Open Access Article
Open Access ArticleA fluorogenic peptide-based smartprobe for the detection of neutrophil extracellular traps and inflammation†
Maria R.
Rios
 a,
Gloria
Garoffolo
b,
Giulia
Rinaldi
c,
Alicia
Megia-Fernandez
a,
Gloria
Garoffolo
b,
Giulia
Rinaldi
c,
Alicia
Megia-Fernandez
 a,
Silvia
Ferrari
b,
Calum T.
Robb
c,
Adriano G.
Rossi
c,
Maurizio
Pesce
a,
Silvia
Ferrari
b,
Calum T.
Robb
c,
Adriano G.
Rossi
c,
Maurizio
Pesce
 b and
Mark
Bradley
b and
Mark
Bradley
 *a
*a
aEaStCHEM School of Chemistry, University of Edinburgh, David Brewster Road, EH9 3FJ Edinburgh, UK. E-mail: Mark.Bradley@ed.ac.uk
bTissue Engineering Research Unit, Centro Cardiologico Monzino, IRCCS, Milan, Italy
cCentre for Inflammation Research, Queen's Medical Research Institute, University of Edinburgh, UK
First published on 8th December 2020
Abstract
A highly sensitive optical probe for the detection of activated neutrophils and Neutrophil Extracellular Traps (NETs) is reported. It is based on a triple-quenched, super-silent tri-branched probe that generates >20 fold increase in fluorescence upon cleavage. The probe was highly specific for human neutrophil elastase, a protease that mediates a variety of inflammatory diseases, and detected NETosis and neutrophil activation in in vitro differentiated neutrophils and isolated human neutrophils.
Neutrophils (phagocytic leukocytes) are the most abundant immune cells circulating in human blood and amongst the first cells to be recruited to inflammatory sites, providing a first line of immune defence against pathogens.1 However, activation of neutrophils is associated with many inflammatory diseases and has become a cellular biomarker for a broad range of inflammation-mediated diseases.2 Neutrophils have a wide range of defence mechanisms against invading microorganisms, including phagocytosis, degranulation and reactive oxygen species (ROS) production. In 2004, Brinkmann et al.3 reported a novel process by which activated neutrophils extrude chromatin studded with antimicrobial proteins in the extracellular environment to entrap and kill invading pathogens, such as bacteria,3 virus4 and fungi.5 The structures formed are named Neutrophil Extracellular Traps (NETs).3 These extracellular traps are not restricted to neutrophils as they can be produced by many cell types in a broad range of organisms and indeed are believed to be an evolutionary preserved defence mechanism.6 Despite their beneficial role in host defence NETs are known to play pivotal roles in many inflammatory diseases7 and aberrant NET formation is linked to pulmonary diseases like acute respiratory distress syndrome (ARDS), acute lung injury (ALI)8 and more recently, COVID-19.9 Thus, neutrophils isolated from COVID-19 patients that exhibit pneumonia-associated ARDS are more susceptible to form NETs; a response that correlates with severity and mortality of the disease.8 NETs forming in arteries and veins are thought to be important in initiating thrombosis in the vasculature, again of relevance in the immunopathology in COVID-19 patients.10 Detection and visualisation of NETs has therefore become a potential tool for diagnosis of NET-associated pathologies7 however, to date, this has largely been based on immuno-staining of NET markers using antibodies.
Activated serine proteases are present on NETs. Human neutrophil elastase (hNE) is one of the four active neutrophil serine proteases. In fact, it is the most abundant serine protease stored in neutrophil granules at millimolar concentration. This protease is present in NETs and found bound to DNA, the key scaffold component of NETs, due to hNE high binding affinity to chromatin. hNE is thus a good target for detection and visualisation of NETs. Several inhibitors for hNE have been developed and some of them used clinically including the inhibitor sivelestat,11 which is used to treat COVID-19 patients.12 When it comes to protease sensing most substrates rely on chromogenic sensing or fluorescence, and for hNE the vast majority are based on the peptide sequence AAPV which is also cleavable by the closely related serine protease Proteinase 3 (PR3).13 Only one highly specific sensor for hNE has been validated as a preclinical fluorescent activated sensor and this was based on the sequence PMAVVQSVP.14 Activity based probes for elastase, that bind covalently to the enzyme have been reported for hNE, however, signal amplification is not possible as the enzyme is generally inhibited irreversibly following the enzymatic reaction.15 Thus, there is a need for sensing methods that show high specificity, sensitivity and high signal-to-noise ratios. Peptide based “dendritic” or multi-branched sensors offer a promising approach to overcome these limitations as they potentially provide significantly higher signal amplification and lower background noise when compared to their linear counterparts as well as offering high sensitivity.16 Our group previously reported activatable probes for serprocidins17 (hNE, PR3 and CG) and elastase18 sensing but they proved unsuccessful for NETs visualisation, while their high background fluorescence, made them unsuitable for sensing and in vivo applications. Herein, we present a novel fluorogenic sensor, specific for hNE that allows detection and visualisation of NETs – the first molecular probe developed that detects hNE in NETs and inflammatory induced conditions.
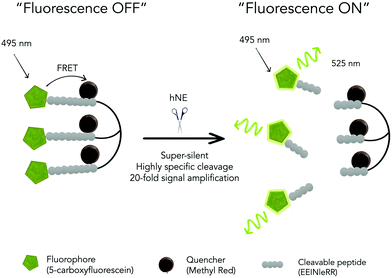
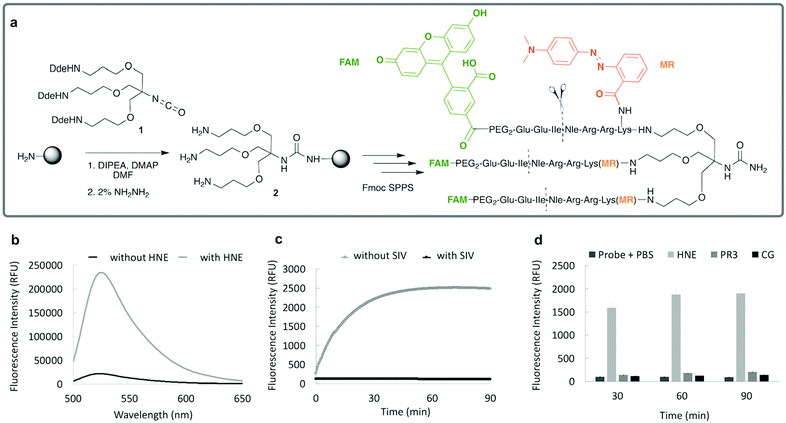
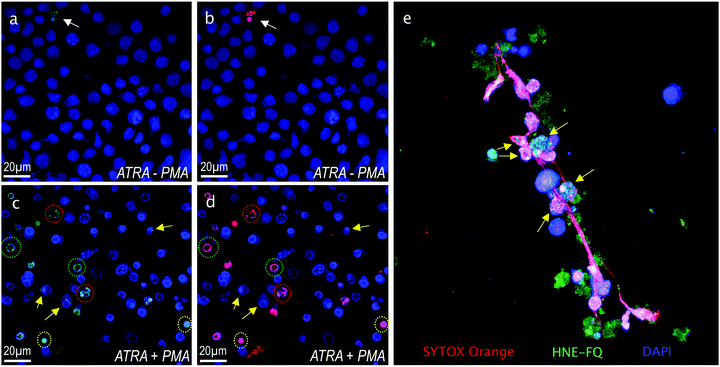
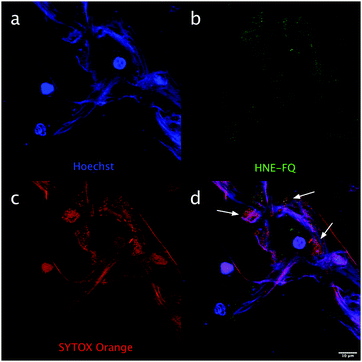
Importantly this sensor provides an OFF/ON fluorescent signal upon cleavage by hNE in activated neutrophils and NETs. The sensor was based on a trivalent scaffold combined with FRET-labelled peptides (Scheme 1). The trivalent structure provides amplification of signal upon substrate (peptide) cleavage and enhanced self-quenching capacity by linear quenching between the fluorophores and the quenchers.19 The sequence (Glu–Glu–Ile↑Nle–Arg–Arg) was adapted from a peptide20 that is cleaved specifically by hNE, where modification of methionine in the reported sequence with norleucine provides in vivo stability. The peptide was labelled with the fluorophore 5-carboxyfluorescein (5-FAM) and quencher Methyl Red (MR) as the FRET pair. Methyl red was incorporated as Fmoc-Lys(MR)-OH (4) (ESI†), avoiding the need for orthogonal deprotection21 on the lysine, which would complicate the synthesis of these branched peptides due to incomplete deprotection on one or more branches. A bis-ethylene glycol spacer was introduced between the last amino acid and the fluorophore, to increase hydrophilicity and promote aqueous solubility. The probe was synthesised on an aminomethyl ChemMatrix resin functionalised with the Fmoc-Rink Amide linker. The starting trivalent isocyanate (1) was synthesized following a previously reported procedure22 and the isocyanate coupled to the resin using DIPEA and DMAP. Removal of the Dde (N-(1-(4,4-dimethyl-2,6-dioxocyclohexylidene)ethyl)) protecting group with 2% hydrazine in DMF gave the scaffold (2) for the synthesis of the FRET-peptides using Fmoc solid-phase peptide synthesis (SPPS) with DIC/Oxyma as the coupling combination with 5-carboxyfluorescein capping the three peptide branches (Fig. 1a) (ESI†). Interestingly the use of the N-hydroxysuccinimide ester of 5-carboxyfluorescein diacetate in the final coupling step gave final crude products that were considerably purer than those generated using DIC/Oxyma/5-carboxyfluorescein (see Fig. S1, ESI†). The probe was optically silent, but rapidly de-quenched within seconds by hNE with a 20-fold increase in signal following exposure of the probe (5 μM) to the enzyme (Fig. 1b) with a KM of 2.7 × 10−6 M and a catalytic efficiency (kcat/KM) of 1.5 × 104 M−1 s−1 (ESI†). Cleavage specificity was confirmed by MALDI-TOF MS and LC-MS (Fig. S2 and S3, ESI†), while pre-incubation with sivelestat (100 μM) resulted in no increase of fluorescence (Fig. 1c). The closely related neutrophil serine proteases Cathepsin G and Proteinase 3, showed negligible cleavage demonstrating that the probe was specific for hNE (Fig. 1d). The fluorescence signal of the activated probe was not affected by incubation in a range of different biological environments (ESI†) and was stable between pH 6–8.5. As expected, due to the pH dependent character of carboxyfluorescein, fluorescence was reduced at lower pHs (ESI†). The human promyelocytic cell line HL-60 is a valid cell culture model to study neutrophil differentiation, activation and NETs generation.23 To validate the probe, cells were stimulated with all-trans-retinoic acid (ATRA, 2 μM) for 4 days to induce a neutrophil-like phenotype. A quantitative nitro-blue tetrazolium (NBT) assay was performed in order to assess neutrophil differentiation efficiency in terms of phagocytic functionality (ESI†). For NETs formation, differentiated HL-60 cells were seeded into 8-well chamber slides coated with poly-L-lysine, and treated with phorbol 12-myristate 13-acetate (PMA; 100 nM) and probe HNE-FQ (5 μM) for 3 h. PMA is a potent stimulator of NET formation, since it triggers neutrophil NADPH oxidase activation in neutrophil activation and inflammation.24 To verify whether the treatments induced NETosis, cells were treated with all-trans-retinoic acid (ATRA) ± PMA and incubated with micrococcal nuclease which disrupts and releases the DNA of NETs into the supernatant. DNA release was monitored by a SYTOX Orange assay (ESI†). An increase in NET-associated DNA after cell stimulation with PMA was observed (Fig. S4, ESI†). In order to visualize NETs using HNE-FQ, cells were stained with probe (5 μM), 4′,6-diamidine-2′-phenylindole dihydrochlorideI (DAPI; 0.2 mM) and the DNA intercalating dye (SYTOX Orange; 5 μM). Morphologically, ATRA/PMA treated cells exhibited four different stages of NETosis, based on nuclear morphology and hNE distribution (Fig. 2). Early stages of neutrophil activation exhibited a small and round nucleus with hNE colocalized with condensed chromatin (stage 1; yellow circles).24 At later stages, chromatin decondensation led to spherical (stage 2; green circles) or more “cloud-like spread” shapes (stage 3; red circles). In the final stage, neutrophils formed extracellular chromatin filaments composed by cytoplasmic granules and hNE (stage 4; Fig. 2e).25 NETosis was induced in isolated primary human neutrophils from healthy volunteers (isolated using a protocol approved by the Accredited Medical Regional Ethics Committee (AMREC, reference number 15-HV-013)) following a similar procedure producing well-defined NETs (ESI†) (Fig. 3).
Activation of HL-60 neutrophils was analyzed by flow cytometry, using an antibody directed against CD11b antigen, a well-known marker of neutrophil differentiation,26 and the probe HNE-FQ (Fig. S5, ESI†). Under these conditions, we observed an increase in the fluorescence intensity of cells labeled with the probe in parallel to the increase in fluorescence intensity of CD11b+ HL-60-derived neutrophils when treated with PMA.
In conclusion, we have developed a tri-branched activatable fluorescent hNE sensor that allows the rapid, specific and sensitive detection of hNE in activated neutrophils and NETs. The data show that HNE-FQ is highly specific for staining of human neutrophil activation and highlights its potential as a diagnostic tool to detect and quantify NETosis and activated neutrophils, with current efforts focused on the synthesis of a near infrared (NIR) variant of the sensor.
We would like to thank Engineering and Physical Sciences Research Council (EPSRC, UK). Interdisciplinary Research Collaboration grants EP/K03197X/1 and EP/R005257/1. Medical Research Council grant MR/N02995X/1. CTR and AGR were funded by the Medical Research Council (MR/K013386/1). GR was funded by EPSRC/MRC CDT and OPTIMA (EP/L016559/1). M. P., G. G. and S. F. were funded by Ricerca Corrente and 5 per mille (Italian Ministry of Health). Thanks to Matthew Owens for cell lysate experimental advice.
Conflicts of interest
There are no conflicts to declare.References
- E. Kolaczkowska and P. Kubes, Nat. Rev. Immunol., 2013, 13, 159–175 CrossRef CAS PubMed.
- T. N. Mayadas, X. Cullere and C. A. Lowell, Annu. Rev. Phytopathol., 2014, 9, 181–218 CrossRef CAS PubMed.
- V. Brinkmann, U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D. S. Weiss, Y. Weinrauch and A. Zychlinsky, Science, 2004, 303, 1532–1535 CrossRef CAS PubMed.
- G. Schonrich and M. J. Raftery, Front. Immunol., 2016, 7, 366 Search PubMed.
- E. Guiducci, C. Lemberg, N. Kung, E. Schraner, A. P. A. Theocharides and S. LeibundGut-Landmann, Front. Immunol., 2018, 9, 1573 CrossRef PubMed.
- C. T. Robb, E. A. Dyrynda, R. D. Gray, A. G. Rossi and V. J. Smith, Nat. Commun., 2014, 5, 4627 CrossRef CAS PubMed.
- V. Papayannopoulos, Nat. Rev. Immunol., 2018, 18, 134–147 CrossRef CAS.
- A. Caudrillier, K. Kessenbrock, B. M. Gilliss, J. X. Nguyen, M. B. Marques, M. Monestier, P. Toy, Z. Werb and M. R. Looney, J. Clin. Invest., 2012, 122, 2661–2671 CrossRef CAS PubMed.
- B. J. Barnes, J. M. Adrover, A. Baxter-Stoltzfus, A. Borczuk, J. Cools-Lartigue, J. M. Crawford, J. Dassler-Plenker, P. Guerci, C. Huynh, J. S. Knight, M. Loda, M. R. Looney, F. McAllister, R. Rayes, S. Renaud, S. Rousseau, S. Salvatore, R. E. Schwartz, J. D. Spicer, C. C. Yost, A. Weber, Y. Zuo and M. Egeblad, J. Exp. Med., 2020, 217, e20200652 CrossRef CAS.
- T. A. Fuchs, A. Brill and D. D. Wagner, Arterioscler., Thromb., Vasc. Biol., 2012, 32, 1777–1783 CrossRef CAS PubMed.
- S. Matsumoto, S. Hidaka, K. Goto, S. Hagiwara, C. Shingu, H. Iwasaka and T. Noguchi, J. Anesth., 2009, 23, 288–291 CrossRef PubMed.
- Z. Jin, J. Y. Liu, R. Feng, L. Ji, Z. L. Jin and H. B. Li, Eur. J. Pharmacol., 2020, 883, 173326 CrossRef CAS PubMed.
- N. Jugniot, P. Voisin, A. Bentaher and P. Mellet, Contrast Media Mol. Imaging, 2019, 2019, 7417192 Search PubMed.
- T. Kalupov, M. Brillard-Bourdet, S. Dade, H. Serrano, J. Wartelle, N. Guyot, L. Juliano, T. Moreau, A. Belaaouaj and F. Gauthier, J. Biol. Chem., 2009, 284, 34084–34091 CrossRef CAS PubMed.
- A. C. Schulz-Fincke, A. S. Tikhomirov, A. Braune, T. Girbl, E. Gilberg, J. Bajorath, M. Blaut, S. Nourshargh and M. Gütschow, Biochemistry, 2018, 57(5), 742–752 CrossRef CAS PubMed.
- M. L. Ternon, J. J. Diaz-Mochon, A. Belsom and M. Bradley, Tetrahedron, 2004, 60, 8721–8728 CrossRef CAS.
- T. H. Craven, N. Avlonitis, N. McDonald, T. Walton, E. Scholefield, A. R. Akram, T. S. Walsh, C. Haslett, M. Bradley and K. Dhaliwal, Sci. Rep., 2018, 8, 13490 CrossRef PubMed.
- N. Avlonitis, M. Debunne, T. Aslam, N. McDonald, C. Haslett, K. Dhaliwal and M. Bradley, Org. Biomol. Chem., 2013, 11, 4414–4418 RSC.
- J. M. Ellard, T. Zollitsch, W. J. Cummins, A. L. Hamilton and M. Bradley, Angew. Chem., Int. Ed., 2002, 41, 3233–3236 CrossRef.
- B. Korkmaz, S. Attucci, M. A. Juliano, T. Kalupov, M. L. Jourdan, L. Juliano and F. Gauthier, Nat. Protoc., 2008, 3, 991–1000 CrossRef CAS PubMed.
- J. J. Díaz-Mochón, L. Bialy and M. Bradley, Org. Lett., 2004, 6, 1127–1129 CrossRef PubMed.
- S. Lebreton, S. E. How, M. Buchholz, B. E. Yingyongnarongkul and M. Bradley, Tetrahedron, 2003, 59, 3945–3953 CrossRef CAS.
- R. Tasseff, H. A. Jensen, J. Congleton, D. Dai, K. V. Rogers, A. Sagar, R. P. Bunaciu, A. Yen and J. D. Varner, Sci. Rep., 2017, 7, 14327 CrossRef PubMed.
- R. S. Keshari, A. Verma, M. K. Barthwal and M. Dikshit, J. Cell Biol., 2013, 114, 532–540 CAS.
- T. Kawakami, J. He, H. Morita, K. Yokoyama, H. Kaji, C. Tanaka, S. Suemori, K. Tohyama and Y. Tohyama, PLoS One, 2014, 9, e84704 CrossRef PubMed.
- R. Spijkerman, L. Hesselink, C. Bertinetto, C. C. Bongers, F. Hietbrink, N. Vrisekoop, L. P. Leenen, M. T. Hopman, J. J. Jansen and L. Koenderman, J. Leukocyte Biol., 2020, 1–10 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc07028a |
| This journal is © The Royal Society of Chemistry 2021 |