Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
First-generation shaped gel reactors based on photo-patterned hybrid hydrogels†
Phillip R. A.
Chivers
,
Jamie A.
Kelly
,
Max J. S.
Hill
and
David K.
Smith
 *
*
Department of Chemistry, University of York, Heslington, York, YO10 5DD, UK. E-mail: david.smith@york.ac.uk
First published on 9th May 2020
Abstract
This paper reports the development of first-generation photo-patterned ring-shaped gel reactors that catalyse the hydrolysis of para-nitrophenol phosphate using a phosphatase enzyme. Encapsulation of alkaline phosphatase within a hybrid gel combining a low-molecular-weight gelator (LMWG) and a polymer gelator (PG) does not appear to inhibit its ability to turn over this reaction. The PG enables photo-patterning of the gel, facilitated by the pre-formed LMWG network which limits convection and diffusion during the patterning step. Furthermore, the rheologically weak, reversible LMWG network can be washed away, allowing straightforward release of the patterned gel reactor. By tailoring reactor design and reaction conditions, and changing the enzyme to acid phosphatase, the distribution of substrate and product between the gel and the different solution phases could be controlled. Although these first generation gel reactors have some limitations, in particular with the gel adsorbing significant amounts of substrate/product, a number of design criteria emerge with regard to choice of gelator, enzyme and assembly technique. These insights will inform the future development of this approach to reaction engineering.
Introduction
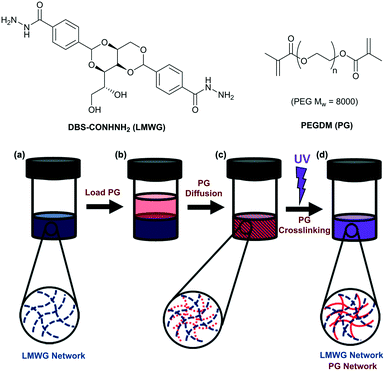
Compartmentalisation of reactive components is an effective strategy in biology to maintain the integrity of functional biomolecules.1 In a chemical setting, the separation of catalyst and product is a key factor in the design of efficient reaction processes.2 The drive towards sustainable synthesis has seen enzymatic processes gain increased importance.3 Enzyme immobilisation is a common strategy in membrane bio-reactors.4 More recently, there has been increasing interest in incorporating enzymes in gels that self-assemble from low-molecular-weight gelators (LMWGs) via non-covalent interactions.5 Such gels are suitable for catalysis as a result of their excellent solvent compatibility and the ability of small molecules to diffuse through the gel network, with larger molecules being entrapped.6 In pioneering work, Xu and co-workers demonstrated enhanced oxidative activity of haemoglobin when physically encapsulated in an amino acid-based LMWG hydrogel.7 Indeed, a number of enzymes and biological units retain their activity within a LMWG gel matrix.8 This class of bioactive material has potential as reaction additives, biosensors, separation matrices and therapeutic systems.9 Despite progress, the mechanical weakness of most self-assembled gels may limit use as sensing/reactor devices, because spatial control and patterning is important.10 In this regard, there has been interest in using polymer gels to create smart bioreactors,11 but this work has not extended to LMWGs.Combining LMWG hydrogels with a mechanically robust polymer gelator (PG) is an emerging strategy to overcome the poor mechanical properties of LMWG gels.12 Through careful choice of the PG, spatially resolved hybrid gels can be formed using techniques such as photo-patterning,13 3D-printing,14 wet-stamping15 or controlled diffusion.16 Enzymes have also been used to mediate hybrid gel assembly.17 We have reported hybrid gels that combine a PG with an LMWG based on 1,3;2,4-dibenzylidene-D-sorbitol (DBS).18 Using poly(ethylene glycol)dimethacrylate (PEGDM) with either DBS-COOH19 or DBS-CONHNH2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 enhanced the mechanical properties, whilst the unique properties of the LMWG network were retained.13 UV-initiated formation of the PEGDM network with a photomask gives spatial resolution.13 In this study, we aimed to combine enzymes with this class of hybrid gels, and explore different new strategies for the development of gel-based shaped bioreactors. We reason the soft biocompatible nature of hydrogel materials, combined with the ability to introduce patterning, makes them well-suited to this type of application.
20 enhanced the mechanical properties, whilst the unique properties of the LMWG network were retained.13 UV-initiated formation of the PEGDM network with a photomask gives spatial resolution.13 In this study, we aimed to combine enzymes with this class of hybrid gels, and explore different new strategies for the development of gel-based shaped bioreactors. We reason the soft biocompatible nature of hydrogel materials, combined with the ability to introduce patterning, makes them well-suited to this type of application.
Results and discussion
Synthesis of gelators and gels
The synthesis of DBS-CONHNH2 (Fig. 1) was performed as described previously.13 PEG8000 was used for the synthesis of PEGDM as reported previously.20 Gels of DBS-CONHNH2 (6–8 mM, 0.285–0.38% wt/vol) were prepared by sonication followed by a heat/cool cycle. Gels based on PEGDM, were prepared at concentrations between 3 and 60% wt/vol via irradiation with long-wavelength UV in the presence of a photoinitiator (PI, Irgacure 2959, 0.05% wt/vol). Hybrid LMWG/PG gels were prepared by gelling a solution of DBS-CONHNH2 (0.5 mL, 6 mM, 0.285% wt/vol), then loading a solution (0.5 mL) of PEGDM (known wt/vol) and PI (0.05% wt/vol) on top, and standing for three days to allow diffusion of PEGDM and PI into the gel. The PG was then polymerised by application of long-wavelength UV to yield the dual-network hybrid gel. Characterisation of this hybrid LMWG/PG gel has previously been reported by us.13Diffusion in the gels
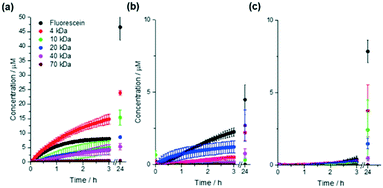
Ideally enzyme reactors should retain larger molecules such as enzymes while allowing diffusion of smaller reagents and products, and we therefore initially assessed gel permeability. Gels (0.5 mL, height 0.5 cm) were prepared in fluorescence cuvettes and 0.5 mL of a 50 μM solution containing fluorescein or a fluorescein isothiocyanate–dextran (FITC–dextran) of known molar mass was pipetted on top. The fluorescence intensity at the base of the gel was recorded over time (Fig. 2), allowing quantification of diffusion through the gel (Table 1). Initial diffusion through DBS-CONHNH2 was faster than through the hybrid LMWG/PG gel, which in turn was faster than through PEGDM PG hydrogels. Diffusion of FITC–dextrans through DBS-CONHNH2 was, as would be expected, dependent on size, with 4 kDa being most mobile, and 70 kDa being fully immobile. It was perhaps surprising that there is greater diffusion through the hybrid PG/LMWG gel than 10% PEGDM, as it may have been expected that with two networks present, there should be a less porous gel and therefore less diffusion than when using the PG alone. This observation therefore suggests that the presence of the self-assembled LMWG network may have somewhat limited the crosslinking of the PG network during gel fabrication, hence meaning the network remains somewhat more porous. However, it should also be noted that in the hybrid gel, although initial diffusion was faster, the total amount that diffused to the bottom of the vial over 24 hours was somewhat less, suggesting different rates of diffusion over time, and perhaps suggesting a degree of direct interaction between the diffusing species and the gel networks (see below). Indeed there was some evidence for example that fluorescein interacted somewhat with DBS-CONHNH2 (Fig. 2a) leading to an unexpectedly lower rate of diffusion nonetheless, the key observation from these studies was that there was effective diffusion of low-molecular-weight fluorescein through the hybrid gel and near total exclusion of 70 kDa FITC–dextran, suggesting that the hybrid gel was suitable for further study as it could limit the mobility of enzymes whilst allowing the mobility of smaller molecules.| Dye | Initial diffusion rate μM cm−1 s−1 (×10−3) | Concentration at base of sample after 24 h, μM | ||||
|---|---|---|---|---|---|---|
| LMWG | Hybrid | PG | LMWG | Hybrid | PG | |
| a It is not clear why the data for this system is an outlier, it is possible that the gels were damaged during formation leading to more diffusion than expected based on molecular weight. | ||||||
| Fluorescein | 168 | 28 | 10 | 47 | 4.5 | 7.8 |
| FITC-4 kDa | 202 | 6.8 | 4.3 | 24 | 2.2 | 3.8 |
| FITC-10 kDa | 64 | 1.4 | 2.8 | 15 | 0.3 | 2.4 |
| FITC-20 kDa | 56 | 25a | 3.3 | 8.6 | 2.7a | 1.5 |
| FITC-40 kDa | 56 | 2.5 | 0.7 | 5.3 | 0.8 | 0.5 |
| FITC-70 kDa | 5.4 | 0.3 | 0.1 | 0.5 | 0.02 | 0.04 |
Incorporation of enzyme
We chose to use alkaline phosphatase (ALP), a homodimeric metalloenzyme with molecular weight 140 kDa. ALP catalyses the hydrolysis of para-nitrophenol phosphate (pNPP) into para-nitrophenol (pNP). At pH 9 (ALP's optimum pH), the product is deprotonated, affording a bright yellow colour (λmax = 405 nm), in contrast with the starting material (λmax = 315 nm) (Fig. S1–S6†). Given that ALP will not be able to diffuse freely through these gels, it was necessary to introduce the enzyme during initial assembly of the LMWG network. DBS-CONHNH2 (6 mM) was suspended in a 0.2 U mL−1 solution of ALP (0.5 mL) and gel formation initiated via a heat/cool cycle as previously described. After cooling, the surface of the gel was washed with pH 9 buffer. A buffered solution of pNPP was then pipetted on top of the gel, but no colour change was observed over 24 h, indicating total enzyme denaturation during the heat–cool cycle (Fig. S7†). To preserve enzyme activity, ALP was thus introduced only after cooling of the hot DBS-CONHNH2 solution to 80 °C (just above the Tgel). After ALP addition, the solution was immediately cooled to 0 °C, resulting in very rapid gel assembly. The development of a bright yellow colour in both gel and solution on treatment with pNPP suggested ALP retained its catalytic activity (Fig. S8†).The compatibility of ALP with UV-initiated PEGDM crosslinking was then studied. ALP showed no apparent sensitivity to UV light (0.5 h exposure), but when irradiated in a solution of the PI, total loss of activity was observed. Interestingly, however, when the same ratio of enzyme and PI was exposed to UV light in the presence of PEGDM, ALP activity was preserved (Fig. S9†). The presence of the PG appears to prevent significant radical-induced inactivation of the enzyme.
We then confirmed that pNPP hydrolysis was catalysed by ALP encapsulated in the gel, rather than enzyme leaching into the solution phase. ALP-loaded DBS-CONHNH2, PEGDM and hybrid gels were prepared as described above and pH 9 buffer pipetted on top. This supernatant was removed after 24 h and diluted with pNPP solution. The rate of hydrolysis was determined by UV-vis spectroscopy and very little active ALP (<1%) was found to be present. No enzyme release was seen on subsequent washes, confirming that the active ALP is gel-encapsulated. This fabrication procedure, whilst less mild than many in the literature,8 is nonetheless appropriate for encapsulation of active enzymes such as ALP within the hybrid gel matrix. We note that less thermally-stable enzymes may not survive this treatment, however, increasingly there is interest in the use of extremophile enzymes in the field of biocatalysis to ensure catalytic performance can survive more extreme conditions.21
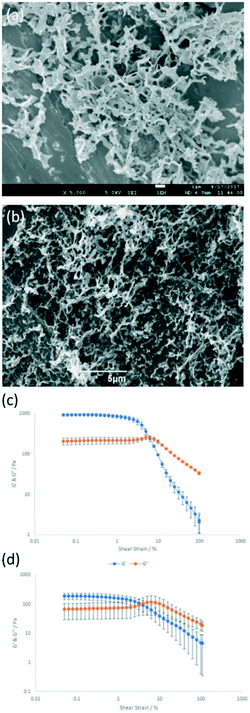
SEM and TEM images of freeze-dried gels were similar whether or not ALP was present (Fig. 3a and b, Fig. S10–S12†), with characteristic fibrous and sheet morphologies seen for DBS-CONHNH2 and PEGDM networks respectively (Fig. S10–S12†). No change in thermal stability (Tgel >100 °C for all samples) was observed in the presence of ALP. IR spectra of xerogels were not altered by the presence of ALP (Fig. S19–S22†), suggesting enzyme immobilisation is due to physical encapsulation and not gel network binding. Although there were no apparent differences in the rheology of either DBS–CONHNH2 or PEGDM hydrogels that incorporated ALP, the mechanical strength of the enzyme-loaded hybrid gel decreased from a G′ of ca. 1000 Pa to ca. 200 Pa in the presence of ALP (Fig. 3c and d and Fig. S13–S18†). We suggest subtle differences in the ALP-loaded DBS–CONHNH2 gel, possibly caused by rapid cooling or the presence of ALP, alter PEGDM uptake or crosslinking, impacting on gel stiffness. Nonetheless, ALP was immobilised and the hybrid gels were reasonably robust.
Ring-shaped reactor with enzyme in ring
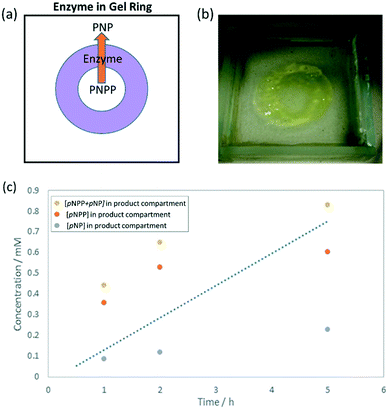
To make use of the DBS-CONHNH2/PEGDM hybrid gel and catalyse pNPP hydrolysis, we then explored shaping the ALP-loaded gel. A ring-shaped ‘reactor’ was designed (Fig. 4a) where the inner compartment would be loaded with substrate, which must diffuse through the enzyme-loaded ring to progress to the outer compartment. It was reasoned that the larger volume of the outer compartment would help drive diffusion of substrate through the reactive gel via a concentration gradient, and that the enzyme should convert pNPP to pNP during diffusion.The ring-shaped hybrid gel was fabricated by adapting our previous method.13 After adding ALP to the DBS-CONHNH2 sol at 80 °C, the mixture was transferred to a glass mould (5 × 5 × 1 cm) and immediately cooled to 0 °C, resulting in rapid formation of an ALP-loaded LMWG hydrogel. An aqueous solution (10 mL) of PEGDM (10% wt/vol) and PI (0.05% wt/vol) was pipetted on top and left for 3 days, after which time the supernatant was removed. A laser-printed photomask was fixed over the mould and the sample exposed to UV irradiation for 30 min. A ring (0.8 cm diameter) of robust hybrid PG/LMWG gel formed. The presence of the pre-assembled LMWG enhances the resolution of this patterning step.13 The surrounding pre-assembled soft and reversible LMWG matrix has the advantage that it can be easily washed away using a gentle stream of water. In this way, the LMWG reversibility facilitates the release of the patterned gel – a key advantage of this hybrid gel approach.
The reagent, pNPP (10 mM, 0.3 mL), was loaded into the inner compartment and pH 9 buffer (2 mL) into the outer compartment (Fig. 4a). At known time-points the contents of the outer compartment were removed and the UV-vis spectrum recorded to determine [pNP] and [pNPP] (Fig. 4c). Surprisingly, a greater proportion of pNPP (0.6 mM) was present in the outer compartment than hydrolysed pNP product (0.2 mM). A similar substrate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) product ratio was found in the inner compartment. However, a yellow colour developed in the hybrid gel ring during the reaction (Fig. 4b), suggesting conversion was taking place, but the product remained in the gel after reaction. Indeed, ca. 56% of total substrate/product remained in the gel after 5 h.
product ratio was found in the inner compartment. However, a yellow colour developed in the hybrid gel ring during the reaction (Fig. 4b), suggesting conversion was taking place, but the product remained in the gel after reaction. Indeed, ca. 56% of total substrate/product remained in the gel after 5 h.
Ring-shaped reactor with enzyme in outer compartment
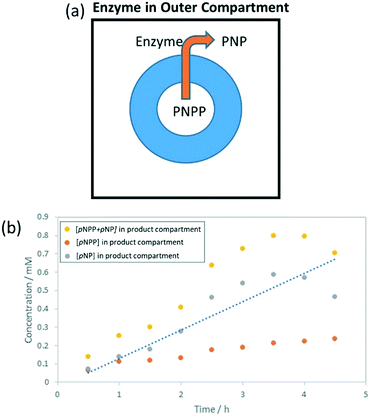
To further investigate this, a ring-shaped hybrid gel was again prepared, but in this case ALP (26 U mL−1) was loaded into the outer compartment to give a reactor with a different design (Fig. 5a) – diffusion of pNPP through the gel is required before it comes into contact with the enzyme. Again, the gel ring took on a significant yellow colour over the course of the reaction (Fig. S23†); ca. 75% of the pNPP/pNP was in the gel after 5 h. More pNP and significantly less pNPP was seen in the outer compartment compared to when ALP was in the gel ring (Fig. 5b). This suggests that pNPP reaching the outer compartment is rapidly turned over by ALP, and that this is more effective than the requirement for pNPP to contact the enzyme during diffusion through the gel ring. After 3.5 h, the concentration of pNP in the outer compartment decreased, which we propose is the result of the pNP product partitioning into the gel; supported by the gel ring taking on a bright yellow colour. To test this hypothesis, the uptake of pNPP and pNP by the hybrid gel was studied (Fig. S24†). Under alkaline conditions, it was found that pNP is preferentially absorbed into these hybrid gels. Control experiments indicate DBS-CONHNH2 is responsible.To understand this further, we recorded a 1H NMR spectrum of a DBS-CONHNH2 hydrogel in the presence of pNP (10 mM). It was found that pNP is mobile within the gel, suggesting its uptake is not the result of specific interactions with the solid-like gel nanofibres (Fig. S25†). The reasons for partitioning of pNP into the gel are therefore not clearly understood, but this effect is likely due to polarity, and not unique to this gel. In previous reports of reactive matrices, similar yellowing of gels has been reported.8d Substrate/product retention is an important but seldom discussed factor in gel bioreactor design.
Using our data on the gel uptake of pNP and/or pNPP (Fig. S24†), the total conversions of the ring-shaped enzyme bioreactors with ALP either in the gel ring or the outer compartment could be calculated as ca. 67% and 98% respectively (Table 2). These conversions are much higher than those reported from a similar gel-immobilised ALP (ca. 3%), despite using a lower concentration of enzyme (0.4 U mL−1 compared to 90 U mL−1).8d However, the majority of pNP product (>50%) is being partitioned into the gel, and this clearly requires improvement in designing a second generation of gel reactor (see Conclusions and Outlook section).
| Experiment | Compartment | pNPP (×10−8) mol | pNP (×10−8) mol | pNPP + pNP (×10−8) mol | % total in sol | % pNP in sol |
|---|---|---|---|---|---|---|
| ALP in gel ring | Inner | 38 | 10 | 131 | 44% | 11% |
| Outer | 60 | 23 | ||||
| ALP in outer compartment | Inner | 2.3 | 39 | 76 | 25% | 23% |
| Outer | 4.4 | 30 | ||||
| AP in outer compartment | Inner | 12 | 25 | 203 | 68% | 26% |
| Outer | 114 | 52 |
Use of acid phosphatase
In a preliminary additional experiment, we aimed to enhance reactor efficiency by altering the reaction conditions. Relatively low uptake of pNP into the hybrid gels was observed under acidic conditions (Fig. S25†). We therefore reasoned that lowering the pH would limit gel uptake of the product. However, ALP is denatured under acidic conditions, but a similar enzyme, acid phosphatase (AP), can catalyse this reaction at lower pH values. AP (26 U mL−1) was therefore loaded into the outer compartment. Greater quantities of pNPP substrate and pNP product were seen in the outer compartment, and it was calculated that in this experiment, >50% of the total substrate/product remained in solution rather than partitioning into the gel (Table 2), a marked improvement over the ALP reactor in which >50% ended up in the gel. These observations suggest that modifying reaction conditions can address product partitioning issues.Conclusions
In summary, we report the development of first-generation photo-patterned phosphatase bioreactors which catalyse the hydrolysis of pNPP. Encapsulation of the enzyme within the gel does not appear to inhibit its ability to turn over this reaction. By tailoring reactor design and reaction conditions, the distribution of substrate and product between the gel and solution phases could be changed. In this system, the PG plays a key role in enabling the shaping and patterning of the gel, facilitated by the pre-formed LMWG network which limits convection and diffusion during the patterning step. Furthermore, the rheologically weak and reversible nature of the LMWG network enables this part of the multi-domain material to be easily washed away releasing the patterned gel reactor.Although these first generation gel reactors have some limitations, a number of key design criteria have emerged from this study that will help inform the future evolution of this innovative concept.
1. It is important to use hybrid gels that allow rapid diffusion of small molecules and prevent the diffusion of the biocatalyst. This was achieved in the current design.
2. The gel should not favourably interact with or partition substrate or product. Indeed, it would be desirable for the gel to specifically disfavour interaction with the product. In this study, we demonstrated that pH could influence these factors but did not fully solve the problems for this system. Ideally gelators should be chosen that are non-interactive – DBS-CONHNH2 may not be the best gelator in this regard given its ability to interact with different species.20
3. The gel must be formed using conditions that are compatible with the biocatalyst. In this case, using a thermally-triggered gelator, we chose to achieve this by using relatively thermally-stable enzymes AP and ALP. This approach may also be suitable for extremophile enzymes of increasing importance in biocatalysis.21 To incorporate more thermally-sensitive enzymes, it would be necessary to use an LMWG such as DBS-COOH that could be triggered using a different stimulus, (e.g. pH change).19
Future research in our labs will therefore focus on identifying optimal combinations of LMWG, PG, enzyme and reaction conditions to increase isolable product yields, and encourage greater accumulation of product in a single reactor compartment. We also suggest that in the future, the LMWG network embedded within the reactor may also play an active role in catalysis, either by being catalytically-active in its own right or by binding to catalytically active units.6
This paper represents a first innovative step in the development of 2D and 3D flow bioreactors based on photo-patterned hybrid gels. This is a key target, and is a potentially transformative reaction engineering technology.22
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge financial support from EPSRC and University of York through the DTA funding scheme (PRAC).Notes and references
- P. L. Urban, New J. Chem., 2014, 38, 5135–5141 RSC.
- D. J. Cole-Hamilton, Science, 2003, 299, 1702–1706 CrossRef CAS PubMed.
- (a) J. M. Woodley, Trends Biotechnol., 2008, 26, 321–327 CrossRef CAS PubMed; (b) P. Fernandes, Enzyme Res., 2010, 2010, 862537 Search PubMed.
- (a) G. M. Rios, M. P. Belleville, D. Paolucci and J. Sanchez, J. Membr. Sci., 2004, 242, 189–196 CrossRef CAS; (b) E. P. Cipolatti, A. Valério, R. O. Henriques, D. E. Moritz, J. L. Ninow, D. M. G. Freire, E. A. Manoel, R. Fernandez-Lafuente, D. de Oliveira and E. Csöregi, RSC Adv., 2016, 6, 104675–104692 RSC.
- (a) R. G. Weiss, J. Am. Chem. Soc., 2014, 136, 7519–7530 CrossRef CAS PubMed; (b) E. R. Draper and D. J. Adams, Chem, 2017, 3, 390–410 CrossRef CAS.
- (a) B. Escuder, F. Rodríguez-Llansola and J. F. Miravet, New J. Chem., 2010, 34, 1044–1054 RSC; (b) D. Díaz-Díaz, D. Kühbeck and R. J. Koopmans, Chem. Soc. Rev., 2011, 40, 427–448 RSC; (c) W. Fang, Y. Zhang, J. Wu, C. Liu, H. Zhu and T. Tu, Chem. – Asian J., 2018, 13, 712–729 CrossRef CAS PubMed.
- Q. Wang, Z. Yang, L. Wang, M. Ma and B. Xu, Chem. Commun., 2007, 1032–1034 RSC.
- (a) Q. Wang, Z. Yang, X. Zhang, X. Xiao, C. K. Chang and B. Xu, Angew. Chem., Int. Ed., 2007, 46, 4285–4289 CrossRef CAS PubMed; (b) Q. Wang, Z. Yang, Y. Gao, W. Ge, L. Wang and B. Xu, Soft Matter, 2008, 4, 550–553 RSC; (c) P. W. J. M. Frederix, R. Kania, J. A. Wright, D. A. Lamprou, R. V. Ulijn, C. J. Pickett and N. T. Hunt, Dalton Trans., 2012, 41, 13112–13119 RSC; (d) J. Wang, X. Miao, Q. Fengzhao, C. Ren, Z. Yang and L. Wang, RSC Adv., 2013, 3, 16739–16746 RSC; (e) C. Hickling, H. S. Toogood, A. Saiani, N. S. Scrutton and A. F. Miller, Macromol. Rapid Commun., 2014, 35, 868–874 CrossRef CAS PubMed; (f) Y. Mao, T. Su, Q. Wu, C. Liao and Q. Wang, Chem. Commun., 2014, 50, 14429–14432 RSC; (g) L. A. Solomon, J. B. Kronenberg and H. C. Fry, J. Am. Chem. Soc., 2017, 139, 8497–8507 CrossRef CAS PubMed; (h) Y. Wang, Y. Zhang, M. Wang, Y. Zhao, W. Qi, R. Su and Z. He, RSC Adv., 2018, 8, 6047–6052 RSC; (i) N. Falcone, T. Shao, R. Rashid and H.-B. Kraatz, Molecules, 2019, 24, 2884 CrossRef CAS PubMed.
- (a) S. Kiyonaka, K. Sad, I. Yoshimura, S. Shinkai, N. Kato and I. Hamachi, Nat. Mater., 2004, 3, 58–64 CrossRef CAS PubMed; (b) S. Mizrahi, D. Rizkov, N. Hayat and O. Lev, Chem. Commun., 2008, 2914–2916 RSC; (c) J. H. Kim, S. Y. Lim, D. H. Nam, J. Ryu, S. H. Ku and C. B. Park, Biosens. Bioelectron., 2011, 26, 1860–1865 CrossRef CAS PubMed; (d) H. Shigemitsu, T. Fujisaku, S. Onogi, T. Yoshii, M. Ikeda and I. Hamachi, Nat. Protoc., 2016, 11, 1744–1756 CrossRef CAS PubMed; (e) Q. Wei, S. Jiang, R. Zhu, X. Wang, S. Wang and Q. Wang, iScience, 2019, 14, 27–35 CrossRef CAS PubMed.
- P. R. A. Chivers and D. K. Smith, Nat. Rev. Mater., 2019, 4, 463–478 CrossRef CAS.
- (a) V. N. Luk, L. K. Fiddes, V. M. Luk, E. Kumacheva and A. R. Wheeler, Proteomics, 2012, 12, 1310–1318 CrossRef CAS PubMed; (b) L. K. Fiddes, V. N. Luk, S. H. Au, A. H. C. Ng, V. Luk, E. Kumacheva and A. R. Wheeler, Biomicrofluidics, 2012, 6, 014112 CrossRef PubMed; (c) Z. Yi, Y. Zhang, S. Kootala, J. Hilborn and D. A. Ossipov, ACS Appl. Mater. Interfaces, 2015, 7, 1194–1206 CrossRef CAS PubMed; (d) M. Maier, C. P. Radtke, J. Hubbuch, C. M. Niemeyer and K. S. Rabe, Angew. Chem., Int. Ed., 2018, 57, 5539–5543 CrossRef CAS PubMed; (e) D. Simon, F. Obst, S. Haefner, T. Heroldt, M. Peiter, F. Simon, A. Richter, B. Voit and D. Appelhans, React. Chem. Eng., 2019, 4, 67–77 RSC; (f) F. Obst, D. Simon, P. J. Mehner, J. W. Neubauer, A. Beck, O. Stroyuk, A. Richter, B. Voit and D. Appelhans, React. Chem. Eng., 2019, 4, 2141–2155 RSC.
- D. J. Cornwell and D. K. Smith, Mater. Horiz., 2015, 2, 279–293 RSC.
- (a) D. J. Cornwell, B. O. Okesola and D. K. Smith, Angew. Chem., Int. Ed., 2014, 53, 12461–12465 CAS; (b) P. R. A. Chivers and D. K. Smith, Chem. Sci., 2017, 8, 7218–7227 RSC.
- Q. Wei, M. Xu, C. Liao, Q. Wu, M. Liu, Y. Zhang, C. Wu, L. Cheng and Q. Wang, Chem. Sci., 2016, 7, 2748–2752 RSC.
- M. Lovrak, W. E. J. Hendriksen, C. Maity, S. Mytnyk, V. van Steijn, R. Eelkema and J. H. van Esch, Nat. Commun., 2017, 8, 15317 CrossRef CAS PubMed.
- C. C. Piras, P. Slavik and D. K. Smith, Angew. Chem., Int. Ed., 2020, 59, 853–859 CrossRef CAS PubMed.
- Q. Wei, Y. Chang, G. Ma, W. Zhang, Q. Wang and Z. Hu, J. Mater. Chem. B, 2019, 7, 6195–6201 RSC.
- B. O. Okesola, V. M. P. Vieira, D. J. Cornwell, N. K. Whitelaw and D. K. Smith, Soft Matter, 2015, 11, 4768–4787 RSC.
- D. J. Cornwell, B. O. Okesola and D. K. Smith, Soft Matter, 2013, 9, 8730–8736 RSC.
- (a) B. O. Okesola and D. K. Smith, Chem. Commun., 2013, 49, 11164–11166 RSC; (b) E. J. Howe, B. O. Okesola and D. K. Smith, Chem. Commun., 2015, 51, 7451–7454 RSC.
- (a) L. Kumar, G. Awasthi and B. Singh, Biotechnol., 2011, 10, 121–135 CrossRef CAS; (b) J. A. Littlechild, Front. Bioeng. Biotechnol., 2015, 3, 161 Search PubMed; (c) M. A. Cabrera and J. M. Blamey, Biol. Res., 2018, 51, 37 CrossRef CAS PubMed.
- (a) E. Peris, O. Okafor, E. Kulcinskaja, R. Goodridge, S. V. Luis, E. Garcia-Verdugo, E. O'Reilly and V. Sans, Green Chem., 2017, 19, 5345–5349 RSC; (b) L. Tamborini, P. Fernandes, F. Paradisi and F. Molinari, Trends Biotechnol., 2017, 36, 73–88 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Including materials, methods, characterisation and assay data. See DOI: 10.1039/d0re00109k |
| This journal is © The Royal Society of Chemistry 2020 |