Open Access Article
Open Access ArticleAn RGB-emitting molecular cocktail for the detection of bacterial fingerprints†
Sheng
Hong‡
 a,
Di-Wei
Zheng‡
a,
Qiu-Ling
Zhang
a,
Wei-Wei
Deng
b,
Wen-Fang
Song
a,
Si-Xue
Cheng
a,
Di-Wei
Zheng‡
a,
Qiu-Ling
Zhang
a,
Wei-Wei
Deng
b,
Wen-Fang
Song
a,
Si-Xue
Cheng
 a,
Zhi-Jun
Sun
b and
Xian-Zheng
Zhang
a,
Zhi-Jun
Sun
b and
Xian-Zheng
Zhang
 *a
*a
aKey Laboratory of Biomedical Polymers of Ministry of Education, Department of Chemistry, Wuhan University, Wuhan 430072, P. R. China. E-mail: xz-zhang@whu.edu.cn
bDepartment of Oral Maxillofacial Head Neck Oncology, School and Hospital of Stomatology, Wuhan University, Wuhan 430072, P. R. China
First published on 2nd April 2020
Abstract
Accumulating evidence indicates that colonized microbes play a crucial role in regulating health and disease in the human body. Detecting microbes should be essential for understanding the relationship between microbes and diseases, as well as increasing our ability to detect diseases. Here, a combined metabolic labeling strategy was developed to identify different bacterial species and microbiota by the use of three different fluorescent metabolite derivatives emitting red, green, and blue (RGB) fluorescence. Upon co-incubation with microbes, these fluorescent metabolite derivatives are incorporated into bacteria, generating unique true-color fingerprints for different bacterial species and different microbiota. A portable spectrometer was also fabricated to automate the colorimetric analysis in combination with a smartphone to conveniently identify different bacterial species and microbiota. Herein, the effectiveness of this system was demonstrated by the identification of certain bacterial species and microbiota in mice with different diseases, such as skin infections and bacteremia. By analyzing the microbiota fingerprints of saliva samples from clinical patients and healthy people, this system was proved to precisely distinguish oral squamous cell carcinoma (OSCC, n = 29) samples from precancerous (n = 10) and healthy (n = 5) samples.
Introduction
The human body is a diverse ecosystem that harbors up to 1014 bacterial cells.1 With our in-depth understanding of the critical relationship between microbes and diseases, the study of commensal microbes is currently in the spotlight.2–7 For example, Fusobacterium nucleatum, a bacterium that exists in the gut, can be viewed as a biomarker for colorectal cancer (CRC) diagnosis.8 Some specific oral bacteria, including Capnocytophaga gingivalis, Prevotella melaninogenica, and Streptococcus mitis, have been found to predict 80% of oral squamous cell carcinoma (OSCC) cases.9 Apart from the changes in specific types of bacteria, the occurrence and progression of disease is also accompanied by alteration of the microbial community structure. By using genome sequencing, analysis of the microbial community gives a classification accuracy of 80.3% in identifying CRC patients.10–12 Considering the close relationship between microbes and diseases, detection methods for microbes should be essential for understanding the interactions of microbes and diseases as well as furthering our ability to detect disease with increasing sophistication.There are various techniques that have been developed to analyze microbes. Among those, genome sequencing, protein mass spectrum detection, and fluorescence in situ hybridization (FISH) are the three most commonly used techniques.13,14 The first two are superior in their high throughput, but their shortcomings include high cost and complex operation, which largely hinder their pervasive application. Although FISH has been broadly used in cellular imaging and pathogenic diagnosis due to its high sensitivity and satisfactory specificity, it is a tedious procedure with too many false negative results, which limits its application prospects. Additionally, the limitation of using one specific probe for only one bacterial species also greatly restricts its capacity in multiple-bacteria analysis and microbiota identification.15 Given that the interaction of microbes and diseases is a complex relationship involving diverse bacterial species, it is quite necessary to develop techniques that can not only recognize specific bacterial species, but also identify the distinct fingerprint between different microbiota.
Metabolic labeling, depending on the biosynthetic mechanism to assimilate functionalized metabolites, has recently emerged as a promising method for studying the metabolic processes of living systems.16–18 Peptidoglycan, one of the major components of bacterial cell walls, is a multi-layer network structure composed of glycans and short peptides, and it has been labeled to study the synthesis of bacterial cell walls.19–23 Modified monosaccharides and amino acids have been shown to be incorporated into the peptidoglycan construction without affecting the normal growth, metabolism, or multiplication of bacteria, and have been used to efficiently label bacteria for the purpose of distinguishing them. Specifically, fluorophore-modified D-amino acids have been developed for the metabolic labeling of bacteria.24–27 In addition, previous studies have reported the labeling of bacteria with derivatives of monosaccharides, such as N-acetylmuramic acid, N-acetylglucosamine, N-acetylgalactosamine, and arabinose.28–31 However, the metabolic incorporation strategy to use only one functionalized metabolite is imprecise and is insufficient for bacterial recognition in a complex environment such as the microbiota.
Taking these into account, we attempted to develop a strategy to more accurately distinguish specific bacterial species and even different microbiota using a combined metabolic labeling strategy. Based on the fact that the metabolic characteristics vary between bacteria, and different bacterial species incorporate different metabolites for biosynthesis, the labeling of bacteria through the modification of different metabolites with different reporters might be a more effective strategy to distinguish bacteria.
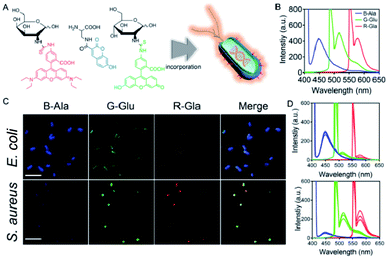
Here, we chose three metabolites, D-amino acids, N-acetylglucosamine, and N-acetylgalactosamine, that are essential for bacterial biosynthesis and modified them with blue fluorescent coumarin, green fluorescent fluorescein, and red fluorescent rhodamine B, respectively (Fig. 1A). The metabolic incorporation capacity of these metabolite derivatives varies in different bacteria. Thus, different bacteria will incorporate different modified metabolites to exhibit different red, green, and blue (RGB) fluorescence and generate distinguishable true-color patterns that can be used to conveniently and effectively identify different bacterial species or even different microbiota.32 We also fabricated a portable spectrometer by 3D printing technology to intuitively observe the RGB fluorescence, and be able to directly read the RGB fluorescence values with a commercialized mobile application (App) on a smartphone (coined the “Microcolor system”).
To verify its effectiveness, the efficiency of the Microcolor system in distinguishing different bacterial species was first studied. Samples collected from complex environments (skin infection and bacteremia) were also tested by the Microcolor system to demonstrate its robustness. After that, we extended this strategy to detect diseases that are accompanied by changes in microbiota fingerprints. Saliva specimens of OSCC (n = 29), precancerous (n = 10) cases, and healthy volunteers (n = 5) were analyzed by the Microcolor system to confirm its potential to detect diseases.
Results and discussion
To verify the effectiveness of our combined metabolic labeling strategy, three fluorophore-conjugated metabolites, including blue fluorescent coumarin labeled-alanine (B-Ala), green fluorescent fluorescein labeled-glucose (G-Glu), and red fluorescent rhodamine labeled-galactose (R-Gal), were synthesized. The results from both proton nuclear magnetic resonance (1H-NMR) spectroscopy and electrospray ionization-mass spectrometry (ESI-MS) demonstrated the successful synthesis of these modified metabolites (Fig. S1†). After successfully modifying the metabolites, the fluorescence spectra of the three fluorophore-conjugated metabolites were measured. As shown in Fig. 1B, B-Ala, G-Glu, and R-Gal exhibited characteristic blue, green, and red fluorescence, respectively, under excitation. Then, two common bacterial species, Escherichia coli and Staphylococcus aureus, were chosen to co-culture with these fluorescent metabolite derivatives to verify whether the modified metabolites could be incorporated into the bacteria as expected.The fluorescent images in Fig. 1C indicated the successful incorporation of the modified metabolites into the two bacterial species, in which the E. coli showed quite strong blue fluorescence and S. aureus showed obvious green and red fluorescence. The distinctly different fluorescence characteristics of E. coli and S. aureus demonstrated the potential of this combined metabolic labeling strategy to distinguish between bacterial species. The fluorescence spectra of the two bacterial species in Fig. 1D also demonstrated the same result. Additionally, whether the labeling procedure would influence the growth of bacteria was also investigated, and it was proved to have no effect on bacterial growth (Fig. S2†).
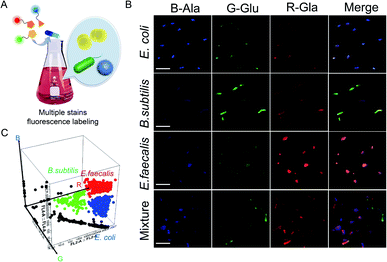
Furthermore, whether this combined metabolic labeling strategy could specifically label multiple bacteria in a mixed culture system was investigated (Fig. 2A). As shown in Fig. 2B, images from fluorescence microscopy indicated that Enterococcus faecalis, E. coli, and Bacillus subtilis exhibited distinct fluorescence signatures not only in single cultivation but also in mixed cultivation. After labeling, it was observed that E. faecalis, E. coli, and B. subtilis displayed obvious orange, green, and blue fluorescence, respectively. Even in a triple mixture, the different species could still be accurately identified by their unique fluorescence emission, indicating that this strategy could also specifically label multiple bacteria in a mixed system. Here, flow cytometry was also used to analyze the different bacterial species in a mixed culture system, and the bacteria were also successfully divided into three groups, which was consistent with the result obtained for the combined metabolic labeling strategy (Fig. 2C). Different bacterial species in a complex mixture were separately labeled with combined metabolic labeling, indicating that this strategy may provide a valuable tool for investigating the communication, competition, and cooperation within a microbial ecosystem.33
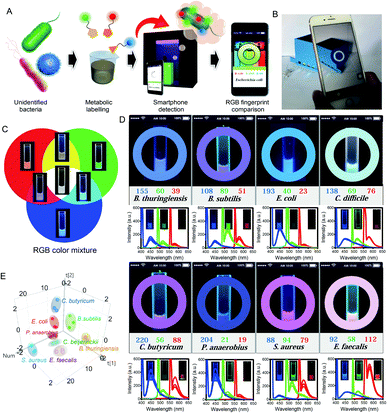
In order to intuitively observe the RGB fluorescence of samples, we then fabricated a portable spectrometer with three tunable band-pass filters (blue, green, and red optical filters) by using 3D printing technology (Fig. S3†).34 A commercialized mobile application (App) on a smartphone was further used for conveniently and directly reading the RGB fluorescence values of the samples by connecting it to the portable spectrometer (Fig. 3A and B). When the samples are labeled by modified metabolites and then analyzed by this portable spectrometer, the color of the fluorescence emitted by the sample can be observed under excitation with a portable flashlight. The RGB fluorescence values of the samples can be directly read with the App through the filters at three specific wavelengths (coined the “Microcolor system”), and this provides great convenience for detection of the fluorescent samples.
In order to verify that the Microcolor system will provide a direct readout of RGB values, the RGB fluorescence of these metabolite derivatives was tested. When these RGB fluorescent metabolites were mixed in different proportions, a characteristic fluorescence emission was observed by the Microcolor system (Fig. 3C). Then, the Microcolor system was used for identifying various evolutionarily distant bacterial species. Eight bacterial species, consisting of Bacillus thuringiensis, B. subtilis, E. coli, Clostridium difficile, Clostridium butyricum, Peptostreptococcus anaerobius, S. aureus, and E. faecalis, were all labeled by RGB fluorescent metabolites and detected by the Microcolor system. As expected, after being cultured with RGB probes, each bacterial species displayed a specific fluorescent profile with distinguishing RGB values that matched with the fluorescence spectra of the bacteria (Fig. 3D). Fluorescence imaging was also used to confirm the validity of the Microcolor system in identifying the bacteria, and was found to be consistent with the result measured by the Microcolor system (Fig. S4†). Furthermore, the RGB values produced by the App were further analyzed by principal component analysis (PCA).35Fig. 3E reveals that the analysis achieved a meaningful classification of bacterial species based on their unique fluorescent fingerprints. These results indicated that the Microcolor system effectively and conveniently distinguished between bacterial species and might be a useful tool for bacterial detection.
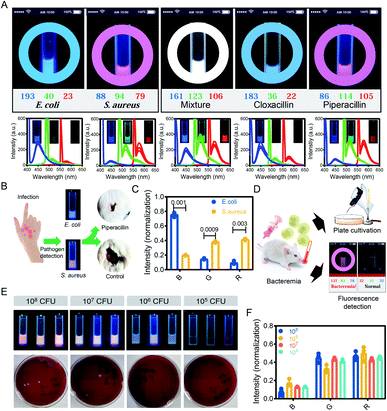
After demonstrating that the Microcolor system can identify between bacterial species in vitro, we further explored whether this system can detect bacterial species in complex environments. First, the emission of two pathogenic bacteria, E. coli and S. aureus, was tested by the Microcolor system. A blue and pink color was observed for E. coli and S. aureus, respectively, which coincided with the results of the fluorescence spectra and fluorescence imaging (Fig. 4A). Accordingly, a mixture of the two bacteria displayed an overlay of the two colors appearing as a white color, which was the same as that of the fluorescence spectra. Interestingly, when the mixture of the two bacteria was treated with piperacillin (one of the most effective antibiotics against E. coli)36 and analyzed by the Microcolor system, the blue color of E. coli almost disappeared, and the color of the mixture changed to the pink color previously shown by S. aureus. Similarly, after being treated with cloxacillin (a narrow-spectrum antibiotic for S. aureus infection),37 the mixture exhibited a color change from white to blue. The same results were also obtained via fluorescence spectra and fluorescence imaging (Fig. 4A and S5†). These results demonstrated that the Microcolor system was able to distinguish different bacterial species in mixed culture.
Here, the Microcolor system was applied to detect bacterial infection in murine skin infection and bacteremia models.38E. coli- and S. aureus-mediated skin infection models were established using mice, and pus samples were collected from infected mice and further analyzed by the Microcolor system. After labeling and analysis by the Microcolor system, the pathogenic bacteria were identified by comparing their emitted color with the pre-established fluorescent fingerprint data (Fig. 4B, C and S6†). The identification of pathogens is an essential prerequisite for the rational and correct use of antibiotics. Benefiting from the guidance of the Microcolor system, the mice receiving symptomatic antimicrobial therapy displayed accelerated wound closures (65.4% of mice treated with antimicrobial therapy versus 18.1% of untreated mice). Because the entire process was carried out in a bacterial culture medium, the cellular components from the human body in the samples might not affect this method.
Bacteremia is a dangerous disease associated with severe mortality. Clinically, detection based on blood culture takes up to 2 days, and this may delay antimicrobial treatments.39S. aureus-mediated bacteremia models were established using mice, and blood samples were collected from mice with bacteremia and further analyzed by the Microcolor system. As shown in Fig. 4D, the Microcolor system accurately diagnosed S. aureus-mediated bacteremia in whole-blood samples within 12 h. As compared with the blood culture method, our method achieved a relatively low detection limit. Specifically, the Microcolor system detected bacteremia with 105 colony-forming units (CFU) of S. aureus per mouse. In sharp contrast, even at an injection dose of 107 CFU, the blood culture method still exhibited a false negative result (Fig. 4E). The RGB values detected by the Microcolor system in mouse bacteremia models were further normalized and analyzed, and there was no significant difference between mice infected with different bacterial concentrations (Fig. 4F and S7†). Through the above research, the Microcolor system was proven to detect bacteria in samples from living organisms, indicating that the Microcolor system possesses the potential to diagnose diseases that are accompanied by changes in bacterial fingerprints in a complex environment. With the capacity to innocuously label each species of bacteria in a complex environment, the Microcolor system might be able to identify unique microbiota consisting of different proportions of bacteria.
To verify whether the Microcolor system can diagnose diseases by identifying the structure of the microbiota, we implemented a proof-of-principle study to evaluate its clinical utility. Head and neck squamous cell carcinoma (HNSCC) is one of the most common cancer types worldwide. OSCC, as one of the key members of HNSCC, has been reported to have a close relationship with oral microbiota alterations.40,41 Indeed, bacteria including Capnocytophaga gingivalis, Prevotella melaninogenica, and Streptococcus mitis have been recognized as important markers for the early diagnosis of OSCC.9
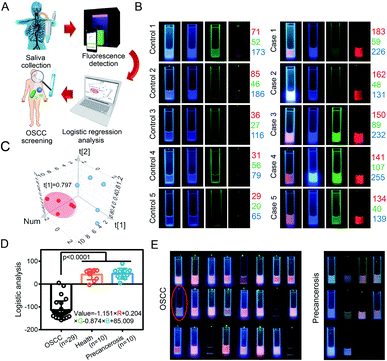
In this study, saliva samples from healthy volunteers (n = 10), OSCC patients (n = 29), and patients with precancerous lesions (n = 10) were collected and analyzed by the Microcolor system (Fig. 5A). Saliva samples from 5 healthy volunteers and 5 OSCC patients were initially used to establish the mathematical modeling (Fig. S8†). As shown in Fig. 5B, the fluorescence patterns between these two groups displayed obvious differences. Strong red fluorescence was found in saliva samples from all 5 OSCC patients. In contrast, enhanced green fluorescence was only observed in 2 OSCC cases. Overall, the comprehensive consideration of RGB values enabled the accurate identification of cancerous cases. The fluorescent fingerprints of saliva from healthy individuals varied from each other. However, OSCC cases showed a similar fluorescence pattern. This result was also confirmed by PCA analysis (Fig. 5C). Although it is obvious that saliva samples from healthy people and OSCC patients were significantly different, a quantitative criterion is still required for the diagnosis.
We further developed a logistic regression model that connected RGB values to the different types of cases.42 A regression equation for use with the Microcolor system that would optimally predict the risk of developing OSCC is listed in Fig. 5D. In this regression model, the more negative the result, the higher the probability of a OSCC diagnosis. Additionally, it was noted that red fluorescence accounted for a much larger proportion of the result than the other variables included in the model. This regression equation was used to predict the state of 24 other samples (Fig. 5E and S9†). By using a threshold value of 0, 23 of 24 patients were successfully identified as having OSCC, and this result led to a relatively high accuracy rate of 95.8%. Only one false negative result was obtained during this small-scale investigation.
To verify the effectiveness of the Microcolor system for OSCC prediction, the change in the oral microbiome between OSCC patients and healthy people was measured with polymerase chain reaction (PCR) technology. As shown in Fig. S10,† significantly increased Streptococcus and Prevotella, which are the important markers for OSCC, were observed in the oral microbiome of OSCC patients. This result indicated that the Microcolor system is an effective approach for OSCC diagnosis. Precancerous lesions at the early stage may be misdiagnosed as cancers before they progress to OSCC.43 To address this issue, we also tested whether this logistic regression model could distinguish between the saliva samples from patients with OSCC or precancerous lesions (Fig. S11†). As shown in Fig. 5D and E, most patient samples of precancerous lesions or benign tumors (10 of 10) displayed an emission color that was similar to that of healthy volunteers. In consistence with morphological observation, the logistic regression model also suggested that the lesions of these 10 patients were unlikely to be malignant tumors.
These results demonstrated that the Microcolor system is an effective approach for OSCC diagnosis. This method might be further developed so that it can provide valuable guidance for cancer screening.
Conclusions
In this study, we have proposed a method based on pattern recognition for diagnosing diseases that are accompanied by changes in the microbiota structure. Three bioorthogonal metabolites, B-Ala, G-Glu, and R-Gal, were synthesized. Based on the fact that metabolites are incorporated into bacteria through biosynthetic pathways, these metabolites are utilized differently by different types of bacteria. Thus, different bacteria will exhibit characteristic RGB fluorescence emission and produce a unique and distinguishable true color pattern. By combining this method with different detection methods, such as portable spectrometry, fluorescence spectrophotometry, flow cytometry, and fluorescence microscopy, there is the potential for this method to be flexibly used for various medical applications. It can be expected that the combination of the Microcolor system and other clinical technologies might lead to an update of the current methods for studying host–microbial interactions and disease diagnosis.Ethical statement
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the Animal Experiment Center of Wuhan University (Wuhan, China). All mouse experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People's Republic of China.Saliva samples were obtained from patients at the School and Hospital of Stomatology of Wuhan University. The School and Hospital of Stomatology of Wuhan University Medical Ethics Committee approved this study, and informed consent was obtained from the patients before they underwent surgery. The clinical stages of their HNSCC were classified according to the guidelines of the International Union Against Cancer (UICC 2002).
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2019YFA0905603) and the National Natural Science Foundation of China (51690152, 21721005 and 51833007).Notes and references
- K. Z. Coyte, J. Schluter and K. R. Foster, Science, 2015, 350, 663 CrossRef CAS PubMed.
- J. A. Gilbert, R. A. Quinn, J. Debelius, Z. Z. Xu, J. Morton, N. Garg, J. K. Jansson, P. C. Dorrestein and R. Knight, Nature, 2016, 535, 94 CrossRef CAS PubMed.
- S. V. Lynch and O. Pedersen, N. Engl. J. Med., 2016, 375, 2369–2379 CrossRef CAS PubMed.
- M. O. Din, T. Danino, A. Prindle, M. Skalak, J. Selimkhanov, K. Allen, E. Julio, E. Atolia, L. S. Tsimring, S. N. Bhatia and J. Hasty, Nature, 2016, 536, 81 CrossRef CAS PubMed.
- O. Felfoul, M. Mohammadi, S. Taherkhani, D. de Lanauze, Y. Zhong Xu, D. Loghin, S. Essa, S. Jancik, D. Houle, M. Lafleur, L. Gaboury, M. Tabrizian, N. Kaou, M. Atkin, T. Vuong, G. Batist, N. Beauchemin, D. Radzioch and S. Martel, Nat. Nanotechnol., 2016, 11, 941 CrossRef CAS PubMed.
- D.-W. Zheng, Y. Chen, Z.-H. Li, L. Xu, C.-X. Li, B. Li, J.-X. Fan, S.-X. Cheng and X.-Z. Zhang, Nat. Commun., 2018, 9, 1680 CrossRef PubMed.
- J.-X. Fan, Z.-H. Li, X.-H. Liu, D.-W. Zheng, Y. Chen and X.-Z. Zhang, Nano Lett., 2018, 18, 2373–2380 CrossRef CAS PubMed.
- D.-W. Zheng, X. Dong, P. Pan, K.-W. Chen, J.-X. Fan, S.-X. Cheng and X.-Z. Zhang, Nat. Biomed. Eng., 2019, 3, 717–728 CrossRef CAS PubMed.
- D. L. Mager, A. D. Haffajee, P. M. Devlin, C. M. Norris, M. R. Posner and J. M. Goodson, J. Transl. Med., 2005, 3, 27 CrossRef CAS PubMed.
- A. M. Thomas, P. Manghi, F. Asnicar, E. Pasolli, F. Armanini, M. Zolfo, F. Beghini, S. Manara, N. Karcher, C. Pozzi, S. Gandini, D. Serrano, S. Tarallo, A. Francavilla, G. Gallo, M. Trompetto, G. Ferrero, S. Mizutani, H. Shiroma, S. Shiba, T. Shibata, S. Yachida, T. Yamada, J. Wirbel, P. Schrotz-King, C. M. Ulrich, H. Brenner, M. Arumugam, P. Bork, G. Zeller, F. Cordero, E. Dias-Neto, J. C. Setubal, A. Tett, B. Pardini, M. Rescigno, L. Waldron, A. Naccarati and N. Segata, Nat. Med., 2019, 25, 667–678 CrossRef CAS PubMed.
- J. Wirbel, P. T. Pyl, E. Kartal, K. Zych, A. Kashani, A. Milanese, J. S. Fleck, A. Y. Voigt, A. Palleja, R. Ponnudurai, S. Sunagawa, L. P. Coelho, P. Schrotz-King, E. Vogtmann, N. Habermann, E. Niméus, A. M. Thomas, P. Manghi, S. Gandini, D. Serrano, S. Mizutani, H. Shiroma, S. Shiba, T. Shibata, S. Yachida, T. Yamada, L. Waldron, A. Naccarati, N. Segata, R. Sinha, C. M. Ulrich, H. Brenner, M. Arumugam, P. Bork and G. Zeller, Nat. Med., 2019, 25, 679–689 CrossRef CAS PubMed.
- M. S. Shah, T. Z. DeSantis, T. Weinmaier, P. J. McMurdie, J. L. Cope, A. Altrichter, J.-M. Yamal and E. B. Hollister, Gut, 2018, 67, 882 CrossRef CAS PubMed.
- H. J. Chung, C. M. Castro, H. Im, H. Lee and R. Weissleder, Nat. Nanotechnol., 2013, 8, 369 CrossRef CAS PubMed.
- S. Sauer and M. Kliem, Nat. Rev. Microbiol., 2010, 8, 74 CrossRef CAS PubMed.
- J. Kuczynski, C. L. Lauber, W. A. Walters, L. W. Parfrey, J. C. Clemente, D. Gevers and R. Knight, Nat. Rev. Genet., 2011, 13, 47 CrossRef PubMed.
- F. Natalio, R. Fuchs, S. R. Cohen, G. Leitus, G. Fritz-Popovski, O. Paris, M. Kappl and H.-J. Butt, Science, 2017, 357, 1118 CrossRef CAS PubMed.
- A. D. Radkov, Y.-P. Hsu, G. Booher and M. S. VanNieuwenhze, Annu. Rev. Biochem., 2018, 87, 991–1014 CrossRef CAS PubMed.
- H. Wang, R. Wang, K. Cai, H. He, Y. Liu, J. Yen, Z. Wang, M. Xu, Y. Sun, X. Zhou, Q. Yin, L. Tang, I. T. Dobrucki, L. W. Dobrucki, E. J. Chaney, S. A. Boppart, T. M. Fan, S. Lezmi, X. Chen, L. Yin and J. Cheng, Nat. Chem. Biol., 2017, 13, 415 CrossRef CAS PubMed.
- H. Lam, D.-C. Oh, F. Cava, C. N. Takacs, J. Clardy, M. A. de Pedro and M. K. Waldor, Science, 2009, 325, 1552 CrossRef CAS PubMed.
- D. K. Ranjit and K. D. Young, J. Bacteriol., 2013, 195, 2452 CrossRef CAS PubMed.
- E. Kuru, C. Lambert, J. Rittichier, R. Till, A. Ducret, A. Derouaux, J. Gray, J. Biboy, W. Vollmer, M. VanNieuwenhze, Y. V. Brun and R. E. Sockett, Nat. Microbiol., 2017, 2, 1648–1657 CrossRef CAS PubMed.
- Y.-P. Hsu, E. Hall, G. Booher, B. Murphy, A. D. Radkov, J. Yablonowski, C. Mulcahey, L. Alvarez, F. Cava, Y. V. Brun, E. Kuru and M. S. VanNieuwenhze, Nat. Chem., 2019, 11, 335–341 CrossRef CAS PubMed.
- C. Jiang, P. J. B. Brown, A. Ducret and Y. V. Brun, Nature, 2014, 506, 489 CrossRef CAS PubMed.
- G. W. Liechti, E. Kuru, E. Hall, A. Kalinda, Y. V. Brun, M. VanNieuwenhze and A. T. Maurelli, Nature, 2013, 506, 507 CrossRef PubMed.
- S. E. Pidgeon, J. M. Fura, W. Leon, M. Birabaharan, D. Vezenov and M. M. Pires, Angew. Chem., Int. Ed., 2015, 54, 6158–6162 CrossRef CAS PubMed.
- E. Kuru, H. V. Hughes, P. J. Brown, E. Hall, S. Tekkam, F. Cava, M. A. de Pedro, Y. V. Brun and M. S. VanNieuwenhze, Angew. Chem., Int. Ed., 2012, 51, 12519–12523 CrossRef CAS PubMed.
- W. Wang, L. Lin, Y. Du, Y. Song, X. Peng, X. Chen and C. J. Yang, Nat. Commun., 2019, 10, 1317 CrossRef PubMed.
- P. J. Calabretta, H. L. Hodges, M. B. Kraft, V. M. Marando and L. L. Kiessling, J. Am. Chem. Soc., 2019, 141, 9262–9272 CrossRef PubMed.
- R. Sadamoto, T. Matsubayashi, M. Shimizu, T. Ueda, S. Koshida, T. Koda and S.-I. Nishimura, Chem.–Eur. J., 2008, 14, 10192–10195 CrossRef CAS PubMed.
- K. E. DeMeester, H. Liang, M. R. Jensen, Z. S. Jones, E. A. D'Ambrosio, S. L. Scinto, J. Zhou and C. L. Grimes, J. Am. Chem. Soc., 2018, 140, 9458–9465 CrossRef CAS PubMed.
- A. Dumont, A. Malleron, M. Awwad, S. Dukan and B. Vauzeilles, Angew. Chem., Int. Ed., 2012, 51, 3143–3146 CrossRef CAS PubMed.
- J. E. Kwon, S. Park and S. Y. Park, J. Am. Chem. Soc., 2013, 135, 11239–11246 CrossRef CAS PubMed.
- S. R. Scott, M. O. Din, P. Bittihn, L. Xiong, L. S. Tsimring and J. Hasty, Nat. Microbiol., 2017, 2, 17083 CrossRef CAS PubMed.
- K. Pardee, S. Slomovic, P. Q. Nguyen, J. W. Lee, N. Donghia, D. Burrill, T. Ferrante, F. R. McSorley, Y. Furuta, A. Vernet, M. Lewandowski, C. N. Boddy, N. S. Joshi and J. J. Collins, Cell, 2016, 167, 248–259 CrossRef CAS PubMed.
- J. W. Lee, J.-S. Lee and Y.-T. Chang, Angew. Chem., Int. Ed., 2006, 45, 6485–6487 CrossRef CAS PubMed.
- P. N. A. Harris, P. A. Tambyah, D. C. Lye, Y. Mo, T. H. Lee, M. Yilmaz, T. H. Alenazi, Y. Arabi, M. Falcone, M. Bassetti, E. Righi, B. A. Rogers, S. Kanj, H. Bhally, J. Iredell, M. Mendelson, T. H. Boyles, D. Looke, S. Miyakis, G. Walls, M. Al Khamis, A. Zikri, A. Crowe, P. Ingram, N. Daneman, P. Griffin, E. Athan, P. Lorenc, P. Baker, L. Roberts, S. A. Beatson, A. Y. Peleg, T. Harris-Brown and D. L. Paterson, J. Am. Med. Assoc., 2018, 320, 984–994 CrossRef CAS PubMed.
- P. M. Colavite, L. L. W. Ishikawa, S. F. G. Zorzella-Pezavento, L. R. C. d. Oliveira, T. G. D. França, L. C. da Rosa, F. Chiuso-Minicucci, A. E. Vieira, C. F. Francisconi, M. d. L. R. d. S. da Cunha, G. P. Garlet and A. Sartori, Cell. Microbiol., 2016, 18, 998–1008 CrossRef CAS PubMed.
- C. Mao, Y. Xiang, X. Liu, Z. Cui, X. Yang, K. W. K. Yeung, H. Pan, X. Wang, P. K. Chu and S. Wu, ACS Nano, 2017, 11, 9010–9021 CrossRef CAS PubMed.
- A. Pechorsky, Y. Nitzan and T. Lazarovitch, J. Microbiol. Methods, 2009, 78, 325–330 CrossRef CAS PubMed.
- L. Rao, L.-L. Bu, L. Ma, W. Wang, H. Liu, D. Wan, J.-F. Liu, A. Li, S.-S. Guo, L. Zhang, W.-F. Zhang, X.-Z. Zhao, Z.-J. Sun and W. Liu, Angew. Chem., Int. Ed., 2018, 130, 998–1003 CrossRef.
- L. Mao, W. K. Hong and V. A. Papadimitrakopoulou, Cancer Cell, 2004, 5, 311–316 CrossRef CAS PubMed.
- K. G. Shah, V. Singh, P. C. Kauffman, K. Abe and P. Yager, Anal. Chem., 2018, 90, 6967–6974 CrossRef CAS PubMed.
- B. W. Neville and T. A. Day, Ca-Cancer J. Clin., 2002, 52, 195–215 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc01704c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |