Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Influence of short- and long-term exposure on the biodegradation capacity of activated sludge microbial communities in ready biodegradability tests†
Joost A.
Dalmijn‡
a,
Baptiste A. J.
Poursat‡
 *ab,
Rob J. M.
van Spanning
c,
Bernd W.
Brandt
d,
Pim
de Voogt
*ab,
Rob J. M.
van Spanning
c,
Bernd W.
Brandt
d,
Pim
de Voogt
 ae and
John R.
Parsons
a
ae and
John R.
Parsons
a
aInstitute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Science Park 904, 1098 XH Amsterdam, The Netherlands
bSub-Department of Environmental Technology, Wageningen University & Research, P.O. Box 47, 6700 AA Wageningen, The Netherlands. E-mail: baptiste.poursat@wur.nl
cDepartment of Molecular Cell Biology, Vrije Universiteit, de Boelelaan 1108, 1081 HZ Amsterdam, The Netherlands
dDepartment of Preventive Dentistry, Academic Centre for Dentistry Amsterdam, Gustav Mahlerlaan 3004, 1081 LA, Amsterdam, The Netherlands
eKWR Water Research Institute, Nieuwegein, The Netherlands
First published on 14th November 2020
Abstract
Ready biodegradability tests (RBTs) are extensively used to screen the potential of chemicals to be biodegraded. The use of RBT protocols often results in large variations of test results that may lead to wrong interpretations. The present study aims to obtain a fundamental understanding of this variability. For this, we subjected the compounds 4-chloroaniline (4CA), carbamazepine (CBZ), metformin (MET), and N-methylpiperazine (NMP) to a variety of different test conditions. Inocula from five local wastewater treatment plants (WWTPs) were used in an attempt to enhance the Organisation for Economic Co-operation and Development (OECD) 310 biodegradability tests. The biodegradation capacity in RBTs, community composition and adaptation of the communities were compared after one week of pre-exposure in batch and four months exposure in chemostat. The results confirm that none of the test compounds is readily biodegradable in the standard OECD 310 RBT. However, when pre-exposure under either batch or chemostat conditions was included, 4CA was degraded in some cases and less variability among different inocula was observed for the transformation of MET. Bacterial communities from the five locations were found to be significantly different in composition from one another. In addition, pre-treatment performed before the RBT significantly changed the composition of each community. Results of this experiment show that short-term pre-exposure may increase the absolute number of degraders and deserves to be further investigated as a potential method to reduce the outcome variability of RBTs.
Water impactWith this manuscript we aimed to provide additional input on the necessity to modify and enhance the current ready biodegradability tests. These tests are extremely valuable, however, they suffer several problems. We explored the feasibility, advantages and limitation of pre-exposure of the inoculum prior such a test, as one of the solution to their drawbacks. |
1 Introduction
Variability in removal rates of organic contaminants between different wastewater treatment plants (WWTPs) has been observed in many studies.1–4 This variability could partly be explained i) by the different operating conditions between WWTPs, ii) by differences between the physico-chemical characteristics of the influent5 and iii) by the microbial composition of the activated sludge. The latter is also influenced by external and environmental factors, such as temperature, precipitation and contents of carbon and free energy sources and essential nutrients, all of which may vary seasonally6,7 and geographically.8 Most importantly, due to differences in the metabolic capacities of microbes, the composition of the microbial community in the activated sludge of WWTP's may have a dramatic effect on the removal efficiency of chemicals.5 It is well known that exposure to a certain chemical often leads to adaptation of the microbial community towards degrading that chemical, thereby enhancing its removal rates.1,2,9Environmental parameters can influence the outcome of ready biodegradability tests (RBTs),10 as these tests often use activated sludge as an environmental inoculum.11 RBTs are extensively used in order to screen substances for ready biodegradability. RBTs are conducted under aerobic conditions and rely on measurement of general metabolic activities, such as the O2 consumption or CO2 production to quantify mineralization of the test chemical. According to the OECD 310 guideline,11 a chemical must show at least 60% of mineralization within a 10 d window after the end of the lag-phase to be considered as readily biodegradable. If a compound fails this test, it may be subjected to further research in order to establish its persistence according to the European Chemicals Agency (ECHA). Due to large variations in outcomes and erratic behaviour of some substances in these tests, a large number of false negatives are reported.12–17 This leads to extra efforts and costs being directed towards further testing and research on the biodegradation potential of novel substances.18 Improving RBTs by decreasing their variability and increasing their relevance will decrease the number of unnecessary (eco)toxicology tests performed on biodegradable compounds. This will have many financial and ethical advantages as it reduces the costs and the number of test animals during an extended environmental risk assessment.16 Care must be taken, however, that the enhancement of RBT protocols does not increase the probability of false positives, as this could possibly lead to omission of further testing and the introduction of persistent substances to the environment. Multiple methods have been proposed to improve current ready biodegradability testing protocols with the aim to lower the number of false negatives and test variability, without having an impact on the probability of a false positive result. These methods include i) pre-exposure,9,14,19 ii) increasing the inoculum density,20,21 iii) increasing the test duration14,20 and iv) increasing the volume of test flasks.18,22
Pre-exposure, at test concentration, is thought to reduce the differences between inocula caused by their differing origins and to reduce the differences in lag phases at the start of the test.19 The source of the inoculum is one of the major factors causing variability in RBTs.13,19,23,24 Furthermore, as mentioned previously, the constant and long term exposure to some organic contaminants in a WWTP leads to an adaptation of the local community to these specific organic contaminants.9 This can influence the biodegradation activity of the communities in WWTPs2,5,25 as well as in RBTs.1 Hence, pre-exposing the inoculum to a test concentration has been proposed as a way to normalize the community biodegradation capacity to the test chemical and reduce the variability of the test, with limited effect on the general community composition and activity.14 However, using an artificially pre-exposed inoculum is not allowed under the current ECHA guidelines for biodegradability testing.11 Pre-exposing the inocula to a chemical at the test concentration may change the biodegradation kinetics of some compounds without lowering the relevance of RBTs, as such tests do not use kinetic information obtained at environmentally relevant concentrations to classify chemicals.
Different methods exist for exposing a microbial community to a test substance, which can range from simple batch cultures to more complex continuous flow systems, such as a chemostat bioreactor. It remains questionable how the different exposure methods compare in terms of substance removal, impact on microbial community composition and environmental relevance. More complex systems, such as continuous bioreactors, could mimic an environmentally realistic situation, such as a WWTP, but could also lead to the formation of communities that are not representative of the original ecosystem. It is, therefore, important to assess if the exposed communities are representative of an environmentally realistic exposure situation and thus comparable with the original activated sludge community.
A previous study showed the effects of sampling location and exposure history on the biodegradation of a specific chemical in RBTs.1 However, Itrich et al. (2015)1 compared inocula coming from two different countries located on different continents and subjected to different climates (the U.S.A and The Netherlands). The present study aimed to obtain a fundamental understanding of the impact of pre-exposure on the microbial community and biodegradation of chemicals in RBTs. More specifically, we aimed to compare activated sludge communities located in a similar geographical area and subject to comparable climatic conditions, in order to reduce the number of variables that might influence the community response to pre-exposure. We hypothesised that the different inoculum origins are reflected by significant differences in bacterial community composition and, therefore, variable biodegradation capacities and rates of the tested substances when following the OECD 310 protocol. Furthermore, it is hypothesized that these variabilities decrease when pre-exposing the inocula to the test substances.
We used four organic chemicals, 4-chloroaniline (4CA), carbamazepine (CBZ), metformin (MET) and N-methylpiperazine (NMP) to test the biodegradation capacity of the different natural and exposed communities. All these test substances are reported to be not readily biodegradable according to the OECD RBTs.12,26–28 No readily biodegradable compounds were selected for this experiment because we targeted chemicals with erratic behaviours in RBTs or with known induced adaptation. NMP was selected based on previous experiments that showed that long term exposure can lead to the biodegradation of this chemical in RBTs.28 In addition, MET and 4CA were selected for their erratic behaviour in RBTs before and after long term exposure in chemostats.28,29 Finally, CBZ was selected for its prevalence and persistence in WWTPs.30 CBZ and MET are both pharmaceuticals produced on a large scale. MET is biotransformed into a metabolite, guanylurea (GUA), which is thought to be even more environmentally persistent,27 while recent studies succeed to degrade this product under laboratory conditions.29,31 4CA is a building block used in the synthesis of pesticides, pharmaceuticals and dyes. It is also an intermediate produced of the microbial transformation of chlorinated pesticides and has been detected in the aquatic environment and wastewater.32 NMP is a building block used in the chemical industry and no information is available about its prevalence and behaviour in the aquatic environment.
In this project, these chemicals were not quantified in the sampled WWTP and activated sludge, but metformin and carbamazepine have been monitored by the Dutch National Institute for Public Health and the Environment (RIVM) in the influent and effluent of certain WWTPs (Appendix A). However, 4-chloroaniline and N-methylpiperazine were not monitored by the RIVM and the presence of these compounds was not reported in the local WWTPs, hence, their environmental concentration is unknown.
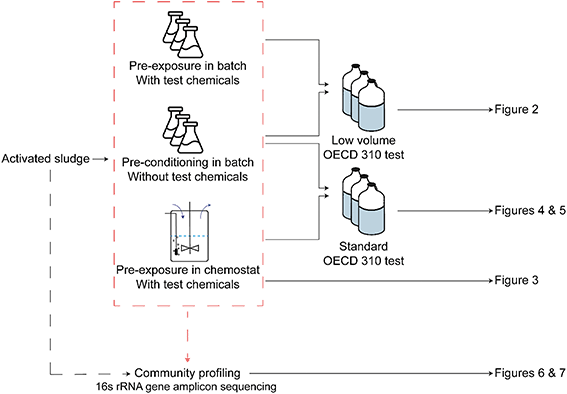
The experimental approach was to use inocula from five different locations, all of which were used in a biodegradation test under the following conditions: i) after pre-conditioning in batch for six days without test chemical, ii) after pre-exposure in batch, for six days, or in chemostat bioreactors, for up to 4 months, to the test chemicals and iii) with two different test volumes. In an attempt to miniaturize the test and increase the number of incubations, two tests with different incubation volumes were developed. A low-volume test, that used 20 ml incubation bottles, was compared with the standard test, which used 120 ml bottles as recommended by the OECD 310 guideline.11
Biodegradation was measured indirectly by CO2 production in time, following the OECD 310 guideline,11 and directly by quantification by LC-MS/MS. In addition, community composition and dynamics was assessed by 16S rRNA gene amplicon sequencing as to correlate succession behaviour of the community to breakdown capacities in time and to compare the exposed communities with the natural ones.
2 Materials and methods
A schematic representation of experiment and the associated figures can be found in Fig. 1.2.1 Chemicals, inocula and exposure treatments
The research and reference substances (4CA, aniline (ANL), CBZ, MET & NMP) were all of >99% purity and acquired from Sigma Aldrich.The inocula used in this research were derived from activated sludge which was sampled at five different WWTPs in the Netherlands: Amstelveen (AMV), Amsterdam (AMS), Utrecht (UTR), Bennekom (BNK) and Eindhoven (EIN). The capacity of these WWTPs is 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 1
000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 014
014![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 530
000, 530![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 25
000, 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 750
000 and 750![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitant equivalents (Appendix A), respectively. The AMV, AMS and UTR WWTPs are expected to receive mainly urban influent, because of their location in large cities, while BNK and EIN are expected to have a relatively higher amount of agricultural and industrial influent respectively. Due to logistical and practical limitations, sampling took place at all plants in two rounds in February (winter) and October (autumn) 2018. Weather conditions during winter sampling were air temperatures close to zero degrees Celsius, which may influence the community composition and activity,33 while in autumn 2018, sampling took place after a relatively warm period, with ambient air temperatures around 20 degrees Celsius. Unfortunately, the effect of the weather during sampling on the activated sludge activity and performance could not be assessed, as the temperature of the activated sludge was not measured. Whilst reported water temperature by the WWTP of Utrecht are 12.5 °C in February and 15.5 °C in October 2018, this information is not available for the other sampled WWTPs. However, it should be mentioned that even if every sampled activated sludge is sourced from outdoor facilities, they are all temperature controlled by the operators, which should limit the influence of the air temperature on the activated sludge.
000 inhabitant equivalents (Appendix A), respectively. The AMV, AMS and UTR WWTPs are expected to receive mainly urban influent, because of their location in large cities, while BNK and EIN are expected to have a relatively higher amount of agricultural and industrial influent respectively. Due to logistical and practical limitations, sampling took place at all plants in two rounds in February (winter) and October (autumn) 2018. Weather conditions during winter sampling were air temperatures close to zero degrees Celsius, which may influence the community composition and activity,33 while in autumn 2018, sampling took place after a relatively warm period, with ambient air temperatures around 20 degrees Celsius. Unfortunately, the effect of the weather during sampling on the activated sludge activity and performance could not be assessed, as the temperature of the activated sludge was not measured. Whilst reported water temperature by the WWTP of Utrecht are 12.5 °C in February and 15.5 °C in October 2018, this information is not available for the other sampled WWTPs. However, it should be mentioned that even if every sampled activated sludge is sourced from outdoor facilities, they are all temperature controlled by the operators, which should limit the influence of the air temperature on the activated sludge.
For the conditioning of the activated sludge samples the OECD 310 mineral buffer medium was used in all treatments (pre-conditioning, pre-exposure and chemostats), in which the sludge was suspended at 30 mg l−1 dry matter.11 Prior to the inoculation of the test flasks, pre-conditioning of activated sludge was performed according to the OECD 310 protocol. Activated sludge was suspended in mineral buffer medium at 30 mg l−1 dry matter and incubated for 6 d in a 500 ml Erlenmeyer flask with a permeable paper stopper. The pre-exposure treatment differed from the pre-conditioning treatment in that the tested substance was included with the sludge in the Erlenmeyer flasks at the same concentration as in the biodegradability test (20 mg l−1) in which the resulting inoculum would be used. As such, the pre-exposed inocula were only exposed to the same substance concentration with which the biodegradability test was carried out. RBTs are typically performed at high concentration (mg l−1) in order to ensure that CO2 production is above background and reaches the quantification limit of the analytical apparatus.
The chemostat treatment consisted of five aerobic bioreactors with OECD 310 medium,11 magnetic stirrers, sterile air supply and pressure-equilibrated flow which were inoculated with autumn activated sludge from the different cities at 30 mg l−1 dry matter, so that each city had an associated chemostat. In addition, all four test substances were added to the inflow medium reservoirs at 5 mg l−1 each. Medium reservoirs were replaced every two weeks with fresh sterile medium that also contained 5 mg l−1 of the test substances. A static equilibrium volume of 0.5 l was maintained in the reactor, with a flow rate of 4 ml h−1 and a corresponding dilution rate of 0.008 h−1 (equivalent to a hydraulic retention time of 125 h). This relatively low dilution rate was chosen because earlier tests had shown that high dilution rates lead to a loss of biomass and lower degradation capacities.28,29 The chemostats were kept running for 4 months in total and multiple liquid and DNA samples were taken during this period. RBTs were performed with the unexposed activated sludge and with the inocula from each chemostat after 2 and 4 months of incubation.
2.2 Biodegradation tests
Biodegradation tests on the chemicals with all the different types of inocula were carried using biological triplicates, following the OECD 310 protocol. This protocol describes the use of closed bottles in which a substance is dissolved in a mineral buffer medium as the only carbon and free energy source (20 mg l−1). These solutions were inoculated with activated sludge or exposed microbial communities at a density of 3 mg l−1 dry matter. Aerobic mineralization of the compound was calculated by sampling and measuring the CO2 in the headspace of the bottle at fixed time intervals over a period of 28 d. The bottle headspace was initially filled with artificial air (80% N2, 20% O2) and samples were taken from the headspace using a syringe with an electric pump and transferred to analysis vials, which were flushed with helium. Subsequently, the bottles were opened and liquid samples were taken, after which the bottles were closed again and repressurized with artificial air. This sampling procedure took place at five time points after 3, 7, 14, 21 and 28 d of testing. Furthermore, a liquid sample of the initial solutions at the beginning of each experiment (T0) was taken, in order to establish the initial substance concentrations.In an attempt to miniaturize the test and increase the number of incubations two different test volumes were used. The first set of biodegradation tests with natural and pre-exposed activated sludge treatment were carried out in 20 ml bottles and are referred to as a low-volume test. Activated sludge samples used in the low-volume test were collected during the winter sampling campaign. The second set of tests included unexposed activated sludge and chemostat inocula and were performed with 120 mL bottles, which is recommended by the OECD 310 standard guideline. Activated sludges samples used in the standard tests were collected during the second sampling campaign, in autumn.
The potential abiotic degradation of the test compounds was assessed in parallel by using an autoclaved inoculum (121 °C, 211 kPa, 21 min), which was diluted to the same inoculum density as the test inocula (30 mg l−1). Abiotic controls followed the exact same procedure that the biodegradation tests. Positive control tests, using aniline as a test chemical, were also performed to assess the mineralization activity of each inoculum.
In all the tests, the introduced inoculum density was 3 mg l−1 of dry matter, which means that the absolute number of cells was lower in the low-volume test. Incubation took place in the dark, at room temperature (22 ± 1 °C) and under constant shaking (150 rpm). pH was measured at the beginning and the end of the test. Initial pH was of 7.4 and due to the used phosphate buffer no significant variation was observed at the end of the test (∼pH 7).
2.3 Analytical chemistry
The CO2 concentration in the headspace of the incubation bottles was measured with a TRACE GC Ultra gas chromatograph (Thermo Fisher, Breda, The Netherlands), equipped with a FID and a Hayesep Q 1/8′′ 80–100 2 m + Hayesep N 1/8′′ 80–100 0.73 m column. The theoretical maximum inorganic carbon (ThIC) production of each bottle was calculated using the nominal and measured concentration of the added test compound. The amount of IC in the blank, that did not contain any test substance, was subtracted from the test results. The corrected IC was subsequently expressed as a percentage of the ThIC.In addition, LC-MS/MS analysis was carried out on triplicate liquid samples of the biodegradation tests and duplicate samples of the chemostats using a Shimadzu LC-20AD HPLC with a C18 (1.6 μm, 50 × 2.0 mm, Shimpack, Shimadzu) for 4CA and CBZ or HILIC (1.7 μm, 50 × 2.1 mm, amide BEH, Waters) column for MET/GUA and NMP, connected to an AB Sciex 4000 QTRAP MS/MS with Turbo Ion Spray electrospray ion source. The eluent for the C18 method consisted of ELGA ultrapure water with 0.1% v/v acetic acid (eluent A) and HPLC-grade methanol (eluent B). The eluent for the HILIC method consisted of ELGA ultrapure water with 5 mM NH4CO3 and 0.075% v/v formic acid (eluent A) and acetonitrile with 5 mM NH4CO3 and 0.075% v/v formic acid (eluent B). Both methods had a duration of four minutes. The flow rates for the C18 and HILIC methods were 0.3 ml min−1 and 0.4 ml min−1, respectively. Test compounds were ionized in positive mode. Multiple reaction monitoring (MRM) parameters and fragment masses of all analysed substances are shown in Table 1. Data acquisition and analysis were performed using AB SCIEX Analyst software (Ver. 1.5.1). Analysis of the parent compound is a deviation from the OECD 310 guideline and does not assess mineralization of the test compound but does give information about the removal of the test chemical and potential accumulation of metabolites.
| Name | Q1 (m/z) | Declustering potential (V) | Quantifier (TR1) | Qualifier (TR2) | LC method | Rt (min) | Detection limits in samples (ng ml−1) | ||
|---|---|---|---|---|---|---|---|---|---|
| Q3 (m/z) | Collision energy (V) | Q3 (m/z) | Collision energy (V) | ||||||
| 4CA | 128 | 60 | 93 | 27 | 75 | 48 | C18 | 2.50 | 0.0195 |
| CBZ | 237 | 91 | 194 | 29 | 192 | 29 | 2.67 | 0.0450 | |
| NMP | 101 | 116 | 58 | 27 | 44 | 33 | HILIC | 2.28 | 0.586 |
| MET | 130 | 47 | 71 | 32 | 60 | 19 | 1.88 | 0.154 | |
| GUA | 103 | 37 | 86 | 13 | 60 | 16 | 1.80 | 0.164 | |
2.4 Community profiling
Total DNA was extracted from each sample using a MoBio Powersoil kit (QIAGEN, Benelux B.V.) according to the supplier's protocol and stored at −80 °C until analysis. The total DNA concentration was determined with a Qubit dsDNA HS Assay kit and a Qubit fluorometer (Thermo Fischer Scientific, Waltham, MA, USA). The used primers, which were selected from,34 targeted the V3–V4 region of bacterial 16S rRNA genes. PCR products were purified using an Agencourt AMPure XP magnetic bead (Beckman Coulter Nederland B.V.). Agarose gel was used to control the amplification and purification steps. Sequencing of amplicons was performed on an Illumina MiSeq platform using a MiSeq Reagent kit at the Cancer Center Amsterdam, VUmc, Amsterdam (The Netherlands). Illumina sequencing raw data have been deposited on the NCBI sequence read archive database and can be accessed using the BioProject ID: PRJNA643142.2.5 Data analyses
Data analyses of the biodegradation tests were performed using the software program R (ver. 3.5.1).35 Sequencing reads from the Illumina MiSeq platform were processed into an operational taxonomic unit (OTU) table using USEARCH, as described in ref. 36 with the following differences. After merging, and before clustering, all sequences were additionally quality-filtered using a maximum expected error rate of 0.005, no ambiguous bases allowed. Next, clustering was carried out as described previously36 on sequences passing a maximum expected error rate of 0.002. Finally, the sequences previously passing the 0.005 threshold were mapped to the OTU centroids. For taxonomic assignments SILVA (ver. 1.3.2),37 instead of SILVA 119, was used. The sequences were trimmed to the V3–V4 16S rDNA region as described previously.36The OTU tables and phylogenic trees were then analyzed in R using the package Phyloseq (ver. 1.26.0).38 Subsequent analyses, such as diversity analysis, and data visualization were achieved using the R packages Phyloseq (ver. 1.26.0),38 Vegan (ver. 2.5-3)39 and ggplot2 (ver. 3.1.0).40 A negative binomial transformation was applied to the abundance data of all samples using the DESeq function from the R package DESeq2 (ver. 1.22.1),41 before performing beta-diversity analyses. Cluster analyses of the chemostat cultures data were performed separately from the rest of the data, but following the same procedure. Beta-diversity analyses, such as Bray-dissimilarity, weighted and unweighted Unifrac distance analyses were calculated on the transformed data using the package Phyloseq (ver. 1.26.0).38
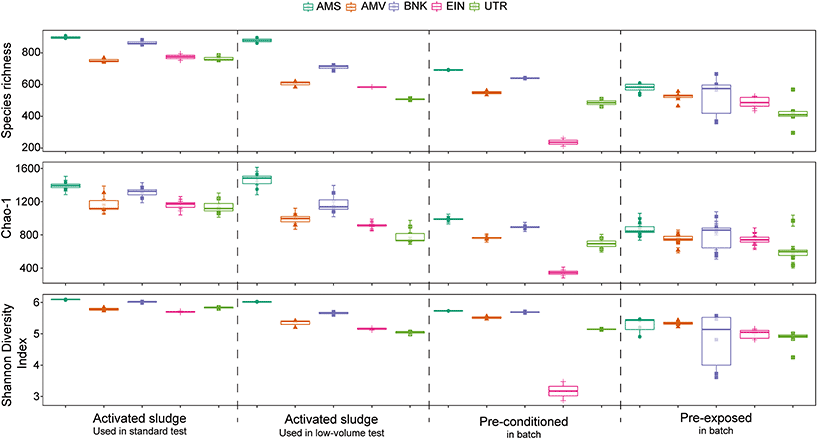
Alpha diversity indices, such as the observed species richness, which represents here the number of observed OTU in each sample, Chao-1 estimated species richness and Shannon diversity index were calculated, after rarefaction at an even depth (3500 reads), using the R package Phyloseq (ver. 1.26.0) and Vegan (ver. 2.5-3).
Statistical comparison of the bacterial communities was performed using a PERMANOVA analysis (1000 permutations) that implements the Adonis function in the package Vegan (ver. 2.5-3). Statistical comparison of the alpha diversity indices was performed using ANOVA and Kruskal–Wallis tests using R (ver. 3.5.1).
3 Results
3.1 Impact of short- term pre-exposure on biodegradation in low volume tests
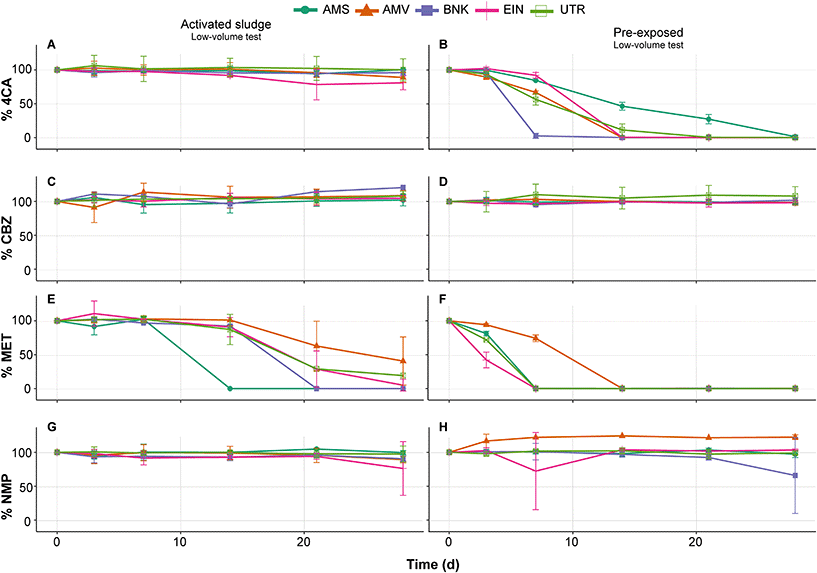
The results of the primary degradation in biodegradability tests measured via LC-MS/MS indicate that only MET is efficiently removed by the unexposed inoculum originating from activated sludge in the low volume test (Fig. 2E). Furthermore, comparison of the different inocula shows variations in their removal rate of MET. These variations decreased after pre-exposure in batch culture, but did not completely disappear (Fig. 2F). Pre-exposure also increases the degradation rate of 4CA for all inocula as no removal is observed without this treatment (Fig. 2A and B). Probably due to the low volume used during the tests and to the low amount of carbon in the medium, very little CO2 production, if at all, was measured regardless of whether inocula were pre-exposed or unexposed. With regard to NMP and CBZ, no significant removal of these two compounds is observed either before and after pre-exposure (Fig. 2C, D, G and H).Furthermore, it should be noted that none of the chemicals were removed in the abiotic controls (<5% degradation), regardless of the test (Appendix C, Fig. S1†). This means that observed degradation (>10% removal) is the result of the inoculum activity.
3.2 Impact of long-term exposure in chemostats on standard biodegradation test outcomes
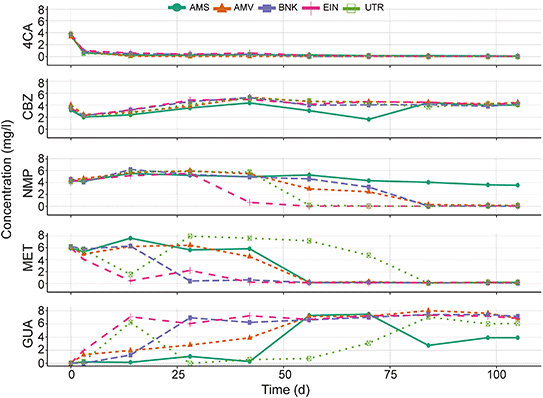
4CA is rapidly removed in the chemostat cultures regardless of the origin of the activated sludge (Fig. 3). CBZ on the other hand is not degraded, except for the AMS bioreactor that responds after 42 d of operation. Removal of MET and transformation to GUA occurs in all bioreactors, although the transformation rate differs between the different chemostats.Removal rates of MET in the UTR bioreactor are fluctuating over time. Initially, there is rapid removal, but after 14 d the removal capacity seems to decrease and the MET concentration rises again. Some removal of GUA could be observed in the AMS chemostat between 70 to 81 d of incubation, after which the GUA concentration stabilizes. Adaptation and elimination of NMP starts between 28 and 56 d in all bioreactors except the one with the AMS inoculum. The latter showed little catabolic activity both in the chemostat and the biodegradability tests.
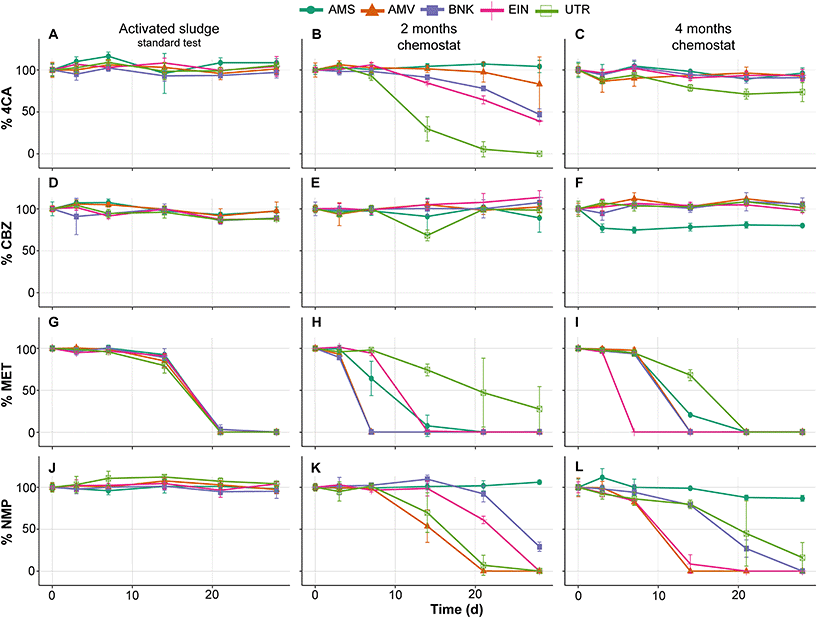
Similar to the tests performed with activated sludge in the low-volume test, those from the standard OECD 310 test did not show removal of 4CA, CBZ and NMP during biodegradability testing (Fig. 4A, D and J). MET is removed by all inocula and with comparable removal rates, which is in contrast with the low-volume test (compare Fig. 4G and 2E). No or very little CO2 is produced in these tests conducted with the activated sludge from autumn (see Fig. 5).
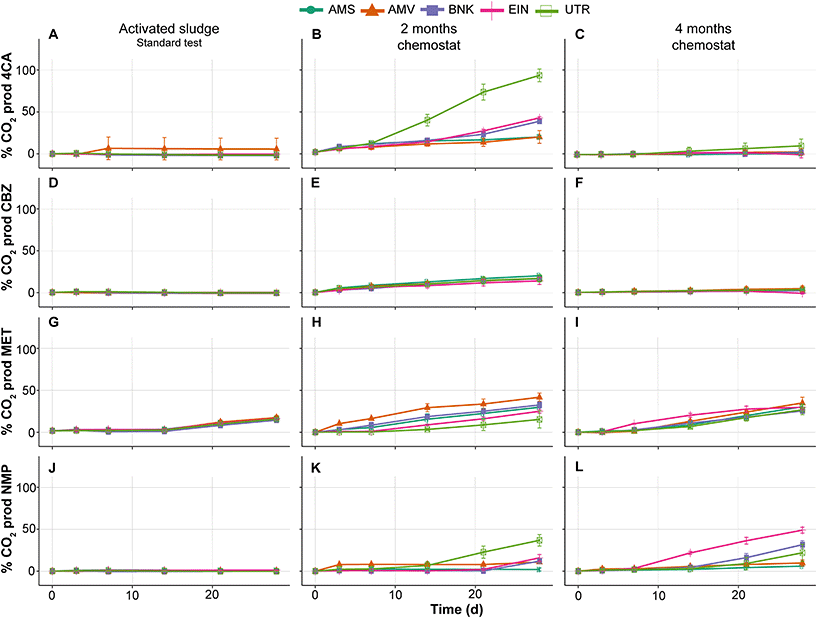 | ||
| Fig. 5 % of CO2 production of 4CA (A–C), CBZ (D–F), MET (G–I) and NMP (J–L) by activated sludge (autumn) and chemostat communities in biodegradability tests. | ||
As expected, biodegradability tests performed with inocula that were exposed to the test substances for two months in the chemostats showed increased removal rates. The UTR inoculum completely removed 4CA and produced CO2, while the one from AMS did not (Fig. 4B and 5B). Furthermore, MET removal rates diverged with faster rates by the AMV, AMS, BNK and EIN inocula, and slower rates by the UTR inoculum (Fig. 4H). All inocula except AMS are able to remove NMP within 2 months of exposure under these conditions (Fig. 4K).
After four months of exposure in the chemostats, the UTR inoculum only partly removed 4CA in the RBTs, and produced very little CO2. All inocula completely removed MET, though with variable rates (Fig. 4I). NMP was also removed by all inocula, except by AMS (Fig. 4L). RBTs performed with inocula pre-exposed for four months produced almost the same amount of CO2 as those pre-exposed for two months (Fig. 5), with the difference that less variation between inocula was observed for the mineralization of MET. Furthermore, EIN produced more CO2 for NMP than the rest of the inocula after the same time of pre-exposure.
CBZ was not removed in any of our RBTs, with the exception of the test bottle inoculated with AMS after four months (Fig. 4F). It should be noted, however, that CO2 was not produced at all in this case.
Despite the mineralization of aniline in some inocula, they were unable to meet the OECD 310 guideline requirement (Appendix C). Most of the activated sludge inocula either reached 60% of CO2 production more than 10 days after the end of the lag phase (10 d window) or did not reach this threshold at all. In contrast, some communities exposed in chemostats for two and four months conformed to the requirement. Overall, CO2 production from ANL is inconsistent and might reflect a loss of biodegradation capacity for this specific compound due to the chemostat conditions during which slow growing species are washed out of the culture. The limited CO2 production of ANL shows that our mineralization data are not representative of the natural mineralization potential of the communities. Even if mineralization was limited, the produced data are still reliable and may highlight a community dysfunction. Results indicate that the cultivated communities lost part of their mineralization capacity but not of their biotransformation activity, which is in accordance with the results presented in Fig. 4 and 5. According to the OECD guidelines, such data would not be accepted in an official RBT and could require additional tests with a different positive control, such as sodium benzoate.
3.3 Bacterial community analysis
The Illumina MiSeq generated a total number 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 163
163![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 877 reads. The OTU table included 7384 OTUs and 5
877 reads. The OTU table included 7384 OTUs and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600
600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 504 reads. The number of reads per sample ranged from 110, for the negative controls, to 190
504 reads. The number of reads per sample ranged from 110, for the negative controls, to 190![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 reads, with a median at 34
260 reads, with a median at 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 745 and a mean at 36
745 and a mean at 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 367 reads (SD 25
367 reads (SD 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 489). A bar plot representing the most abundant OTUs (>0.5% of relative abundance) in the sampled activated sludge, can be founded in the ESI† (Appendix E, Fig. S3).
489). A bar plot representing the most abundant OTUs (>0.5% of relative abundance) in the sampled activated sludge, can be founded in the ESI† (Appendix E, Fig. S3).
Comparison of the alpha diversity indices (Fig. 6) reveals significant differences between the different activated sludge samples (Shannon: p < 0.001, S.Obs: p < 0.05, Chao1: p < 0.001). Post hoc analysis shows that among the activated sludge samples, Amsterdam (AMS) is the only one significantly different from the other samples in both sampling campaigns. Comparison of the sampling season for each location also shows significant differences of the alpha diversity indices (Shannon: p < 0.001, S.Obs: p < 0.01, Chao1: p < 0.001). In general, activated sludge samples collected in winter and used in the low-volume test, were less diverse than activated sludge samples collected in autumn that were used in the standard test.
Pre-conditioning and pre-exposure also have an influence on the alpha diversity of the activated sludge communities, as both of them displayed significantly lower diversity index values. However, changes in diversity after pre-exposure are not correlated with the degradation kinetics of the tested chemical, as no significant difference is observed between them. Finally, cultivation in chemostats resulted in an important loss of diversity over time in comparison with the original activated sludge inoculum. Alpha diversity indices of the communities from chemostats over time are displayed in the ESI† (Appendix F, Fig. S4).
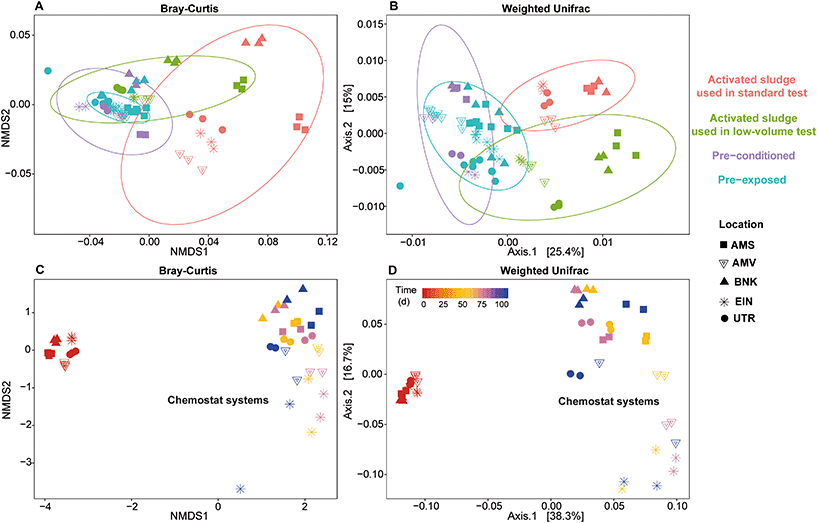
Both Bray–Curtis dissimilarity analysis and weighted Unifrac distance analysis revealed clear differences between the different communities (Fig. 7). These observations are supported by the PERMANOVA analysis that confirmed that all activated sludge communities were significantly different from each other. Further analysis of weighted (quantitative) and unweighted (qualitative) Unifrac showed that the difference between winter and autumn communities for Amsterdam and Bennekom were due to changes in the abundance of pre-existing taxa, while for the other locations, the presence or absence of taxa had more implications on the community dissimilarity.
As described earlier, pre-exposing or pre-conditioning of the winter communities led to significant changes in its composition and abundance. Furthermore, a comparison between pre-exposed and pre-conditioned communities showed significant differences. As observed with the alpha diversity analysis, long term exposure in chemostats led to a significant change in community composition and structure over time (Fig. 7C and D). Moreover, even if each chemostat was fed with an identical medium, each cultivated community remained significantly different from the others throughout the whole experiment (PERMANOVA, p < 0.001, F = 3.3146).
4 Discussion
4.1 Origin of the inocula
The geographical origin of an inoculum influences the biodegradability of organic chemicals in laboratory studies42 and can also influence their biodegradation in (ready) biodegradability tests.1,23,43,44 This is likely, as such communities share a small bacterial community core8 that is linked to activated sludge performance and might differ in biodegradation function and activity. Our results indicate that when performing the RBTs following the standard OECD 310 protocol no differences are observed in the degradation rates between the inocula of the five different WWTPs (Fig. 4 and 5). It should be noted that, although the inocula came from locations relatively close to each other, community profiling results revealed significant taxonomic divergence between the communities. Significant community dissimilarity among geographical distant location is known and is expected to increase at a regional level.8 However, our study did not produce enough data to provide a distance–decay relationship. This relationship assumes that community similarity decreases as the geographical distance between communities increases.8 The two most geographically distant samples are AMS and EIN, however, the dissimilarity between these two communities is not stronger than with the others. The most frequently detected OTUs in all activated sludge samples were associated with the bacterial phyla Proteobacteria, Bacteroidetes and Nitrospirae. These phyla are known to be among the most common detected bacterial phyla in activated sludges, even in WWTPs operated under different conditions.8,45 However, alpha diversity indices showed that inocula sampled during the first campaign, in winter, were significantly less diverse in terms of observed OTUs (AMV, UTR, EIN) or abundance (AMS, BNK) than the ones sampled in autumn (Fig. 6). This observation is consistent with the literature, as a lower diversity in winter has already been reported,33 without clear influence on the community activity. This is the case in the present investigation, in which the divergence of diversity and composition of the inocula did not have any effect on the degradation pattern of the test substances, in the standard RBT (Fig. 4), while divergence was observed for the degradation of MET in the low volume test (Fig. 2). Communities with a higher diversity should be more likely to contain key degrading strains, which would result in a positive association between diversity and biodegradation.46,47 This observation has been shown to be true for some but not all micropollutants,46 as some were even negatively correlated.48 However, more diverse communities most likely have functional genes redundancy,47 increasing the biodegradation chances of a specific chemical.In the present study, the biodegradation kinetics between taxonomically different communities are quite similar. Here it should be noted that the differences in community structure may be the consequence of fluctuation in the number of taxa with a low abundance, which may not be involved in the biodegradation pathway of the test chemicals. Thereby, the test concentrations used in the OECD 310 protocol are higher (mg l−1 range) than can be considered typical micropollutant (ng L−1 to μg L−1) range and this protocol only tests primary (aerobic) degradation. Therefore, micropollutant degradation pathways which could vary between the inocula, such as co-metabolism or nitrification are not considered in this study. Further investigation should be considered to confirm whether sampling location and/or season have a significative influence on RBTs outcome.
4.2 Low-test volume induced variability
The only observed variation between community biodegradation capacity was observed in the low-volume test performed on MET (Fig. 2E). It has been shown that a low test volume could increase the lag phase period and the chance of false negatives, due to the lower transferred biomass and therefore a lower chance to introduce competent degraders.49–53 This “biodegradation lottery” is well known and several solutions, such as increasing the inoculum density20,21 or the test volume12 has been proposed. In the present study, pre-exposing the inocula reduced the MET biodegradation variation and thus reduced the effect of the biodegradation lottery. The absolute abundance of MET degraders in the test was directly influenced by the volume of the test. Pre-exposure allowed the MET degrader population to increase and reduced the original variation in relative abundance between inocula. It should be noted that the chemical concentration used in biodegradability tests and during the pre-exposure treatment are not environmentally relevant and were specifically selected to promote growth-linked degradation, as recommended by the OECD 310 guideline.114.3 Exposure treatments
Deliberate pre-exposure has been proposed as a way to reduce the variability in the outcomes of RBTs in a number of studies9,14,19,22 Pre-exposure is thought to achieve this reduction by two mechanisms. Firstly, it shortens and equalizes the lag-phase before the onset of degradation, thereby reducing the number of false negatives that occur due to the degradation falling out of the 28 day window of the test. Secondly, by having a larger number of inoculum cells exposed to the chemical before the starting the RBT, the chances of sufficient competent degraders being transferred with the inoculum and therefore the chances of biodegradation could increase. Including pre-exposure in RBTs might lead to a better prediction of the environmental biodegradability of some organic chemicals, such as metformin.A short pre-exposure of six days increased the biodegradation capacity of the communities, even if no variation was observed before the treatment. All pre-exposed inocula showed elimination of 4CA during the OECD 310 RBT (Fig. 2B), which confirms that pre-exposure leads to an enhanced elimination of this compound.22 However, it remains the question to what extent the observed kinetic differences are the consequence of inherent characteristics of the original microbial communities. In that respect, it should be noted that 4CA is rapidly eliminated by all inocula in the chemostat bioreactors (Fig. 3) while it is not completely mineralized by the BNK inoculum in the subsequent RBT (Fig. 5). Therefore, the influence of inoculum source is also confounded by the large diversity of the activated sludge samples and by the decrease in diversity after pre-conditioning treatment.54
Our results indicate that both pre-exposure treatment and pre-conditioning, although pre-exposure is not allowed while pre-conditioning is recommended by the OECD 310 guideline,11 have a significant influence on the community composition and diversity (Fig. 6 and 7).54 When comparing the two tested exposure methods, a short batch pre-exposure has a less significant impact on the community composition and function than long term pre-exposure in chemostat bioreactors (Fig. 7). However, the likelihood for a persistent chemical to pass the test after a short pre-exposure is unknown, but it was not observed in the present study. NMP and CBZ, which are classified as persistent, were not degraded after short pre-exposure in batch cultures. This is in contrast to long-term chemostat pre-exposure, which has been shown to lead to the degradation of NMP in a standard RBT.28
As discussed above, pre-exposure also reduced the variability between inocula in the MET test conducted with a lower test volume than recommended. Therefore, pre-exposure mostly reduces the metformin test variability caused by divergence in the abundance of specific degraders. It should be noted that as only four chemicals were used in this study, more research should be conducted in order to draw general conclusions about the effect of pre-exposure on the biodegradation of persistent and inherently biodegradable compounds.
Long exposure using chemostat systems confirmed results of previous research that found that communities in chemostat bioreactors completely shift and diverge in composition and biodegradation capacity from their original activated sludge source communities.28,55 This is most likely explained by the fact that communities in domestic waste water grows on a complex mixture of organic substances, while those in the chemostat cultures are faced with a mineral buffer medium containing only the test substances as carbon and free energy source.54 When communities become adapted to a mixture of test substances, large differences in adaptation time, removal capacity and rate are observed. Furthermore, the observed differences in bacterial community composition between the WWTPs may also impact their behaviour in chemostat systems, as can be observed from the removal patterns of MET and NMP, and can increase their chance to diverge in RBTs. From our results, however, it remains difficult to determine whether these differences are caused by differences in the source influent or by differences in the communities and the probability to introduce specific degraders during inoculation of activated sludge.
Previous investigation related to the biodegradation of MET in chemostat already showed that activated sludge coming for the WWTP of Amsterdam (AMS) contains competent degraders of both MET and GUA.29 However, biodegradation could not be enhanced, even after nine months of exposure in chemostat. Our present results are in apparent contradiction to these results, as biodegradation of MET could be enhanced for most inocula, after two months of exposure in chemostat. Both experiments were conducted at high concentration (mg l−1) but using different dilution rates, which could have limited the loss of degrading biomass in the present study. The inoculum from AMS was also able to remove GUA in chemostat after 70 days of incubation. Very few studies reported the biodegradation of both MET and GUA29,31 in batch or chemostat systems. The catabolic pathway leading to the consumption of GUA is unknown but microbial communities are suspected to use MET and GUA as alternative nitrogen sources under nitrogen limited condition.31 Finally, bacteria from the genus Aminobacter were shown to be involved in the transformation of MET to GUA.29 However, in depth investigation of the inocula composition could not correlate the abundance of Aminobacter related bacteria with the biodegradation level of MET, which means that other members of the community must be responsible for the biodegradation of MET in this experiment.
Furthermore, introduction of variability after exposure and even the loss of degrading capabilities occurred in some cultures that were subjected to long-term pre-exposure in chemostat.29 Therefore, chemostat bioreactors are valuable to test whether a substance is inherently biodegradable and to assess the adaptation capacity of a microbial community to a compound. However, due to the drawbacks mentioned above, they are less suitable than short pre-exposure in batch to be incorporated in RBTs to account for microbial adaptation. Chemostats have the potential to be used to identify potential catabolic pathways under long term culturing. However, as it was highlighted by the aniline results (Appendix D, Fig. S2†), culturing in a chemostat can seriously affect the general community activity and lead to creation of a non-environmentally representative community.
Inconsistent aniline biodegradation by the chemostat communities could be the result of the absence of this compound in the medium and by a drop in the community diversity, resulting in a lower biodegradation capacity47 as it was already observed for metformin.29 We should consider to use an alternative reference chemical, such as acetate or benzoate, in the future. Finally, in accordance with previous research, deliberate pre-exposure of inocula to test substances before performing an RBT could hamper the degradation of the reference substance (e.g. cellulose or aniline in the current study) in the same test.23 This is probably due to activation of additional catabolic enzymes resulting in an inhibition of the enzyme involved in the consumption of the reference chemical. Therefore, this inconvenient side effect of pre-exposure might as well hamper the removal of other readily biodegradable compounds and limit the applicability of the test.
Overall, results of these experiments show that pre-conditioning or pre-exposing the inoculum will influence the community diversity, composition and activity, when compared to the original activated sludge community. Composition and diversity of the treated communities are significantly different from those of the original activated sludge, while their biodegradation activity remain the same for most of the tested chemicals. Pre-exposure could be used to specifically increase the absolute number of degraders and reduce the test variability and the number of false negatives. However, extra care must be taken to not lose the stringency of the test, by increasing the risk of false positives (e.g. ready biodegradation of a persistent chemical), and allowing emission of highly persistent compounds, which could lead to irreversible environmental impacts.56 Enhancing RBTs and understanding the influence of inocula exposure history requires additional work using alternative test chemicals and communities.
5 Conclusions
Ready biodegradability tests (RBTs) commonly use microbial communities from environmental samples, such as activated sludge. The diversity of these inocula can influence the RBTs' outcomes by increasing the variability of results, due to variable abundance of specific degraders. These variabilities and two proposed methods to mitigate these issues, i.e. pre-exposure and long-term exposure in chemostat bioreactors, were investigated in this study. Community profiling showed that the bacterial communities of all activated sludge samples used in this project were significantly different from each other. Although source differences had no clear influences on the biodegradation rate when following the regular OECD 310 protocol, differences between community activity could be observed in chemostat systems. We conclude i) that less variability in removal of metformin and shorter lag-phases are observed when the test is performed with a high cell number or after pre-exposure of the inocula in batch, ii) that long-term exposure, in chemostat systems, has the potential to study in laboratory the behaviour of highly adapted communities toward persistent compounds, such as N-methylpiperazine, and iii) mineralization of the positive control in RBTs is, however, affected by long-term exposure of the inoculum in chemostat, limiting the conclusions of the test.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was funded by the European Chemical Industry Council (Cefic) long-range initiative (LRI project ECO29). The authors would like to acknowledge the useful advice and discussions of the CEFIC LRi ECO29 research liaison team. The authors would also like to thank Paul Eijk (Cancer Center Amsterdam, VUmc, Amsterdam) for operating the Illumina MiSeq. The authors also thank the operators and the staff of the WWTPs of Amsterdam, Amstelveen, Utrecht, Bennekom and Eindhoven for providing activated sludge.References
- N. R. Itrich, K. M. McDonough, C. G. van Ginkel, E. C. Bisinger, J. N. LePage and E. C. Schaefer, et al. Widespread Microbial Adaptation to l-Glutamate-N,N-diacetate (L-GLDA) Following Its Market Introduction in a Consumer Cleaning Product, Environ. Sci. Technol., 2015, 49(22), 13314–13321 CAS.
- S. Kahl, S. Kleinsteuber, J. Nivala, M. van Afferden and T. Reemtsma, Emerging Biodegradation of the Previously Persistent Artificial Sweetener Acesulfame in Biological Wastewater Treatment, Environ. Sci. Technol., 2018, 52(5), 2717–2725 CAS.
- M. Gardner, V. Jones, S. Comber, M. D. Scrimshaw, T. Coello-Garcia and E. Cartmell, et al. Performance of UK wastewater treatment works with respect to trace contaminants, Sci. Total Environ., 2013, 456–457, 359–369 CrossRef CAS.
- J. Rivera, M. Sánchez-Polo, M. Á. Ferro-García, G. Prados-Joya and R. Ocampo-Pérez, Pharmaceuticals as emerging contaminants and their removal from water, A review, Chemosphere, 2013, 93(7), 1268–1287 CrossRef.
- Y. Luo, W. Guo, H. H. Ngo, L. D. Nghiem, F. I. Hai and J. Zhang, et al. A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment, Sci. Total Environ., 2014, 473–474, 619–641 CrossRef CAS.
- F. Ju, F. Guo, L. Ye, Y. Xia and T. Zhang, Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years, Environ. Microbiol. Rep., 2014, 6(1), 80–89 CrossRef CAS.
- Q. Sui, J. Huang, S. Deng, W. Chen and G. Yu, Seasonal Variation in the Occurrence and Removal of Pharmaceuticals and Personal Care Products in Different Biological Wastewater Treatment Processes, Environ. Sci. Technol., 2011, 45(8), 3341–3348 CrossRef CAS.
- L. Wu, D. Ning, B. Zhang, Y. Li, P. Zhang and X. Shan, et al. Global diversity and biogeography of bacterial communities in wastewater treatment plants, Nat. Microbiol., 2019, 4(7), 1183 CrossRef CAS.
- B. A. J. Poursat, R. J. M. Van Spanning, P. de Voogt and J. Parsons, Implications of microbial adaptation for the assessment of environmental persistence of chemicals, Crit. Rev. Environ. Sci. Technol., 2019, 49(23), 2220–2255 CrossRef.
- L. J. Forney, W.-T. Liu, J. B. Guckert, Y. Kumagai, E. Namkung and T. Nishihara, et al. Structure of Microbial Communities in Activated Sludge: Potential Implications for Assessing the Biodegradability of Chemicals, Ecotoxicol. Environ. Saf., 2001, 49(1), 40–53 CrossRef CAS.
- OECD, Test No. 310: Ready Biodegradability - CO2 in sealed vessels (Headspace Test), Organisation for Economic Co-operation and Development, Paris, 2014, Available from: http://www.oecd-ilibrary.org/content/book/9789264224506-en Search PubMed.
- M. Comber and M. Holt, Developing a set of reference chemicals for use in biodegradability tests for assessing the persistency of chemicals. MCC Rep No MCC007, 2010 [cited 2015 Sep 8]; Available from: http://cefic-lri.org/wp-content/uploads/uploads/Project%20publications/MCC_007_Eco12_Final_Report.pdf.
- C. Dick, S. Rey, A. Boschung, F. Miffon and M. Seyfried, Current limitations of biodegradation screening tests and prediction of biodegradability: A focus on fragrance substances, Environ. Technol. Innov., 2016, 5, 208–224 CrossRef.
- ECETOC, Workshop Report 10 - Workshop on Biodegradation and Persistence, Ecetoc, 2007 [cited 2016 Sep 6]. Available from: http://www.ecetoc.org/publication/workshop-report-10-workshop-on-biodegradation-and-persistence/ Search PubMed.
- P. Gerike and W. K. Fischer, A correlation study of biodegradability determinations with various chemicals in various tests: II. Additional results and conclusions, Ecotoxicol. Environ. Saf., 1981, 5(1), 45–55 CrossRef CAS.
- T. J. Martin, A. K. Goodhead, K. Acharya, I. M. Head, J. R. Snape and R. J. Davenport, High Throughput Biodegradation-Screening Test To Prioritize and Evaluate Chemical Biodegradability, Environ. Sci. Technol., 2017, 51(12), 7236–7244 CrossRef CAS.
- N. Nyholm, P. Lindgaard-Jørgensen and N. Hansen, Biodegradation of 4-nitrophenol in standardized aquatic degradation tests, Ecotoxicol. Environ. Saf., 1984, 8(5), 451–470 CrossRef CAS.
- A. Kowalczyk, T. J. Martin, O. R. Price, J. R. Snape, R. A. van Egmond and C. J. Finnegan, et al. Refinement of biodegradation tests methodologies and the proposed utility of new microbial ecology techniques, Ecotoxicol. Environ. Saf., 2015, 111, 9–22 CrossRef CAS.
- G. Thouand, B. Capdeville and J. C. Block, Preadapted Inocula for Limiting the Risk of Errors in Biodegradability Tests, Ecotoxicol. Environ. Saf., 1996, 33(3), 261–267 CAS.
- A. Ott, T. J. Martin, K. Acharya, D. Y. Lyon, N. Robinson and B. Rowles, et al. Multi-laboratory Validation of a New Marine Biodegradation Screening Test for Chemical Persistence Assessment, Environ. Sci. Technol., 2020, 54(7), 4210–4220 CrossRef CAS.
- A. Ott, T. J. Martin, G. F. Whale, J. R. Snape, B. Rowles and M. Galay-Burgos, et al. Improving the biodegradability in seawater test (OECD 306), Sci. Total Environ., 2019, 666, 399–404 CrossRef CAS.
- L. Toräng and N. Nyholm, Biodegradation rates in adapted surface water can be assessed following a preadaptation period with semi-continuous operation, Chemosphere, 2005, 61(1), 1–10 CrossRef.
- V. Mezzanotte, R. Bertani, F. D. Innocenti and M. Tosin, Influence of inocula on the results of biodegradation tests, Polym. Degrad. Stab., 2005, 87(1), 51–56 CrossRef CAS.
- G. A. Vázquez-Rodríguez, F. Garabétian and J.-L. Rols, Inocula from activated sludge for ready biodegradability testing: Homogenization by preconditioning, Chemosphere, 2007, 68(8), 1447–1454 CrossRef.
- T. Chonova, F. Keck, J. Labanowski, B. Montuelle, F. Rimet and A. Bouchez, Separate treatment of hospital and urban wastewaters: A real scale comparison of effluents and their effect on microbial communities, Sci. Total Environ., 2016, 542(Part A), 965–975 CrossRef CAS.
- ECHA, 1-methylpiperazine - Registration Dossier - ECHA-EC 203-639-5, 2013, Available from: https://echa.europa.eu/registration-dossier/-/registered-dossier/13739/5/3/2 Search PubMed.
- C. Trautwein and K. Kümmerer, Incomplete aerobic degradation of the antidiabetic drug Metformin and identification of the bacterial dead-end transformation product Guanylurea, Chemosphere, 2011, 85(5), 765–773 CrossRef CAS.
- B. A. J. Poursat, R. J. M. van Spanning, M. Braster, R. Helmus, P. de Voogt and J. R. Parsons, Long-term exposure of activated sludge in chemostats leads to changes in microbial communities composition and enhanced biodegradation of 4-chloroaniline and N-methylpiperazine, Chemosphere, 2020, 242, 125102 CrossRef CAS.
- B. A. J. Poursat, R. J. M. van Spanning, M. Braster, R. Helmus, P. de Voogt and J. R. Parsons, Biodegradation of metformin and its transformation product, guanylurea, by natural and exposed microbial communities, Ecotoxicol. Environ. Saf., 2019, 182, 109414 CrossRef CAS.
- Y. Zhang, S.-U. Geißen and C. Gal, Carbamazepine and diclofenac: Removal in wastewater treatment plants and occurrence in water bodies, Chemosphere, 2008, 73(8), 1151–1161 CrossRef CAS.
- R. M. Briones, W.-Q. Zhuang and A. K. Sarmah, Biodegradation of metformin and guanylurea by aerobic cultures enriched from sludge, Environ. Pollut., 2018, 243, 255–262 CrossRef CAS.
- A. S. Vangnai and W. Petchkroh, Biodegradation of 4-chloroaniline by bacteria enriched from soil: Biodegradation of 4-chloroaniline by bacteria from soil, FEMS Microbiol. Lett., 2007, 268(2), 209–216 CrossRef CAS.
- J. Johnston, T. LaPara and S. Behrens, Composition and Dynamics of the Activated Sludge Microbiome during Seasonal Nitrification Failure, Sci. Rep., 2019, 9(1), 4565 CrossRef.
- A. Klindworth, E. Pruesse, T. Schweer, J. Peplies, C. Quast and M. Horn, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies, Nucleic Acids Res., 2013, 41(1), e1 CrossRef CAS.
- R core team, R Core Team, 2016. R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/, 2016, Available from: https://www.R-project.org/.
- I. F. Persoon, M. J. Buijs, A. R. Özok, W. Crielaard, B. P. Krom and E. Zaura, et al. The mycobiome of root canal infections is correlated to the bacteriome, Clin. Oral Investig., 2017, 21(5), 1871–1881 CrossRef.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer and P. Yarza, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2013, 41(D1), D590–D596 CrossRef CAS.
- P. J. McMurdie and S. Holmes, phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLoS One, 2013, 8(4), e61217 CrossRef CAS.
- J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre and D. McGlinn, et al. Vegan: Community Ecology Package. R Package version 2.4-2, 2017, Available from: https://CRAN.R-project.org/package¼vegan.
- H. Wickham, Ggplot2: elegant graphics for data analysis, Springer, New York, 2016, p. 212, (Use R!) Search PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15(12), 550 CrossRef.
- H. J. Kool, Influence of microbial biomass on the biodegradability of organic compounds, Chemosphere, 1984, 13(7), 751–761 CrossRef CAS.
- H. Birch, R. Hammershøj, M. Comber and P. Mayer, Biodegradation of hydrocarbon mixtures in surface waters at environmentally relevant levels – Effect of inoculum origin on kinetics and sequence of degradation, Chemosphere, 2017, 184, 400–407 CrossRef CAS.
- S. Kim, K. Rossmassler, C. D. Broeckling, S. Galloway, J. Prenni and S. K. De Long, Impact of inoculum sources on biotransformation of pharmaceuticals and personal care products, Water Res., 2017, 125, 227–236 CrossRef CAS.
- T. Zhang, M.-F. Shao and L. Ye, 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants, ISME J., 2012, 6(6), 1137–1147 CrossRef CAS.
- D. R. Johnson, D. E. Helbling, T. K. Lee, J. Park, K. Fenner and H.-P. E. Kohler, et al. Association of Biodiversity with the Rates of Micropollutant Biotransformations among Full-Scale Wastewater Treatment Plant Communities. Spormann AM, editor, Appl. Environ. Microbiol., 2015, 81(2), 666–675 CrossRef.
- L. B. Stadler, J. D. Vela, S. Jain, G. J. Dick and N. G. Love, Elucidating the impact of microbial community biodiversity on pharmaceutical biotransformation during wastewater treatment, Microb. Biotechnol., 2018, 11(6), 995–1007 CrossRef CAS.
- T. Curtis, M. Polchan, J. Baptista, R. Davenport and W. Sloan, Microbial community assembly, theory and rare functions, Front. Microbiol., 2013, 4, 68 Search PubMed.
- F. Ingerslev and N. Nyholm, Shake-Flask Test for Determination of Biodegradation Rates of 14C-Labeled Chemicals at Low Concentrations in Surface Water Systems, Ecotoxicol. Environ. Saf., 2000, 45(3), 274–283 CrossRef CAS.
- T. J. Martin, A. K. Goodhead, J. R. Snape and R. J. Davenport, Improving the ecological relevance of aquatic bacterial communities in biodegradability screening assessments, Sci. Total Environ., 2018, 627, 1552–1559 CrossRef CAS.
- T. J. Martin, J. R. Snape, A. Bartram, A. Robson, K. Acharya and R. J. Davenport, Environmentally relevant inoculum concentrations improve the reliability of persistent assessments in biodegradation screening tests, Environ. Sci. Technol., 2017, 51(5), 3065–3073 CrossRef CAS.
- G. Thouand, P. Friant, F. Bois, A. Cartier, A. Maul and J. C. Block, Bacterial Inoculum Density and Probability of para-Nitrophenol Biodegradability Test Response, Ecotoxicol. Environ. Saf., 1995, 30(3), 274–282 CrossRef CAS.
- G. Thouand and J. C. Block, Utilisation d'inogula preculttves dans les essais de biodegradabilite the use of precultured inocula for biodegradability tests, Environ. Technol., 1993, 14(7), 601–614 CrossRef CAS.
- A. K. Goodhead, I. M. Head, J. R. Snape and R. J. Davenport, Standard inocula preparations reduce the bacterial diversity and reliability of regulatory biodegradation tests, Environ. Sci. Pollut. Res., 2014, 21(16), 9511–9521 CrossRef CAS.
- M. Carrero-Colón, C. H. Nakatsu and A. Konopka, Microbial community dynamics in nutrient-pulsed chemostats, FEMS Microbiol. Ecol., 2006, 57(1), 1–8 CrossRef.
- I. T. Cousins, C. A. Ng, Z. Wang and M. Scheringer, Why is high persistence alone a major cause of concern?, Environ. Sci.: Processes Impacts, 2019, 21(5), 781–792 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ew00776e |
| ‡ Contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |