Open Access Article
Open Access ArticleG-quadruplex DNA inhibits unwinding activity but promotes liquid–liquid phase separation by the DEAD-box helicase Ded1p†
Jun
Gao‡
 a,
Zhaofeng
Gao‡
b,
Andrea A.
Putnam‡
b,
Alicia K.
Byrd
a,
Zhaofeng
Gao‡
b,
Andrea A.
Putnam‡
b,
Alicia K.
Byrd
 a,
Sarah L.
Venus
b,
John C.
Marecki
a,
Andrea D.
Edwards
a,
Haley M.
Lowe
a,
Eckhard
Jankowsky
*b and
Kevin D.
Raney
*a
a,
Sarah L.
Venus
b,
John C.
Marecki
a,
Andrea D.
Edwards
a,
Haley M.
Lowe
a,
Eckhard
Jankowsky
*b and
Kevin D.
Raney
*a
aDepartment of Biochemistry and Molecular Biology, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA. E-mail: RaneyKevinD@uams.edu
bCenter for RNA Science and Therapeutics, Department of Biochemistry, School of Medicine, Case Western Reserve University, Cleveland, Ohio 44106, USA. E-mail: exj13@case.edu
First published on 7th July 2021
Abstract
G-quadruplex DNA interacts with the N-terminal intrinsically disordered domain of the DEAD-box helicase Ded1p, diminishing RNA unwinding activity but enhancing liquid–liquid phase separation of Ded1p in vitro and in cells. The data highlight multifaceted effects of quadruplex DNA on an enzyme with intrinsically disordered domains.
G-quadruplexes (G4) are four-stranded nucleic acid structures of stacked planar guanine tetrads. The guanines in each plane interact through Hoogsteen hydrogen bonding.1,2 G4 structures play important roles in gene expression, DNA replication, and telomere maintenance.3–5 In addition, G4 structures have been linked to genomic instability and human diseases, including cancer and neurodegenerative disorders.6–9
DEAD-box RNA helicases, characterized by a highly conserved “Asp-Glu-Ala-Asp” (DEAD) motif, are found in all eukaryotes, as well as in many bacteria and archaea.10 These enzymes are involved in most aspects of cellular RNA metabolism and in the formation of stress granules and P-bodies, cellular RNA-protein condensates that form by liquid–liquid phase separation (LLPS).10,11 Many DEAD-box helicases have been linked to diseases, including cancer, neurodegeneration and viral infections.12–14 The DEAD-box RNA helicase Ded1p, an ortholog of human DDX3X, is involved in translation initiation and localizes to stress granules and P-bodies.15–17
We have shown previously that Ded1p binds tightly to, but does not unfold G4DNA.18 To probe possible effects of G4DNA on the function of Ded1p, we tested the impact of G4DNA on the RNA unwinding activity of recombinant Ded1p (Fig. 1). Addition of G4DNA inhibits RNA unwinding of Ded1p, compared to the reaction without DNA (Fig. 1a and b). An equivalent concentration of a DNA (ssDNA) that does not form a G-quadruplex, does not inhibit RNA unwinding of Ded1p to the extent seen with G4DNA (Fig. 1c). We also performed unwinding reactions with increasing concentrations of G4DNA and ssDNA to determine the respective inhibition constants (Ki) (Fig. 1d). For G4DNA, Ki = 4.37 ± 1.24 nM, while for ssDNA a Ki = 276 ± 27 nM reveals a more than 60-fold weaker affinity (Fig. 1d). The results indicate tight binding of G4DNA to Ded1p, consistent with previous equilibrium binding data.18 The data collectively demonstrate that G4DNA inhibits the RNA unwinding activity of Ded1p.
We next examined which domains of Ded1p were involved in the interactions with the G4DNA. In addition to the conserved helicase domain, Ded1p contains an N-terminal unstructured domain and a basic C-terminal tail (Fig. 2a).10 We probed equilibrium binding of Ded1p variants without either the N- or the C-terminal to the G4DNA (Fig. 2b). Compared to full-length Ded1p (Kd = 1.3 ± 0.3 nM), deletion of the C-terminal tail (ΔC Ded1) has little effect on G4DNA binding (Kd = 1.0 ± 0.2 nM). However, deletion of the N-terminal unstructured domain (ΔN Ded1), markedly reduces affinity for G4DNA (Kd = 16 ± 7 nM), compared to full-length Ded1p. These observations suggest that the N-terminal unstructured domain of Ded1p plays a major role in G4DNA binding, whereas the C-terminal domain is largely dispensable. The N-terminal domain of Ded1p contains RGG repeats, which have been shown in other proteins to bind quadruplex structures.18 The helicase domain is also likely involved in G4DNA binding because ΔN Ded1p retains some G4DNA binding activity.
Consistent with the involvement of the N-terminal domain in G4DNA binding, G4DNA inhibits RNA unwinding for both full length Ded1p and ΔC Ded1p (Fig. 2c). The ssDNA does not significantly inhibit unwinding by any of the Ded1p variants at 30 nM. Without the competing DNA, the rate of dsRNA unwinding by ΔN Ded1p is reduced roughly 10-fold, relative to full-length Ded1p, while the rate of duplex RNA unwinding by ΔC Ded1p was reduced more than 40-fold, consistent with previous results.19 Our observations show that both termini are important for efficient unwinding by Ded1p.
DEAD-box helicases with unstructured termini undergo liquid–liquid phase separation (LLPS), a process that underlies stress granule formation.16,20 Stress granules are non-membranous organelles in the cytoplasm composed of translationally repressed mRNA and proteins. Ded1p functions in translation initiation, in part by remodelling RNA secondary structure in 5′-UTRs.15 LLPS and sequestration of Ded1p in stress granules reduce translation of mRNAs that depend on Ded1p, while allowing translation of mRNAs that do not require Ded1p.20 It has been recently reported that G4 structures can also promote phase separation.21–23 We have previously shown that oxidative stress promotes accumulation of G4DNA in the cytoplasm of some human cancer cell lines.24 Furthermore, we found that cytoplasmic G4DNA promotes formation of stress granules through binding to different stress granule proteins such as DHX36, TIA1, and G3BP.24 We thus asked whether G4DNA could promote LLPS of Ded1p in vitro (Fig. 3). Ded1p was labelled with Cy5, and incubated with Cy3-labelled G4DNA. We observed LLPS, including formation of a significant fraction of large (>20 μm) Ded1p granules after 1 hour (Fig. 3a, and ESI,† Fig. S1). LLPS of Ded1p with Cy3-labelled ssDNA or ssRNA did not result in formation of comparable fractions of large granules. In addition, the number of large granules increased with increasing concentrations of G4DNA. In contrast, the number of large granules decreased with increasing concentrations of ssDNA or ssRNA (Fig. 3b).
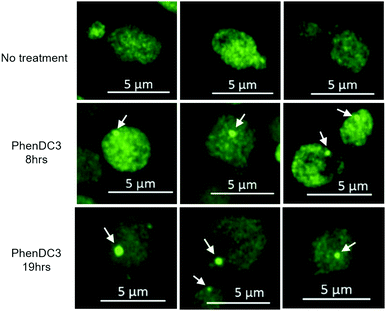
Next, we examined whether G4DNA stimulates LLPS of Ded1p in cells. Since we could not efficiently introduce extra-cellular G4DNA into the cytosol of S. cerevisiae, we tested how Phen-DC3, a well-characterized G4 stabilizer,25 affects Ded1p LLPS in vivo (Fig. 4). S. cerevisiae cells expressing Ded1-GFP from its endogenous chromosomal locus, were grown to early-log phase, then Phen-DC3 was added to the culture to 10 μM. After eight hours of Phen-DC3 treatment, large Ded1-GFP foci are observed. The size of Ded1-GFP foci further increased after 19 hours of Phen-DC3 treatment. A high-resolution 3-dimensional scan of the cell after 19 hours of Phen-DC3 treatment shows that Ded1-GFP forms a non-membrane organelle-like structure at the outside of the nucleus in the cell (ESI,† Video S1). These observations indicate that Phen-DC3 promotes LLPS of Ded1-GFP in cells.
To examine the impact of Phen-DC3 on nucleic acid remodelling by Ded1p, we tested the effect of Phen-DC3 on the ATP-independent G4RNA destabilization by Ded1p in vitro (ESI,† Fig. S2). We found that 0.5 μM Phen-DC3 inhibits G4RNA destabilization by Ded1p, but not its helicase activity on RNA–DNA duplexes (ESI,† Fig. S2 and S3). 1 μM Phen-DC3 can inhibit RNA duplex unwinding by Ded1 (ESI,† Fig. S4). We thus note that inhibition of the helicase activity might contribute to the stimulation of Ded1p LLPS by Phen-DC3 in cells, in addition to the G4 stabilization by Phen-DC3.
In sum, we demonstrated that G4DNA inhibits RNA unwinding activity of DEAD-box helicase Ded1p, and promotes LLPS of Ded1p. Our findings suggest possible new roles for G4 structures and DEAD-box RNA helicases. Many viruses harbor G4 signatures in their genome.26,27 Using G4 DNA or RNA to stimulate LLPS while inhibiting unwinding activities of (a) DEAD-box RNA helicase(s), would be an efficient strategy for a virus to hijack the translation apparatus of the host cell for virus propagation.
This work was supported by National Institutes of Health Grants (R01 GM098922, R01 GM117439, and R35 GM122601 to K. D. R.; R35 GM118088 to E. J.; T32GM007250 and F30CA247347 to S. L. V.), and the UAMS Research Council. The UAMS DNA Sequencing Core is supported by the UAMS Center for Microbial Pathogenesis and Host Inflammatory Responses (National Institutes of Health Grant P20 GM103625), and the UAMS Translational Research Institute (National Institutes of Health Grant UL1TR000039). The UAMS Digital Microscopy Core facility is supported through user fees and university funding.
Conflicts of interest
There are no conflicts to declare.Notes and references
- G. N. Parkinson, M. P. H. Lee and S. Neidle, Nature, 2002, 417, 876–880 CrossRef CAS PubMed.
- H. L. Lightfoot, T. Hagen, N. J. Tatum and J. Hall, FEBS Lett., 2019, 593, 2083–2102 CrossRef CAS PubMed.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627–8637 CrossRef CAS PubMed.
- R. Hänsel-Hertsch, M. Di Antonio and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2017, 18, 279–284 CrossRef PubMed.
- Y. Wu and R. M. Brosh, FEBS J., 2010, 277, 3470–3488 CrossRef CAS PubMed.
- N. Maizels, EMBO Rep., 2015, 16, 910–922 CrossRef CAS PubMed.
- S. Ravichandran, V. K. Subramani and K. K. Kim, Biophys. Rev., 2019, 11, 383–387 CrossRef CAS PubMed.
- P. Kharel, S. Balaratnam, N. Beals and S. Basu, Wiley Interdiscip. Rev.: RNA, 2020, 11, e1568 CAS.
- P. Linder and E. Jankowsky, Nat. Rev. Mol. Cell Biol., 2011, 12, 505–516 CrossRef CAS.
- I. Jarmoskaite and R. Russell, Wiley Interdiscip. Rev.: RNA, 2011, 2, 135–152 CrossRef CAS PubMed.
- F. V. Fuller-Pace, RNA Biol., 2013, 10, 121–132 CrossRef CAS.
- D. Sharma and E. Jankowsky, Crit. Rev. Biochem. Mol. Biol., 2014, 49, 343–360 CrossRef CAS PubMed.
- F. Taschuk and S. Cherry, Viruses, 2020, 12, 181 CrossRef CAS PubMed.
- U. P. Guenther, D. E. Weinberg, M. M. Zubradt, F. A. Tedeschi, B. N. Stawicki, L. L. Zagore, G. A. Brar, D. D. Licatalosi, D. P. Bartel, J. S. Weissman and E. Jankowsky, Nature, 2018, 559, 130–134 CrossRef CAS PubMed.
- M. Hondele, R. Sachdev, S. Heinrich, J. Wang, P. Vallotton, B. M. A. Fontoura and K. Weis, Nature, 2019, 573, 144–148 CrossRef CAS PubMed.
- A. Hilliker, Z. Gao, E. Jankowsky and R. Parker, Mol. Cell, 2011, 43, 962–972 CrossRef CAS PubMed.
- J. Gao, A. K. Byrd, B. L. Zybailov, J. C. Marecki, M. J. Guderyon, A. D. Edwards, S. Chib, K. L. West, Z. J. Waldrip, S. G. Mackintosh, Z. Gao, A. A. Putnam, E. Jankowsky and K. D. Raney, Chem. Commun., 2019, 55, 4467–4470 RSC.
- A. A. Putnam, Z. Gao, F. Liu, H. Jia, Q. Yang and E. Jankowsky, Mol. Cell, 2015, 59, 541–552 CrossRef CAS.
- C. Iserman, C. Desroches Altamirano, C. Jegers, U. Friedrich, T. Zarin, A. W. Fritsch, M. Mittasch, A. Domingues, L. Hersemann, M. Jahnel, D. Richter, U. P. Guenther, M. W. Hentze, A. M. Moses, A. A. Hyman, G. Kramer, M. Kreysing, T. M. Franzmann and S. Alberti, Cell, 2020, 181, 818–831 CrossRef CAS PubMed.
- P. Ivanov, E. O’Day, M. M. Emara, G. Wagner, J. Lieberman and P. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 18201–18206 CrossRef CAS PubMed.
- M. M. Fay, P. J. Anderson and P. Ivanov, Cell Rep., 2017, 21, 3573–3584 CrossRef CAS PubMed.
- Y. Zhang, M. Yang, S. Duncan, X. Yang, M. A. S. Abdelhamid, L. Huang, H. Zhang, P. N. Benfey, Z. A. E. Waller and Y. Ding, Nucleic Acids Res., 2019, 47, 11746–11754 CAS.
- A. K. Byrd, B. L. Zybailov, L. Maddukuri, J. Gao, J. C. Marecki, M. Jaiswal, M. R. Bell, W. C. Griffin, M. R. Reed, S. Chib, S. G. Mackintosh, A. M. MacNicol, G. Baldini, R. L. Eoff and K. D. Raney, J. Biol. Chem., 2016, 291, 18041–18057 CrossRef CAS PubMed.
- W. J. Chung, B. Heddi, F. Hamon, M. P. Teulade-Fichou and A. T. Phan, Angew. Chem., Int. Ed., 2014, 53, 999–1002 CrossRef CAS PubMed.
- M. Metifiot, S. Amrane, S. Litvak and M. L. Andreola, Nucleic Acids Res., 2014, 42, 12352–12366 CrossRef CAS PubMed.
- N. Saranathan and P. Vivekanandan, Trends Microbiol., 2019, 27, 148–163 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed experimental methods, supplementary tables and figures. See DOI: 10.1039/d1cc01479j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |