in the current study.As such, life-style changes, including more healthy diets and dietary supplements, are increasingly seen as efficient alternatives to pharmacological treatment.9 Edible mushrooms have been reported to be effective in combating metabolic disorders including reduction of hyperlipidemia, hypertension, and hyperglycemia, and are a possible resource to isolate their functional ingredients avoiding the side effects of synthetic drugs.10,11 Humans have used edible mushrooms, including Agaricus bisporus, Pleurotus ostreatus, and Lentinula edodes, for hundreds of years because of their health benefits.12–15 White button mushroom (WBM; Agaricus bisporus) is the most produced mushroom worldwide, and could be an interesting source of anti-obesogenic molecules.16,17 It has been shown in rats that a WBM supplement to a high-fat diet resulted in a significant decrease in body weight after 72 days, and inhibited liver accumulation of fat.18 In addition, WBM powder fed to diabetic male Sprague-Dawley rats caused a significant reduction of plasma glucose and triglyceride levels after three weeks. Besides, hypercholesterolemic rats fed with WBM powder showed a significant decrease in plasma total cholesterol and low-density lipoprotein.19 Moreover in a randomized clinical trial in humans, WBM consumption resulted in clear positive effects on body weight and health parameters such as central obesity, blood pressure, and lipid profiles.20
WBM contains a wide range of bioactive compounds, including phenolic compounds, unsaturated and saturated fatty acids, and β-glucans.16 It is unclear however, which components may be responsible for the possible anti-obesogenic effects observed in humans.20 As always, the cause and significance of these health effects are extremely difficult to substantiate in a background of variable and complex food intake. A wide range of these natural compounds, including fatty acids, polyphenols, and flavonoids, have been shown to be ligands of PPARs, thus making these receptors interesting targets to discover natural compounds that are active in the prevention or treatment of metabolic diseases.7,21 To this end, we recently developed a specific panel of human cell-based CALUX® reporter gene assays for all three full-length PPARs and found high PPAR activities in fungi.17 Notably, high levels of PPARδ activation were found in edible mushroom including the common WBM.17 Here, we used effect-directed analysis and chemical identification using liquid chromatography coupled time-of-flight mass spectrometry (LC/MS-TOF) and nuclear magnetic resonance spectroscopy (NMR) to elucidate the identity of the PPAR interacting ligands from WBM.
Materials and methods
WBM powder preparation
WBM was purchased from a local supermarket in The Netherlands. Two kilograms of fresh WBM was cut into small pieces and dehydrated by using a freeze drier. Then, the dehydrated WBM was ground to a powder using an A11 Basic analytical mill (IKA, Staufen, Germany). This standardized batch of WBM powder, used in all experiments, was stored at −80 °C until further use.Chemicals
Rosiglitazone (≥98%, CAS no. 122320-73-4) was purchased from Bio-connect BV (Huissen, The Netherlands). L-0165.041 (≥98%, CAS no. 79558-09-1) and GW7647 (≥98%, CAS no. 25129-71-3) were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands). Methanol (absolute, CAS no. 67-56-1) and acetonitrile (99.9%, CAS no. 75-05-8) were purchased from Biosolve BV (Valkenswaard, TheNetherlands). Ethanol (≥99.5%, CAS no. 64-17-5) was obtained from Thermo Fisher Scientific BV (Breda, The Netherlands). Dimethylsulfoxide (DMSO, 99.9%, CAS no. 67-68-5) was obtained from Thermo Fisher Scientific BV (Breda, The Netherlands). Linoleic acid (LA) (≥99%, CAS no. 60-33-3), conjugated linoleic acid (CLA) 9-cis, 11-trans (≥96%, CAS no. 2540-56-9), and CLA 10-trans, 12-cis (≥96%, CAS no. 2420-56-6) were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands). Water used for extraction and fractionation was purified using a PureLab flex system (ELGA Labwater, High Wycombe, UK).Preparation of WBM extracts
The extraction procedure was a modification of the method developed by Gijsbers et al.;22 in brief: 3 mL of 0.1 M sodium acetate (pH 4.8) and 1 mL of viscozyme L (Sigma-Aldrich, St Louis, MO, USA) were added to 1 g of the dried sample material, and the mixture was incubated at 37 °C for 2 hours. Then, 30 mL of ethanol was added, and the sample mixture was transferred to an ultrasonic bath for 10 minutes and subsequently centrifuged at 3000g for 10 minutes. The supernatant was collected and dried in a GeneVac Rocket evaporation system (Genevac Limited, Ipswich, UK). The WBM dried extract was re-dissolved in 2 mL methanol and filtered. The mixture was kept at −20 °C until further experiments.Cell culture
CALUX cells based on human U2OS osteosarcoma cells (American Type Culture Collection [ATCC], Manassas, USA), expressed a full length PPARα,δ or γ receptor construct, respectively, and a luciferase reporter gene.17 In addition, to assess non-specific luciferase modulation, e.g. due to cytotoxicity, the Cytotox CALUX cell line was used, consisting of U2-OS cells constitutively expressing the luciferase gene.23 CALUX cells were cultured in a DMEM/F12 glutamax medium supplemented with 7.5% fetal calf serum, non-essential amino acids, and penicillin/streptomycin (all from Invitrogen, Breda, The Netherlands) at the final concentrations of 10 U mL−1 and 10 μg mL−1, respectively, as described before.24 Once per week, G418 (Duchefa Biochemie, Haarlem, The Netherlands) was added to the culture medium at a nominal concentration of 200 μg mL−1.CALUX® reporter gene assays
The PPAR activities of sample extracts and fractions were tested by measuring luciferase activity in the PPAR CALUX reporter cells. In parallel, the Cytotox CALUX was used to assess assay interferences. Samples that led to 20% or more signal reduction were excluded from the evaluation.23 PPAR CALUX reporter gene assays were performed as described before.17 In short, PPAR CALUX cells were seeded in 96-well plates (Greiner-Bio-One, Alphen aan den Rijn, The Netherlands) at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in a 100 μL assay medium (DMEM/F12 without phenol red, Thermo Fisher Scientific BV, Breda, The Netherlands). After 24 hours, when the cells formed a monolayer, 100 μL of fresh assay medium supplemented with the sample extract or WBM fractions were added to the wells. When testing the sample, the percentage of DMSO in the exposure medium was kept at 0.1%. On each plate, a dilution series of reference PPAR agonists (rosiglitazone, L-0165.041 and GW7647) was included as a reference curve. After 24 hours of exposure, the light production as relative light units (RLU) of luciferase activity was measured using a TriStar LB 941 luminometer (Berthold Technologies, Bad Wildbad, Germany).
000 cells per well in a 100 μL assay medium (DMEM/F12 without phenol red, Thermo Fisher Scientific BV, Breda, The Netherlands). After 24 hours, when the cells formed a monolayer, 100 μL of fresh assay medium supplemented with the sample extract or WBM fractions were added to the wells. When testing the sample, the percentage of DMSO in the exposure medium was kept at 0.1%. On each plate, a dilution series of reference PPAR agonists (rosiglitazone, L-0165.041 and GW7647) was included as a reference curve. After 24 hours of exposure, the light production as relative light units (RLU) of luciferase activity was measured using a TriStar LB 941 luminometer (Berthold Technologies, Bad Wildbad, Germany).
PPAR CALUX activity in the sample extracts was expressed as a relative induction to the maximum response from the reference compounds. These results were interpolated in the PPAR CALUX bioassay specific calibration curves using the statistical software package GraphPad Prism V5.03 (non-linear regression, variable slope, 4 parameters, robust fit), after which the bioactivity was quantified. For each of the sample extracts, the lowest concentration of the sample extract present in the CALUX exposure medium (μg sample per L medium) causing a relative induction of 10% (PC10) was determined as the lowest observed effect concentration (LOEC).
Sample fractionation and effect-directed analysis
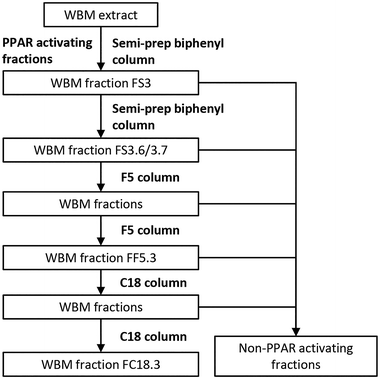
WBM fractionation was carried out using three LC columns (biphenyl semi-prep column, F5 column, and C18 column; Fig. 1), using a HPLC Agilent 1260 Infinity system (Agilent Technologies Netherlands BV, Amstelveen, The Netherlands). First, the WBM extract was fractionated using a biphenyl semi-prep column (Kinetex 5 μM biphenyl, 250 × 10 mm; Phenomenex, Torrance, USA). A gradient solvent system using methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water was used from 20% to 95% for 27 minutes, with a flow rate of 3.8 mL per minute. Subsequent fractions were collected using a Waters fraction collector III (Waters Corporation, Milford, USA) and evaluated for PPAR activities by CALUX assays. WBM fraction 3 (FS3), which was the highest in PPAR activity, was further sub-fractionated using the biphenyl semi-prep column using smaller, one-minute intervals of collection. WBM active fraction FS3.6 and FS3.7 were separated further on an analytical F5 column (Kinetex 1.7 μM F5, 100 × 2.1 mm; Phenomenex, Torrance, USA), using a solvent gradient of methanol
water was used from 20% to 95% for 27 minutes, with a flow rate of 3.8 mL per minute. Subsequent fractions were collected using a Waters fraction collector III (Waters Corporation, Milford, USA) and evaluated for PPAR activities by CALUX assays. WBM fraction 3 (FS3), which was the highest in PPAR activity, was further sub-fractionated using the biphenyl semi-prep column using smaller, one-minute intervals of collection. WBM active fraction FS3.6 and FS3.7 were separated further on an analytical F5 column (Kinetex 1.7 μM F5, 100 × 2.1 mm; Phenomenex, Torrance, USA), using a solvent gradient of methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water from 20% to 100% in 31 minutes, with a flow rate of 0.25 mL per minute, and fractions were tested for bioactivities using PPAR CALUX assays. The WBM active fraction from the F5 column was further sub-fractionated on an F5 column. In the last step of WBM ligand purification, the active fraction collected from the F5 (FF5.3) column was purified by using a C18 column (Kinetex 2.6 μM UXB-C18, 75 × 2.1 mm; Phenomenex, Torrance, USA). A solvent gradient was applied using acetonitrile (0.1% formic acid)
water from 20% to 100% in 31 minutes, with a flow rate of 0.25 mL per minute, and fractions were tested for bioactivities using PPAR CALUX assays. The WBM active fraction from the F5 column was further sub-fractionated on an F5 column. In the last step of WBM ligand purification, the active fraction collected from the F5 (FF5.3) column was purified by using a C18 column (Kinetex 2.6 μM UXB-C18, 75 × 2.1 mm; Phenomenex, Torrance, USA). A solvent gradient was applied using acetonitrile (0.1% formic acid)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (0.1% formic acid) from 70% to 90% in 10 minutes, with a flow rate of 0.25 mL per minute. WBM active fraction from the C18 column was fractionated a second time using the C18 column and the final highly purified PPAR active fraction FC18.3 obtained was dissolved in acetonitrile and kept at −20 °C.
water (0.1% formic acid) from 70% to 90% in 10 minutes, with a flow rate of 0.25 mL per minute. WBM active fraction from the C18 column was fractionated a second time using the C18 column and the final highly purified PPAR active fraction FC18.3 obtained was dissolved in acetonitrile and kept at −20 °C.
LC/MS-TOF analysis and data processing
The WBM active fractions were analyzed by LC/MS-TOF and data were processed by non-target analysis as described before;25 in brief: the fractions were analyzed using a Nexera UHPLC system (Shimadzu, Den Bosch, The Netherlands), coupled to a maXis 4G high-resolution quadrupole time-of-flight HRMS (UHPLC-Q-TOF-MS/MS), equipped with a HD collision cell (Bruker, Leiderdorp, The Netherlands). Liquid chromatography (LC) analysis was performed according to the aforementioned programs for Kinetex F5 column and Kinetex C18 column. 2 mM sodium formate in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol
1 methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water solution was used for the automatic internal calibration of the system. Positive and negative ESI were used to acquire MS- and MS/MS data in the range of 80–1200 m/z. A UV chromatogram (210 nm) was simultaneously recorded to detect the active peak of the samples. The peak and retention times in the UV chromatogram were compared to those found before in the F5 column and C18 column. Non-target analysis (NTA) was performed for processing the LC/MS-TOF data using patRoon, which is an open-source platform that harmonizes various commonly used software tools employed in NTA.26 The NTA workflow has five steps, as follows (see ESI 1†): at first, the LC/MS-TOF raw data were converted to the open mzML format.27 Secondly, feature data were extracted and grouped by OpenMS algorithms.28 Third, the features were filtered by different constraints (see ESI 1†). Fourth, the filtered features were automatically annotated by extracting mass spectral data using the mzR algorithm.29 Formulae were automatically calculated for all the features with both the GenForm and SIRIUS algorithms.30–34 Compound structure annotation was performed with SIRIUS and MetFrag,35 both using the PubChem library.36 Only the top 25 ranked candidates with an explained chemical formula were kept. The last step, “componentization”, was performed to automatically detect the features that are isotopes or MS adducts (using RAMClustR as an algorithm).37
water solution was used for the automatic internal calibration of the system. Positive and negative ESI were used to acquire MS- and MS/MS data in the range of 80–1200 m/z. A UV chromatogram (210 nm) was simultaneously recorded to detect the active peak of the samples. The peak and retention times in the UV chromatogram were compared to those found before in the F5 column and C18 column. Non-target analysis (NTA) was performed for processing the LC/MS-TOF data using patRoon, which is an open-source platform that harmonizes various commonly used software tools employed in NTA.26 The NTA workflow has five steps, as follows (see ESI 1†): at first, the LC/MS-TOF raw data were converted to the open mzML format.27 Secondly, feature data were extracted and grouped by OpenMS algorithms.28 Third, the features were filtered by different constraints (see ESI 1†). Fourth, the filtered features were automatically annotated by extracting mass spectral data using the mzR algorithm.29 Formulae were automatically calculated for all the features with both the GenForm and SIRIUS algorithms.30–34 Compound structure annotation was performed with SIRIUS and MetFrag,35 both using the PubChem library.36 Only the top 25 ranked candidates with an explained chemical formula were kept. The last step, “componentization”, was performed to automatically detect the features that are isotopes or MS adducts (using RAMClustR as an algorithm).37
NMR analysis
1H and 13C{1H} NMR spectra of the dilute solution in the CDCl3 solvent were recorded on a Bruker Avance Neo-500 MHz instrument and calibrated based on residual undeuterated solvent signals as an internal standard. 13C-{1H} NMR spectra of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 transients were measured by broadband decoupling and at a relaxation delay of 2.0 s. 2D spectra were recorded with 2K/256 data points. The 2D homonuclear shift correlation was established using gradient pulses for the selection. The HSQC spectrum probed the 2D 1H-/13C correlation via double inept transfer utilizing sensitivity improvement, phase sensitivity using echo/antiecho-TPPI gradient selection with decoupling during acquisition and multiplicity editing during the selection step.38–42
384 transients were measured by broadband decoupling and at a relaxation delay of 2.0 s. 2D spectra were recorded with 2K/256 data points. The 2D homonuclear shift correlation was established using gradient pulses for the selection. The HSQC spectrum probed the 2D 1H-/13C correlation via double inept transfer utilizing sensitivity improvement, phase sensitivity using echo/antiecho-TPPI gradient selection with decoupling during acquisition and multiplicity editing during the selection step.38–42
Data analysis
PPAR CALUX activities and associated PPAR biological equivalents (BEQs) were expressed as mean ± standard deviation (SD). One-way ANOVA was employed to determine statistical significance. Statistical significance was set at P < 0.05.Results and discussion
Bioassay-guided fractionation of bio-active compounds from WBM
In order to purify and identify the compounds responsible for PPAR activation in WBM extracts,17 we set out to separate and fractionate a standardized sample. Through linkage of fractionated samples with bioactivity, we purified the compound(s) of interest stepwise. In this study, we used HPLC to fractionate the WBM extracts and determined all three PPAR activities using CALUX bio-assays specific for PPARα, PPARδ and PPARγ. First, a standardized WBM extract was prepared to be used in all subsequent experiments. It was extracted and tested for the presence of PPAR ligands using the CALUX assays. The activity profile was similar to the one noted before.17 Again, a high concentration of PPAR agonist activity was measured confirming the quality of the WBM batch to be suitable for further experimentation (Table 1).| PPARα | PPARδ | PPARγ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| LOEC at PC10 (μg L−1 medium) | BEQ (ng GW7647-eq. per g dry WBM) | Maximum PPARα activity of samples (%) | LOEC at PC10 (μg L−1 medium) | BEQ (ng L-165.041 eq. per g) | Maximum PPARδ activity of samples (%) | LOEC at PC10 (μg L−1 medium) | BEQ (ng rosiglitazone eq. per g dry WBM) | Maximum PPARγ activity of samples (%) | |
| The lowest observed effect concentration (LOEC) of samples were expressed as microgram dry WBM per liter medium. PPAR BEQs of samples were expressed relative to GW7647 (PPARα agonist reference compound), L-165.041 (PPARδ agonist reference compound), and rosiglitazone (PPARγ agonist reference compound), respectively. Luciferase activity of samples was expressed as a percentage of the maximum PPAR reference compound response. Data are corrected for solvent control luciferase values and are expressed as mean ± SD (n = 3). *: Significant difference compared with the WBM extract, P < 0.05. | |||||||||
| WBM extract | 190![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
154 ± 8 | 40.1 ± 3.3 | — | 1598 ± 203 | 8.9 ± 0.9 | — | 127 ± 11 | 2.8 ± 0.5 |
| FS3 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 181 181![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 436 436 |
459 ± 54* | 57.9 ± 7.6* | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 478 478![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 967 967 |
3800 ± 407* | 17.0 ± 0.7* | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 517 517![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 214 214 |
660 ± 56* | 16.9 ± 1.7* |
| FS3.6 | 329![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 864 864 |
600 ± 48* | 67.6 ± 7.5* | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 249 249![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 930 930 |
3400 ± 126* | 19.9 ± 1.0* | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 797 797![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 986 986 |
360 ± 30* | 26.1 ± 1.5* |
| FS3.7 | 524![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 406 406 |
620 ± 105* | 49.0 ± 7.8 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 138 138![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 435 435 |
1500 ± 101 | 18.6 ± 1.3* | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 660 660![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
170 ± 3 | 11.6 ± 0.3* |
| FF5.3 | 154![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 387 387 |
300 ± 31* | 61.0 ± 7.3* | 901![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 961 961 |
6000 ± 354* | 23.7 ± 1.2* | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 659 659![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 570 570 |
230 ± 11* | 11.0 ± 0.5* |
| FC18.3 | 155![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 597 597 |
340 ± 12* | 75.4 ± 3.5* | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 218 218![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 990 990 |
6200 ± 403* | 45.4 ± 6.2* | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 295 295![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062 062 |
260 ± 30* | 12.4 ± 1.8* |
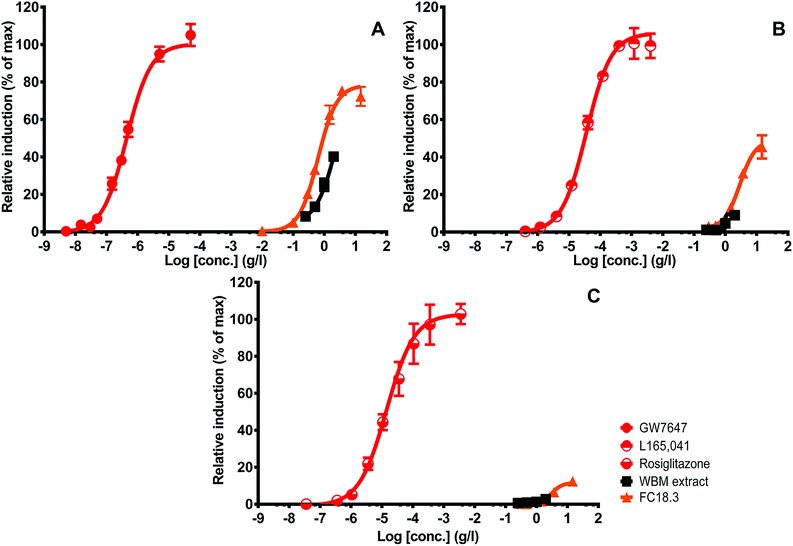
Note that the activity of the PPARδ extract is relatively high as compared to the other PPARs when expressed as biological equivalents (BEQs). This is also because the reference ligand is of lower potency than the available ones for PPARα and PPARγ, which stresses the fact that ligands for PPARδ are less well developed and that there is a great need to search for alternatives,43 as has been done in the current study. The PPAR activities (as expressed as BEQs) tended to be higher in the purified fractions compared to the original extract. In general, concentration response curves were more complete and cytotoxicity was reduced (Fig. 2; note that fewer data points were excluded because of cytotoxicity in the purified fraction).
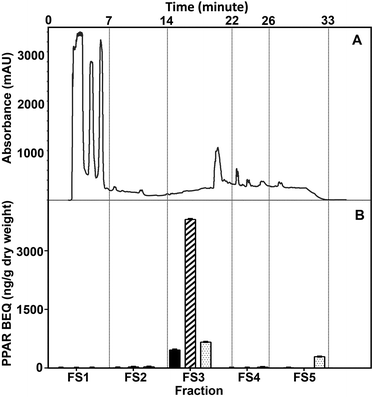
First, the WBM extract was fractionated using the semi-prep biphenyl column and five fractions (FS1 to FS5) were collected (Fig. 3). We chose this column since this seemed to be the most suitable one to separate the various molecules known to be produced by WBM, and since this has been shown to interact with PPARs like polyphenols and lipids.16 Among the five fractions obtained, fraction 3 (FS3) contained the highest PPAR BEQ levels (Fig. 3). The PPAR BEQ values were significantly higher than that of the original unfractionated WBM extract (Table 1), with PPARα levels 3-fold, PPARδ levels 2-fold, and PPARγ levels 5-fold higher, suggesting that all relevant molecules are present in this fraction. The higher activity may be explained by a loss of interfering molecules.
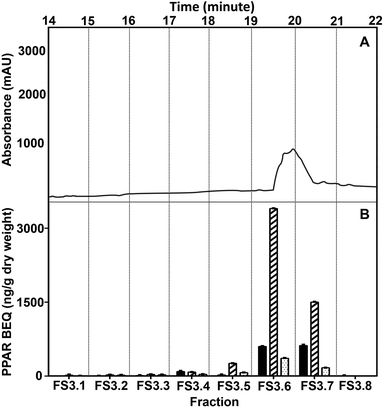
To get a more purified fraction, WBM FS3 was further sub-fractionated using smaller, one-minute intervals of collection. The highest PPAR activity was found in two fractions of one minute, starting from 19 minutes to 21 minutes (FS3.6–FS3.7; Fig. 4), with the highest activity in FS3.6. The chromatogram of the sub-fractionation showed one peak co-eluting with the main activity in FS3, as present in FS3.6 and FS3.7. Therefore, FS3.6 and FS3.7 were pooled to one fraction for further separation.
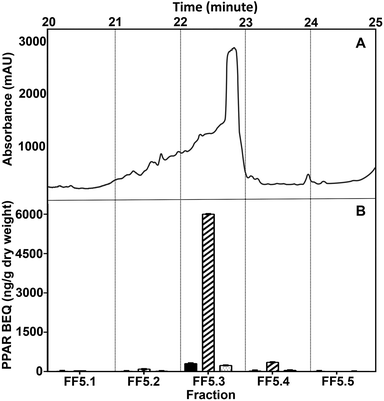
For further purification, we ran the WBM fraction on an analytical F5 column, which provides a different interaction to the semi-prep biphenyl column, resulting in a high efficiency in separation and purity of the identified active fraction. The WBM fractionation process on the F5 column also proceeded, at first, in a small number of relatively large fraction volumes pooled, followed by a more diligent smaller step fractionation. The highest PPAR activity of the biphenyl fraction FS3.6/3.7 separated on the F5 column was in fraction FF5.3 (Fig. 5). At the end of this procedure, the purity of the WBM fraction collected from the F5 column was considered enough for structural elucidation by LC/MS-TOF.
LC/MS-TOF identification of PPAR agonists in the purified WBM extract
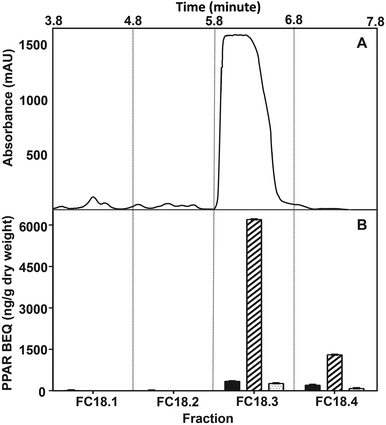
From the LC/MS-TOF analysis of fraction FF5.3, over 30 features were detected (see ESI 2, Table 2-1; 2-2†). Most features (19) were detected with negative ionization, which suggests a predominant presence of acidic compounds. Due to the large number of features, we did not attempt to perform full structural identification for this fraction. Regardless, basic identification of each feature was performed by automatic calculation of chemical formulae. The high hydrogen to carbon ratios of most formulae suggested primarily aliphatic compounds, and from these most formulae only contained the elements C, H, and O, which suggests fatty acid-like structures.To further reduce the complexity of the FF5.3 fraction, subsequent fractionation was performed with a C18 column since this stationary phase is highly suitable for the separation of fatty acid-like compounds.44 FC18.3 showed a single peak, which correlated with high BEQ of PPARα and PPARδ activity (Fig. 6). This further purified fraction WBM FC18.3 was analyzed by LC/MS-TOF, and only a total of 7 features were detected (see ESI 2, Table 2-3†). With positive ionization, the four detected features were of similar intensity; however, negative ionization revealed a single most abundant feature (FC18.3-neg-1, 88% relative intensity). Chemical annotation results from the applied workflow suggested linoleic acid (LA) as the top candidate for the most abundant features detected in both positive and negative ionization; hence, we pursued our studies with this compound.
Fig. 7 shows that the retention times of the FC18.3 active fraction compared to those of LA or its major isoforms CLA 9-cis, 11-trans and CLA 10-trans, 12-cis are close for the latter two and are identical for LA itself.
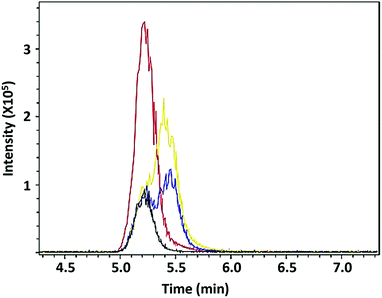 | ||
| Fig. 7 Overlaid extracted ion chromatograms (positive mode) for sample FC18.3 (black) spiked with linoleic acid (LA, red), CLA 10-trans, 12-cis (yellow) and CLA 9-cis, 11-trans (blue). | ||
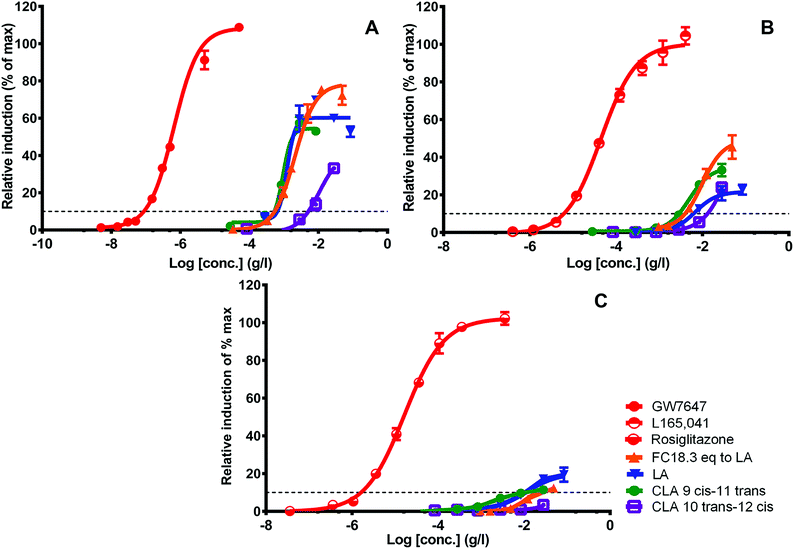
Since LA and isoforms of LA have been reported to interact with PPARs, these are potential candidates that contribute to WBM-derived activity.45,46 Several studies provide evidence of health benefits of LA or its conjugated isoforms (CLAs), including its anti-obesity potential. The most active and well-studied isoforms of LA are CLA 9 cis-11 trans, and CLA 10 trans-12 cis, showing a variety of relevant beneficial biological effects, including reduction of type 2 diabetes and body weight.47,48 We next studied if these compounds could be related to the molecules in the WBM fraction responsible for PPAR activation. We tested them in the panel of PPAR assays in comparison with the activity of WBM FC18.3, assuming that all activity was LA-derived (Fig. 8). The concentration response curves of WBM FC18.3, expressed as LA equivalents, were quite similar to those of LA and CLA 9-cis, 11-trans. On the other hand, the PPAR activity response curve of CLA 10-trans, 12-cis was clearly different compared to the other samples (Fig. 8). Also, when the lowest observed effect concentrations (LOECs) were used, CLA 10-trans, 12-cis was clearly different from the rest, and this isoform seems not to be responsible for PPAR activation by WBM extracts (Table 2).
| PPARα | PPARδ | PPARγ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| LOEC (μg LA L−1) | LOEC relative to FC18.3 | Maximum PPARα activity of samples (%) | LOEC (μg LA L−1) | LOEC relative to FC18.3 | Maximum PPARδ activity of samples (%) | LOEC (μg LA L−1) | LOEC relative to FC18.3 | Maximum PPARγ activity of samples (%) | |
| LOECs were expressed as microgram LA of sample per liter medium. Maximum luciferase activity of samples was expressed as a percentage of the maximum induced by PPAR reference compounds. Data are corrected for solvent control luciferase values and are expressed as mean ± SD (n = 3). *: Significant difference compared with the maximum of FC18.3 PPAR activity, P < 0.05. | |||||||||
| LA of FC18.3 | 539 | 1.0 | 75.0 ± 3.5 | 3957 | 1.0 | 45 ± 6.2 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 899 899 |
1.0 | 12 ± 1.8 |
| LA | 647 | 1.2 | 69.8 ± 3.0 | 6188 | 1.6 | 22.7 ± 2.6* | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 079 079 |
0.5 | 19.5 ± 3.7 |
| CLA 9-cis, 11-trans | 529 | 1.0 | 57.4 ± 5.8* | 2850 | 0.7 | 33.1 ± 3.2* | 9260 | 0.4 | 11.5 ± 1.2 |
| CLA 10-trans, 12-cis | 5115 | 9.5 | 33.1 ± 1.0* | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 102 |
3.3 | 24 ± 0.7* | — | — | 3.4 ± 0.7 |
Fig. 8 shows that there was a difference in shape between the reference curve and the tested samples that could be caused by the fact that the LA-type molecules act at high dosage and never reach their maximum, since at these high dosages cytotoxicity comes into play. Alternatively, the inherent efficacy for receptor activation could be different. In general, the lower end of the concentration response curves of the purest extract (FC18.3) and LA is quite similar, and the curves deviate at the higher dosages (Fig. 8), which suggests that this may be caused by impurities that differ between these samples. When comparing the maximal activity between WBM FC18.3 and the different compounds, again, the LA and CLA 9-cis, 11-trans profiles were again quite similar, while CLA 10-trans, 12-cis was clearly different. An exception seems to be the relatively strong effect of WBM FC18.3 towards PPARδ (Fig. 8 and Table 2).
In conclusion, the LC/MS-TOF results and the candidate molecule testing could not reveal the exact LA isoform and the tested forms are only a minority of the many possible linoleic acid isoforms, all having different biological activities.49 For this reason, further characterization with NMR was pursued to determine the isoform(s) responsible for PPAR activation more precisely.
NMR characterization of PPAR agonists from WBM
The 1H NMR spectrum of the CDCl3 diluted sample shows an almost pure compound with the characteristic features of an unsaturated olefinic acid, i.e. a broad signal of the carboxylic acid proton at a low field and four olefinic protons indicating two double bonds were found to be present. The characteristic methylene signal at 2.80 ppm shows only correlation to these olefinic protons, proving the olefinic acid to be non-conjugated. The NMR spectroscopic data (1H NMR (500 MHz, CDCl3, 298 K) δ 10.73 (broad, 1H, HOOC), 5.38 (m, 4H, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 2.80 (dd, J = 6.6 Hz, 2H,
CH), 2.80 (dd, J = 6.6 Hz, 2H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH –CH2–CH
CH –CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 2.37 (t, J = 7.5 Hz, 2H, HOOC–CH2), 2.07 (dt, J = 6.9 Hz, 4H, CH2–CH2–CH
), 2.37 (t, J = 7.5 Hz, 2H, HOOC–CH2), 2.07 (dt, J = 6.9 Hz, 4H, CH2–CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 1.66 (q, J = 7.5 Hz, 2H, HOOC–CH2–CH2), 1.43–1.26 (m, 14H), 0.91 (t, J = 6.8 Hz, 3H, CH3). 13C{1H} NMR (126 MHz, CDCl3, 298 K) δ 179.23 (HOC
), 1.66 (q, J = 7.5 Hz, 2H, HOOC–CH2–CH2), 1.43–1.26 (m, 14H), 0.91 (t, J = 6.8 Hz, 3H, CH3). 13C{1H} NMR (126 MHz, CDCl3, 298 K) δ 179.23 (HOC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 130.24 (CH), 130.04 (CH), 128.08 (CH), 127.91 (CH), 33.89 (HOOC–CH2), 31.54 (CH2), 29.59 (CH2), 29.36 (CH2), 29.15 (CH2), 29.08 (CH2), 29.03 (CH2), 27.22 (CH2), 27.19 (CH2), 25.64 (CH2), 24.67 (CH2), 22.59 (CH2), 14.08 (CH3)) are comparable to that of a commercially obtained linoleic acid and are in agreement with the literature data of (9Z,12Z)-9,12-octadecanoic acid.50
O), 130.24 (CH), 130.04 (CH), 128.08 (CH), 127.91 (CH), 33.89 (HOOC–CH2), 31.54 (CH2), 29.59 (CH2), 29.36 (CH2), 29.15 (CH2), 29.08 (CH2), 29.03 (CH2), 27.22 (CH2), 27.19 (CH2), 25.64 (CH2), 24.67 (CH2), 22.59 (CH2), 14.08 (CH3)) are comparable to that of a commercially obtained linoleic acid and are in agreement with the literature data of (9Z,12Z)-9,12-octadecanoic acid.50
Linoleic acid is a major poly unsaturated fatty acid (PUFA) and an essential nutrient.51 It is of importance in a number of structural components and physiological processes. While considered generally as beneficial, high intake may be associated with adverse effects, but no recommended daily allowance has yet been established. Being a major component of several vegetable oils, its dietary intake seems rarely insufficient.51,52 LA can pass through the small intestine and is packaged and transported to its targets in chylomicrons. It can also be oxidized enzymatically with its derivatives, like eicosanoids, involved in important cell signaling processes.51 We here report that LA is a major WBM-derived ligand that interacts with PPARs, which is consistent with the earlier reports of it being a PPAR ligand.45,46 Still, it would be difficult to envision that its levels in WBM will be responsible for the reported beneficial health effects, since its dietary intake is generally quite high.51 It is suspected that high dietary levels of LA also can lead to adverse effects.52 While linoleic acid is the main fatty acid in WBM, overall fungi contain low levels of fatty acids,16 which may sufficiently contribute to beneficial anti-obesogenic effects, while avoiding high energy intake counteracting its beneficial effects. In addition, these LA-induced PPAR-mediated effects may be modulated by other WBM-derived molecules. For instance, a range of polyphenols have been reported to modulate PPAR expression in a number of experimental models.53 In our reporter gene assays, we tested for direct PPAR activation specifically and in vivo and there could be more complex cellular models modulating the effects with other WBM compounds like polyphenols, which may alter PPAR expression leading to combined effects. It can also not be totally excluded that other PPAR activating compounds may be involved. Interestingly, in particular, in the case of PPARδ activation, a quite substantial difference was noted between the optimal fold induction by the WBM FC18.3 fraction and that by LA, being twice as high caused by the extract (Fig. 8 and Table 2), which may point towards a yet incomplete view of the PPAR activating ligands in the purified fraction, e.g. due to the presence of a minor component with relatively high biological activity that has a preference for PPARδ activation.
Conclusions
A complex range of compounds is present in WBM, which could contribute to its observed anti-obesity effects.13,16 In this study, a stepwise fractionation was used, directed by activities observed in the bioassays to identify ligands for PPARs, which are considered important in regulating fat metabolism, and important target receptors to select ligands to combat metabolic syndrome.5 To our surprise, we identified a relatively common dietary fatty acid as the main ligand of PPARs in WBM, excluding other candidate molecules. Although LA is known to have relevant beneficial effects, which may be mediated through PPARs,54 high dietary levels of LA can lead to adverse effects.52 While LA is the main fatty acid in WBM, overall fungi contain low levels of fatty acids,16 which may contribute to support beneficial anti-obesogenic effects, while avoiding high energy intake counteracting its beneficial effects. However, it seems more likely that other compounds contribute to these effects. This could include a still to be discovered minor but potent compound involved in PPARδ activation, as indicated by the relatively potent activation by the purified WBM fraction. Alternatively, receptor expression modulating compounds or compounds with completely PPAR unrelated modes of action may contribute. In further studies, we will determine if this is the only molecule responsible for the anti-obesity activities of WBM, or if there are other ones contributing, acting through different mechanisms.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Bio-Based Industries Joint Undertaking under the European Union's Horizon 2020 research and innovation programme [grant number 720720, FUNGUSCHAIN] and by a BE-Basic Foundation-FES grant from the Dutch Ministry of Economic Affairs.References
- K. A. Martin, M. V. Mani and A. Mani, New targets to treat obesity and the metabolic syndrome, Eur. J. Pharmacol., 2015, 763, 64–74 CrossRef CAS.
- S. Patra and S. Nithya, Review of Medicinal Plants for Anti-Obesity Activity, Transl. Biomed., 2015, 6, 1–23 Search PubMed.
- M. Tremmel, U. G. Gerdtham, P. M. Nilsson and S. Saha, Economic burden of obesity: A systematic literature review, Int. J. Environ. Res. Public Health, 2017, 14, 435 CrossRef.
- M. Ahmadian, J. M. Suh, N. Hah, C. Liddle, A. R. Atkins, M. Downes and R. M. Evans, PPARgamma signaling and metabolism: the good, the bad and the future, Nat. Med., 2013, 19, 557–566 CrossRef CAS.
- R. M. Evans, PPARs and the complex journey to obesity, Keio J. Med., 2004, 53, 53–58 CrossRef CAS.
- S. M. Reilly and C.-H. Lee, PPAR delta as a therapeutic target in metabolic disease, FEBS Lett., 2008, 582, 26–31 CrossRef CAS.
- L. Wang, B. Waltenberger, E. M. Pferschy-Wenzig, M. Blunder, X. Liu, C. Malainer, T. Blazevic, S. Schwaiger, J. M. Rollinger, E. H. Heiss, D. Schuster, B. Kopp, R. Bauer, H. Stuppner, V. M. Dirsch and A. G. Atanasov, Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARγ): A review, Biochem. Pharmacol., 2014, 92, 73–89 CrossRef CAS.
- N. Wagner and K. D. Wagner, PPAR Beta/Delta and the Hallmarks of Cancer, Cells, 2020, 9(5), 1133 CrossRef.
- A. Ríos-Hoyo and G. Gutiérrez-Salmeán, New Dietary Supplements for Obesity: What We Currently Know, Curr. Obes. Rep., 2016, 5, 262–270 CrossRef.
- N. Vasudeva, N. Yadav and S. K. Sharma, Natural products: A safest approach for obesity, Chin. J. Integr. Med., 2012, 18, 473–480 CrossRef CAS.
- D. Fang, F. Pei, Q. Hu, L. Zhao, W. Yang and G. Ma, A critical review on the health promoting effects of mushrooms nutraceuticals, Food Sci. Hum. Wellness, 2018, 7, 125–133 CrossRef.
- F. Atila, M. N. Owaid and M. A. Shariati, The nutritional and medical benefits of Agaricus Bisporus: A review, J. Microbiol., Biotechnol. Food Sci., 2017, 7, 281–286 CAS.
- B. B. B. Muszyńska, K. Kała, J. Rojowski, A. Grzywacz and W. Opoka, Composition and Biological Properties of Agaricus Bisporus Fruiting Bodies - A Review, Pol. J. Food Nutr. Sci., 2017, 67, 173–181 CrossRef.
- T. C. Finimundy, A. J. P. Dillon, J. A. P. Henriques and M. R. Ely, A Review on General Nutritional Compounds and Pharmacological Properties of the Lentinula edodes Mushroom, Food Nutr. Sci., 2014, 05, 1095–1105 Search PubMed.
- K. Deepalakshmi and S. Mirunalini, Pleurotus ostreatus : an oyster mushroom with nutritional and medicinal properties, J. Biochem. Technol., 2014, 5, 718–726 Search PubMed.
- M. Ramos, N. Burgos, A. Barnard, G. Evans, J. Preece, M. Graz, A. C. Ruthes, A. Jiménez-Quero, A. Martínez-Abad, F. Vilaplana, L. P. Ngoc, A. Brouwer, B. van der Burg, M. del Carmen Garrigós and A. Jiménez, Agaricus Bisporus and its by-products as a source of valuable extracts and bioactive compounds, Food Chem., 2019, 292, 176–187 CrossRef CAS.
- L. Pham Ngoc, H. Y. Man, H. Besselink, H. Dang Thi Cam, A. Brouwer and B. van der Burg, Identification of PPAR-activating compounds in herbal and edible plants and fungi from Vietnam, Ind. Crops Prod., 2019, 129, 195–200 CrossRef CAS.
- M. Iñiguez, P. Pérez-Matute, María, J. Villanueva-Millán, E. Recio-Fernández, I. Roncero-Ramos, M. Pérez-Clavijo, J.-A. A. Oteo, M. J. Villanueva-Millán, E. Recio-Fernández, I. Roncero-Ramos, M. Pérez-Clavijo and J.-A. A. Oteo, Agaricus Bisporus supplementation reduces high-fat diet-induced body weight gain and fatty liver development, J. Physiol. Biochem., 2018, 74, 635–646 CrossRef.
- S. C. Jeong, Y. Tae Jeong, B. K. Yang, R. Islam, S. Rao Koyyalamudi, G. Pang, K. Y. Cho, C. H. Song, Y. T. Jeong, B. K. Yang, R. Islam, S. R. Koyyalamudi, G. Pang, K. Y. Cho and C. H. Song, White button mushroom (Agaricus Bisporus) lowers blood glucose and cholesterol levels in diabetic and hypercholesterolemic rats, Nutr. Res., 2010, 30, 49–56 CrossRef CAS.
- K. H. Poddar, M. Ames, C. Hsin-Jen, M. J. Feeney, Y. Wang and L. J. Cheskin, Positive effect of mushrooms substituted for meat on body weight, body composition, and health parameters. A 1-year randomized clinical trial, Appetite, 2013, 71, 379–387 CrossRef.
- D. Rigano, C. Sirignano and O. Taglialatela-Scafati, The potential of natural products for targeting PPARα, Acta Pharm. Sin. B, 2017, 7, 427–438 CrossRef.
- L. Gijsbers, H. D. L. M. Van Eekelen, L. H. J. De Haan, J. M. Swier, N. L. Heijink, S. K. Kloet, H. Y. Man, A. G. Bovy, J. Keijer, J. M. M. J. G. Aarts, B. Van Der Burg and I. M. C. M. Rietjens, Induction of peroxisome proliferator-activated receptor γ (PPARγ)-mediated gene expression by tomato (Solanum lycopersicum L.) extracts, J. Agric. Food Chem., 2013, 61, 3419–3427 CrossRef CAS.
- S. C. Van der Linden, A. R. M. von Bergh, B. M. A. van Vught-Lussenburg, L. R. A. Jonker, M. Teunis, C. A. M. Krul and B. van der Burg, Development of a panel of high-throughput reporter-gene assays to detect genotoxicity and oxidative stress, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2014, 760, 23–32 CrossRef CAS.
- L. Gijsbers, H. Y. Man, S. K. Kloet, L. H. J. De Haan, J. Keijer, I. M. Rietjens, B. Van Der Burg and J. M. Aarts, Stable reporter cell lines for peroxisome proliferator-activated receptor γ (PPARγ)-mediated modulation of gene expression, Anal. Biochem., 2011, 414, 77–83 CrossRef CAS.
- B. Collet, B. M. van Vugt-Lussenburg, K. Swart, R. Helmus, M. Naderman, E. D. Rijke, M. Eggesbø, A. Brouwer and B. V. D. Burg, Antagonistic activity towards the androgen receptor independent from natural sex hormones in human milk samples from the Norwegian HUMIS cohort, Environ. Int., 2020, 143, 105948 CrossRef CAS.
- R. Helmus, T. T. Laak, P. d. Voogt, A. v. Wezel and E. Schymanski, Patroon: Open Source Software Platform for Environmental Mass Spectrometry Based Non-target Screening, J. Cheminf., 2020 DOI:10.21203/rs.3.rs-36675/v1.
- L. Martens, M. Chambers, M. Sturm, D. Kessner, F. Levander, J. Shofstahl, W. H. Tang, A. Rompp, S. Neumann, A. D. Pizarro, L. Montecchi-Palazzi, N. Tasman, M. Coleman, F. Reisinger, P. Souda, H. Hermjakob, P. A. Binz and E. W. Deutsch, mzML–a community standard for mass spectrometry data, Mol. Cell. Proteomics, 2011, 10(1) DOI:10.1074/mcp.R110.000133.
- H. L. Röst, T. Sachsenberg, S. Aiche, C. Bielow, H. Weisser, F. Aicheler, S. Andreotti, H. C. Ehrlich, P. Gutenbrunner, E. Kenar, X. Liang, S. Nahnsen, L. Nilse, J. Pfeuffer, G. Rosenberger, M. Rurik, U. Schmitt, J. Veit, M. Walzer, D. Wojnar, W. E. Wolski, O. Schilling, J. S. Choudhary, L. Malmström, R. Aebersold, K. Reinert and O. Kohlbacher, OpenMS: A flexible open-source software platform for mass spectrometry data analysis, Nat. Methods, 2016, 13, 741–748 CrossRef.
- S. N. Bernd Fischer, L. Gatto, Q. Kou and J. Rainer, No TimzR: parser for netCDF, mzXML, mzData and mzML and mzIdentML files (mass spectrometry data), 2019, DOI:10.18129/B9.bioc.mzR, https://bioconductor.org/packages/mzR/.
- M. Meringer, S. Reinker, J. Zhang and A. Muller, MS/MS data improves automated determination of molecular formulas by mass spectrometry, MATCH, 2011, 65, 259–290 CAS.
- S. Böcker, M. C. Letzel, Z. Lipták, A. Pervukhin and O. Troyanskaya, SIRIUS: decomposing isotope patterns for metabolite identification, Bioinformatics, 2009, 25, 218–224 CrossRef.
- K. Dührkop, M. Fleischauer, M. Ludwig, A. A. Aksenov, A. V. Melnik, M. Meusel, P. C. Dorrestein, J. Rousu and S. Böcker, SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information, Nat. Methods, 2019, 16, 299–302 CrossRef.
- S. Böcker and K. Dührkop, Fragmentation trees reloaded, J. Cheminf., 2016, 8, 5 Search PubMed.
- K. Dührkop, H. Shen, M. Meusel, J. Rousu and S. Böcker, Searching molecular structure databases with tandem mass spectra using CSI:FingerID, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12580–12585 CrossRef.
- C. Ruttkies, E. L. Schymanski, S. Wolf, J. Hollender and S. Neumann, MetFrag relaunched: incorporating strategies beyond in silico fragmentation, J. Cheminf., 2016, 8, 3 Search PubMed.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, PubChem 2019 update: Improved access to chemical data, Nucleic Acids Res., 2019, 47, 1102–1109 CrossRef.
- C. D. Broeckling, F. A. Afsar, S. Neumann, A. Ben-Hur and J. E. Prenni, RAMClust: A novel feature clustering method enables spectral-matching-based annotation for metabolomics data, Anal. Chem., 2014, 86, 6812–6817 CrossRef CAS.
- R. D. Boyer, R. Johnson and K. Krishnamurthy, Compensation of refocusing inefficiency with synchronized inversion sweep (CRISIS) in multiplicity-edited HSQC, J. Magn. Reson., 2003, 165, 253–259 CrossRef CAS.
- A. G. Palmer, J. Cavanagh, P. E. Wright and M. Rance, Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy, J. Magn. Reson., 1991, 93, 151–170 CAS.
- C. Zwahlen, P. Legault, S. J. F. Vincent, J. Greenblatt, R. Konrat and L. E. Kay, Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage λ N-peptide/boxB RNA complex, J. Am. Chem. Soc., 1997, 119, 6711–6721 CrossRef CAS.
- W. Willker, D. Leibfritz, R. Kerssebaum and W. Bermel, Gradient selection in inverse heteronuclear correlation spectroscopy, Magn. Reson. Chem., 1993, 31, 287–292 CrossRef CAS.
- J. Schleucher, M. Schwendinger, M. Sattler, P. Schmidt, O. Schedletzky, S. J. Glaser, O. W. Sørensen and C. Griesinger, A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients, J. Biomol. NMR, 1994, 4, 301–306 CrossRef CAS.
- M. Seimandi, G. Lemaire, A. Pillon, A. Perrin, I. Carlavan, J. J. Voegel, F. Vignon, J. C. Nicolas and P. Balaguer, Differential responses of PPARα, PPARδ, and PPARγ reporter cell lines to selective PPAR synthetic ligands, Anal. Biochem., 2005, 344, 8–15 CrossRef CAS.
- S. H. Chen and Y. J. Chuang, Analysis of fatty acids by column liquid chromatography, Anal. Chim. Acta, 2002, 465, 145–155 CrossRef CAS.
- S. A. Kliewer, S. S. Sundseth, S. A. Jones, P. J. Brown, G. B. Wisely, C. S. Koble, P. Devchand, W. Wahli, T. M. Willson, J. M. Lenhard and J. M. Lehmann, Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 4318–4323 CrossRef CAS.
- A. Kennedy, K. Martinez, S. Schmidt, S. Mandrup, K. LaPoint and M. McIntosh, Antiobesity mechanisms of action of conjugated linoleic acid, J. Nutr. Biochem., 2010, 21, 171–179 CrossRef CAS.
- K. Koba and T. Yanagita, Health benefits of conjugated linoleic acid (CLA), Obes. Res. Clin. Pract., 2014, 8, e525–e532 CrossRef.
- I. J. Onakpoya, P. P. Posadzki, L. K. Watson, L. A. Davies and E. Ernst, The efficacy of long-term conjugated linoleic acid (CLA) supplementation on body composition in overweight and obese individuals: A systematic review and meta-analysis of randomized clinical trials, Eur. J. Nutr., 2012, 51, 127–134 CrossRef CAS.
- I. Churruca, A. Fernández-Quintela and M. P. Portillo, Conjugated linoleic acid isomers: Differences in metabolism and biological effects, BioFactors, 2009, 35, 105–111 CrossRef CAS.
- M. Shaaban, H. I. Abd-Alla, A. Z. Hassan, H. F. Aly and M. A. Ghani, Chemical characterization, antioxidant and inhibitory effects of some marine sponges against carbohydrate metabolizing enzymes, Org. Med. Chem. Lett., 2012, 2, 30 CrossRef.
- J. Whelan and K. Fritsche, Linoleic Acid, Adv. Nutr., 2013, 4, 311–312 CrossRef.
- R. J. Jandacek, Linoleic Acid: A Nutritional Quandary, Healthcare, 2017, 5, 25 CrossRef.
- J. A. Domínguez-Avila, G. A. González-Aguilar, E. Alvarez-Parrilla and L. A. de la Rosa, Modulation of PPAR Expression and Activity in Response to Polyphenolic Compounds in High Fat Diets, Int. J. Mol. Sci., 2016, 17, 1002 CrossRef.
- B. Grygiel-Górniak, Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications – a review, Nutr. J., 2014, 13, 17 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0fo02071k |
| This journal is © The Royal Society of Chemistry 2021 |