Open Access Article
Open Access ArticleSynthesis, study of antileishmanial and antitrypanosomal activity of imidazo pyridine fused triazole analogues†
Adinarayana Nandikolla a,
Singireddi Srinivasaraoa,
Banoth Karan Kumar
a,
Singireddi Srinivasaraoa,
Banoth Karan Kumar b,
Sankaranarayanan Murugesanb,
Himanshu Aggarwal
b,
Sankaranarayanan Murugesanb,
Himanshu Aggarwal a,
Louise L. Majorc,
Terry K. Smithc and
Kondapalli Venkata Gowri Chandra Sekhar
a,
Louise L. Majorc,
Terry K. Smithc and
Kondapalli Venkata Gowri Chandra Sekhar *a
*a
aDepartment of Chemistry, Birla Institute of Technology and Science, Pilani, Hyderabad Campus, Jawahar Nagar, Kapra Mandal, Hyderabad – 500078, Telangana, India. E-mail: kvgc@hyderabad.bits-pilani.ac.in; kvgcs.bits@gmail.com; Tel: +91 40 66303527
bMedicinal Chemistry Research Laboratory, Department of Pharmacy, Birla Institute of Technology and Science Pilani, Pilani Campus, Pilani-333031, Rajasthan, India
cSchools of Biology & Chemistry, BSRC, The University, St. Andrews, Fife, Scotland KY16 9ST, UK
First published on 19th October 2020
Abstract
Four groups, thirty-five compounds in total, of novel 1,2,3-triazole analogues of imidazo-[1,2-a]-pyridine-3-carboxamides were designed and synthesized using substituted pyridine, propargyl bromide, 2-azidoethyl 4-methyl benzenesulfonate and substituted acetylenes. These compounds were characterized using 1H NMR, 13C NMR, LCMS and elemental analyses and a crystal structure was obtained for one of the significantly active compounds, 8f. All the synthesized and characterized compounds were screened in vitro for antileishmanial and antitrypanosomal activity against Leishmania major and Trypanosoma brucei parasites, respectively. Among the tested analogues, five compounds (8d, 8f, 8j, 10b and 10d) exhibited significant antileishmanial activity while three compounds (10b, 11a and 11b) showed substantial activity against T. brucei parasite. In silico ADME prediction studies depicted that the essential compounds obeyed Lipinski's rule of five. The predicted in silico toxicity profile suggested that the tested compounds would be non-toxic, which was confirmed experimentally by the lack of cytotoxicity against HeLa cells. Finally, a molecular docking study was also performed, for 10d the most active antileishmanial compound, to study its putative binding pattern at the active site of the selected leishmanial trypanothione reductase target.
1. Introduction
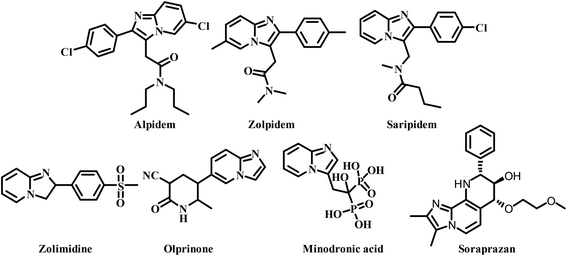
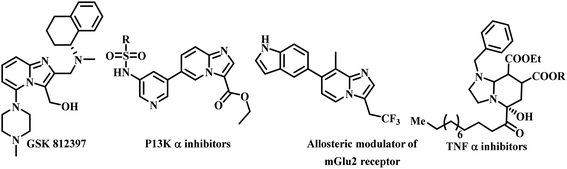
Leishmaniasis is one of the most neglected tropical diseases as identified by the WHO.1 Over twenty species of protozoan parasites cause the disease and it is transmitted to its mammalian host via the bite of ninety different sandfly species. Mostly leishmaniasis affects the developing countries and underprivileged people across the globe. Leishmaniasis was discovered by William Leishman and Charles Donovan in the year 1900 and was observed in the spleen of the patient.2 If a person is infected with Leishmania, treatment is not easy and if it affects immunocompromised patients co-infected with HIV, then the treatment becomes much harder and risky. Leishmania is categorized into four classes: (a) cutaneous leishmaniasis (CL), (b) mucocutaneous leishmaniasis (ML), (c) visercal leishmaniasis (VL) and (d) post-kala-azar dermal leishmaniasis (PKDL).3 The current drugs used for treating leishmaniasis include sodium stibogluconate, itraconazole, paromomycin, miltefosine, amphotericin B, pentamidine, sitamaquine and rifampicin.4 Most of these drugs were discovered in the middle of the last century and use of them is constantly leading to problematic resistance in several Leishmania species. A recent pilot study conducted in India with thirty-eight patients, proved the efficacy of the miltefosine–paromomycin combination in treating PKDL, whose current treatment is lengthy, costly, time consuming and associated with a variety of side-effects.5 Human African trypanosomiasis also known as “sleeping sickness” is a vector-borne disease mainly caused by infection with protozoan parasites belonging to the genus Trypanosoma either Trypanosoma brucei gambiense or Trypanosoma brucei rhodesiense. Out of this, Trypanosoma brucei gambiense is found in twenty-four countries in west and central Africa. This form, currently accounts for 98% of reported cases of sleeping sickness and causes a chronic infection. A person can be infected for months or even years without major signs or symptoms of the disease. Unlike the above, Trypanosoma brucei rhodesiense is found in thirteen countries in eastern and southern Africa. Nowadays, this form represents under 2% of reported cases and causes an acute infection. First signs and symptoms are observed within few months or weeks after infection. In general, it involves two stages, in the first stage (also called haemo-lymphatic stage), the trypanosomes multiply in subcutaneous tissues, blood and lymph. This entails bouts of fever, headaches, enlarged lymph nodes, joint pains and itching. In the second stage (known as the neurological or meningo-encephalic stage), the parasites cross the blood–brain barrier to infect the central nervous system. This is where more obvious signs and symptoms of the disease appear: changes of behaviour, confusion, sensory disturbances and poor coordination as well as disturbance of the sleep cycle. They are transmitted to humans by tsetse fly (Glossina genus) bites. It is endemic in thirty-seven sub-Saharan African countries where it has threatened millions of people. Without treatment, the disease is considered fatal as tsetse flies transmit the disease. Since the number of new human African trypanosomiasis cases reported between 2000 and 2018 dropped by 95%, the WHO neglected tropical diseases road map targeted its elimination as a public health problem by 2020 and eradication by 2030.6The imidazo-[1,2-a]-pyridine moiety already exists in several pharmacologically significant molecules. This core moiety is found in many pharmaceutical compounds that display benzodiazepine receptor agonism, antifungal, antitumor, antiviral, antibacterial, analgesic, and anti-HIV properties.7–9 The imidazo-[1,2-a]-pyridine core is present in some established drugs (Fig. 1), such as alpidem (anxiolytic),10 zolpidem (hypnotic),11 saripidem (sedative and anxiolytic),12 zolimidine (anti-ulcer),13 olprinone (to treat acute heart failure),14 minodronic acid (to treat anxiety, heart failure, osteoporosis),15 and Soraprazan (anti-ulcer).16 It is also present in some biologically active derivatives such as GSK812397 (CXCR4 antagonist),17 PI3K-α inhibitors,18 positive allosteric modulators of mGlu2 receptors,19 TNF-α inhibitors (Fig. 2).20
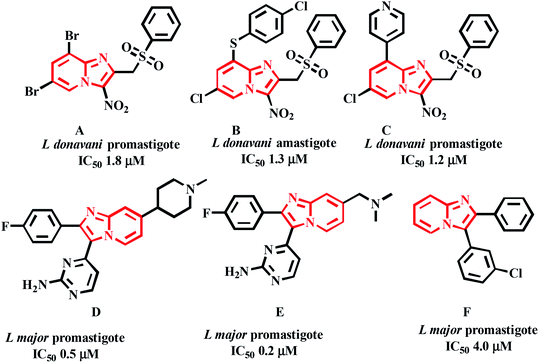
Castera-Ducrosteam reported imidazo-[1,2-a]-pyridine based analogues as antileishmanial agents. From this work, forty-four compounds are tested for Leishmania donovani promastigotes. Four selective hit compounds exhibited IC50 values 12.6, 18.2, 14.0 μM, and compound A (Fig. 3) with an IC50 1.8 μM, was the most active one of the series.21 Cyril Fersing's team reported twenty-nine compounds with good antileishmanial activity against L. donovani, L. infantum and L. major forms of promastigote and amastigote. Compound B (Fig. 3), showed IC50 = 1–2.1 μM and has low cytotoxicity against the human HepG2 cell line (CC50 > 100 μM). The IC50 values of the compound C (Fig. 3) were 1.2 and 2.3 μM against L. donovani of promastigote and intramacrophase of amastigotes, respectively.22 John J. Allocco team did the work on whole-cell growth inhibitors of casein kinase 1 (CK 1) of the L. major and T. brucei parasites. The compounds D and E (Fig. 3) showed growth inhibition with IC50 values of 0.5 and 0.2 μM against L. major while in vitro CK1 activity with IC50 values of 9.0 and 8.0 nM respectively was observed.23 Marhadour et al. reported that the diarylimidazo-[1,2-a]-pyridines exhibited low cytotoxicity against the HeLa cells. Compound F was the most potent one against the promastigote form of L. major with IC50 of 4.0 μM (Fig. 3).24
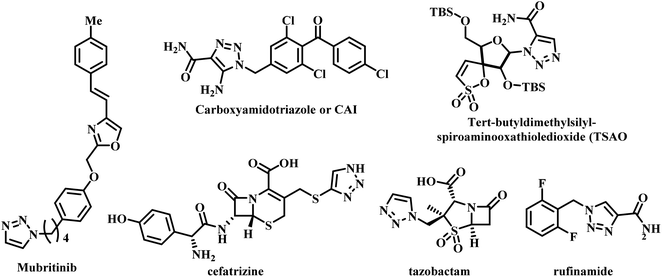
Several triazole containing drugs are currently available in the market. They exhibit pharmacological activities such as antileishmanial, anticancer, antimalarial, antitubercular, etc.25–28 Mubritinib is used for breast, bladder, kidney, and prostate cancers.29 The compound carboxyamidotriazole or CAI is an anticancer drug currently in phase III clinical trials.30,31 tert-Butyldimethylsilyl-spiroaminooxathioledioxide (TSAO) is used as an anti-HIV reverse transcriptase inhibitor.32 Cefatrizine and tazobactam are used as antibiotics,33 while rufinamide is used as an anticonvulsant (Fig. 4).34
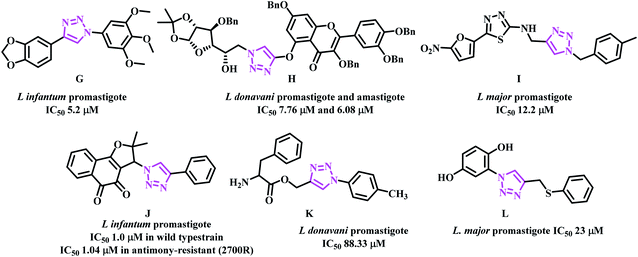
Casemate et al. reported sixteen 1,4-diaryl-1,2,3-triazole analogues. From this work, two compounds showed best in vitro activity against L. amazonensis promastigotes with IC50 values of 1.1 μM and 3.71 μM. Compound G (Fig. 5) showed activity against both L. amazonensis and L. infantum with IC50 values of 7.23 μM and 5.2 μM.35 Dwivedi team reported eighteen triazole based benzylquercetin glycoconjugates that showed activity against the promastigotes and amastigotes of Leishmania donovani. In this work, compound H (Fig. 5) showed the best activity with IC50 values 7.76 μM and 6.08 μM respectively.36 Tahghighi et al. reported fourteen thiadiazol-2-amine containing triazole analogues. From this work, compound I (Fig. 5) was the most active one against the promastigotes forms of L. major with IC50 12.2 μM.37 Guimarães team synthesized lapachone based 1,2,3-triazoles and evaluated in different resistant strains of L. infantum and L. amazonensis. Compound J (Fig. 5) showed activity in wild type with IC50 ∼ 1.00 μM and 1.11 μM against L. infantum and L. amazonensis; in antimony-resistant (2700R) strain IC50 of 1.04 μM and 1.03 μM was observed in both L. infantum and L. amazonensis respectively.38 Mohammad Masood's team reported eighteen amino acid-triazole derivatives. From this work, compound K (Fig. 5) showed moderate activity against the promastigote form of L. donovani (Dd8 strain) with IC50 88.33 μM.39 Chris Marie team synthesized hydroquinone-triazole based eleven compounds and compound L with IC50 23 μM against L. major promastigote form was the most active one. Further, these compounds were found to be non-toxic against human embryonic kidney cells (Fig. 5).40
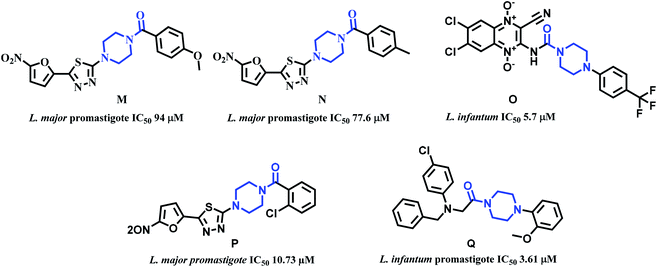
Piperazine is another attractive scaffold that has a multitude of pharmacological effects and is also used as a linker in several drug molecules. It is present in numerous marketed drugs such as ranolazine (antianginal), imatinib (anticancer), buspirone (antidepressant), cinnarazine (antihistamine), flupentixol (antipsychotic), sildenafil (erectile dysfunction), to name a few.41 Literature reports indicate that piperazine is used as a linker in obtaining significantly active antileishmanial compounds. Especially acyl group adjacent to the piperazine N was noticed in several active compounds. Alireza et al. synthesized 1-[5-(5-nitrofuran-2-yl)-1,3,4-thiadiazole-2-yl]-4-benzoyl piperazine derivatives. From this work, compounds M and N (Fig. 6) exhibited good activity with IC50 94 and 77.6 μM values against L. major's promastigote form.42 Carlos Barea et al. synthesized 2-cyano-3-(4-phenylpiperazine-1-carboxamido) quinoxaline 1,4-dioxide derivatives. Compound O (Fig. 6) with IC50 5.7 μM against axenic forms of Leishmania infantum emerged from this work to be the most active compound.43 Fardmoghadam et al. reported furan and thiophene containing 1,3,4-thiadiazole and arylpiperazine based compounds and monitored anti-leishmanial activity against both promastigote and amastigote forms of Leishmania major. In this work, compound P (Fig. 6) displayed significant activity against promastigote form of L. major with IC50 10.73 μM.44 Chandar et al. synthesized various piperazine linked phenyl derivatives and tested them against promastigote and amastigote forms of L. infantum. Compound Q with IC50 values of 3.61 and 44.14 μM against both the forms was the most active compound (Fig. 6).45
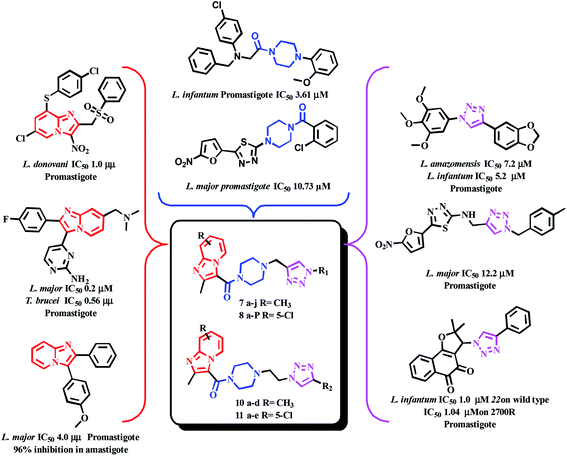
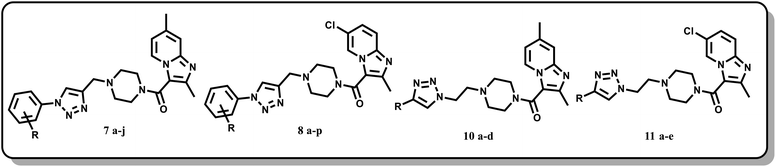
In the current work, we focused mainly on our efforts towards a synthetic approach implicating the combination of the pharmacophore units of imidazo[1,2-a]pyridine, piperazine and triazole (Fig. 7) into one framework. Both imidazo[1,2-a]pyridine and the 1,2,3-triazole moiety were linked together by carbonyl piperazine.
2. Results and discussion
2.1 Synthesis
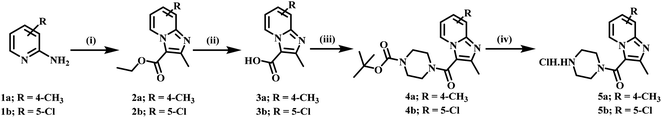
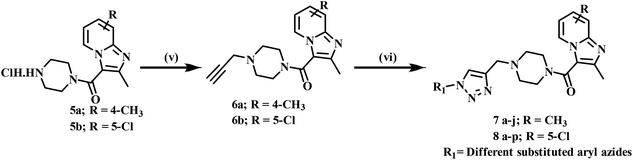
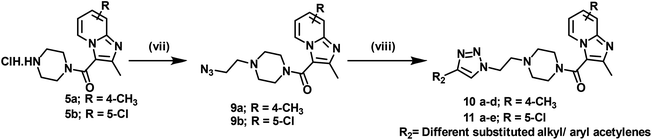
4-Methyl pyridine-2-amine (1a) and 5-chloropyridin-2-amine (1b) on treatment with ethyl 2-chloro-3-oxobutanoate in DME under reflux for 24 h yielded ethyl-2,7-dimethylimidazo[1,2-a]pyridine-3-carboxylate (2a) and ethyl-6-chloro-2-methylimidazo[1,2-a]pyridine-3-carboxylate (2b) respectively.46 Further reaction of ethyl ester with LiOH in ethanol and water at 70 °C for 12 h afforded substituted carboxylic acids (3a–b).46 The acids 3a and 3b on coupling with 1-Boc-piperazine using EDC·HCl, HOBt, and DIPEA in DMF at rt for 12 h yielded 4a and 4b, respectively. The key intermediate 5a and 5b were prepared by BOC deprotection of 4a and 4b using 4 M dioxane–HCl at 0 °C to rt for 4 h, as shown in Scheme 1. We prepared the title compounds 7a–j and 8a–p in two steps, as shown in Scheme 2. First, we performed the SN2 reaction, involving the reaction of 5a and 5b with propargyl bromide, potassium carbonate, and a catalytic amount of potassium iodide in acetone and water at 50 °C for 24 h yielding 6a and 6b. Further, 6a and 6b on the treatment with various substituted azides using CuSO4·5H2O and sodium ascorbate in DMF and water at rt to 55 °C for 12 h afforded the title compounds 7a–j and 8a–p.We prepared the compounds 10a–d and 11a–e in two steps, as shown in Scheme 3. Compounds 5a and 5b on treatment with 2-azidoethyl 4-methylbenzenesulfonate using K2CO3 and a catalytic amount of KI in acetone and water at 50 °C for 24 h yielded 9a and 9b. Finally, 9a and 9b on treatment with various substituted acetylenes using CuSO4·5H2O and sodium ascorbate in DMF and water at rt to 55 °C for 12 h afforded the compounds 10a–d and 11a–e. All the final compounds were purified by column chromatography using 65–95% ethyl acetate in hexane and 2–6% methanol in CH2Cl2 as eluents.
The 1H NMR of final compounds 7a–j, 8a–p, 10a–d, and 11a–e showed one singlet and one multiplet between δ 3.0–4.00 ppm due to methylene protons between the piperazine-triazole nucleus and piperazine (methylene groups) respectively. Another singlet was observed in the range δ 2.00–2.65 ppm due to methyl protons attached to the imidazole ring. In the aromatic region, one singlet appeared between δ 8.50–9.50 ppm due to the single proton of the triazole nucleus of the compounds 7a–j, 8a–p, 10a–d, and 11a–e confirming the formation of products. All the synthesized compounds were confirmed by mass spectrometry, 1H and 13C NMR, and elemental analyses.
2.2 Cytotoxicity evaluation
Initially, all the synthesized final compounds were evaluated for cytotoxicity against HeLa cell lines at 100 μM by an Alamar blue assay, which showed most of these compounds were non-toxic to HeLa cells at tested concentration. Compounds that showed cytotoxicity against HeLa cells were further evaluated to determine cytotoxic concentration (CC50) value. Among the tested derivatives, only compounds 8d and 10b showed moderate cytotoxicity (CC50 58.09 and 62.67 μM, respectively) against HeLa cells.2.3 In vitro antileishmanial and antitrypanosomal activity of the titled compounds
In the current study, the effect of all synthesized title compounds on promastigotes of L. major and trypanocidal properties on bloodstream-form T. brucei was evaluated in vitro initially as a single point. All compounds with IC50 below 75 μM were further tested in replicates of four to obtain a mean and standard deviation. The antileishmanial and antitrypanosomal activities of these novel compounds are shown in Table 1.| S. no. | Comp. code | R | L. major (IC50, μM) | T. brucei (IC50, μM) | CC50, μM | SI (L. major) | SI (T. brucei) |
|---|---|---|---|---|---|---|---|
| a The values are either a single IC50 determination (for compounds with IC50 > 75 μM) or mean ± standard deviations (n = 4) (for compounds with IC50 < 75 μM). | |||||||
| 1 | 7a | 245.7 | 131.10 | >100 | >0.41 | >0.76 | |
| 2 | 7b | 4-NO2 | >500 | >500 | >100 | — | — |
| 3 | 7c | 4-CF3 | 288.3 | 95.90 | >100 | >0.35 | >1.04 |
| 4 | 7d | 4-C2H5 | 74.6 ± 1.4 | 63.2 ± 2.0 | >100 | >1.49 | >1.53 |
| 5 | 7e | 4-Br | 149.2 | >500 | >100 | >0.67 | — |
| 6 | 7f | 3-NO2 | >250 | >500 | >100 | >0.40 | — |
| 7 | 7g | 3-OMe | 179.8 | 259.30 | >100 | >0.56 | >0.39 |
| 8 | 7h | 2-Cl | 211.3 | >500 | >100 | >0.47 | — |
| 9 | 7i | 3-Cl, 4-F | 173.8 | >500 | >100 | >0.58 | — |
| 10 | 7j | 4-OMe, 2-NO2 | 114.0 | 47.7 ± 9.8 | >100 | >0.88 | >2.4 |
| 11 | 8a | H | 192.4 | 116.10 | >100 | >0.52 | >0.86 |
| 12 | 8b | 4-NO2 | >250 | >500 | >100 | >0.40 | — |
| 13 | 8c | 4-CF3 | 62.8 ± 2.4 | 23.2 ± 1.5 | >100 | >1.48 | >4.3 |
| 14 | 8d | 4-C2H5 | 34.5 ± 0.6 | 29.1 ± 1.3 | 58.09 | 1.57 | 1.86 |
| 15 | 8e | 4-F | 88.6 | 94.30 | >100 | >1.13 | >1.06 |
| 16 | 8f | 4-Br | 33.6 ± 0.6 | 28.7 ± 3.1 | >100 | >2.92 | >1.73 |
| 17 | 8g | 3-NO2 | >250 | >500 | >100 | >0.40 | — |
| 18 | 8h | 3-CF3 | 73.6 ± 2.0 | 24.2 ± 1.0 | >100 | >1.43 | >2.98 |
| 19 | 8i | 3-OMe | 111.8 | 50.9 ± 5.7 | >100 | >0.89 | >1.34 |
| 20 | 8j | 3-Cl | 47.1 ± 0.9 | 26.5 ± 1.5 | >100 | >2.22 | >2.36 |
| 21 | 8k | 2-F | 73.1 ± 2.8 | 61.2 ± 2.5 | >100 | >1.38 | >1.66 |
| 22 | 8l | 2-Cl | 91.48 | 33.5 ± 2.7 | >100 | >1.09 | >2.03 |
| 23 | 8m | 3-Cl, 4-F | 212.20 | >500 | >100 | >0.47 | — |
| 24 | 8n | 3,5-CH3 | 70.9 ± 2.8 | 53.90 | >100 | >1.43 | >1.85 |
| 25 | 8o | 3,4-CH3 | 60.9 ± 1.0 | 40.9 ± 2.0 | >100 | >1.43 | >2.42 |
| 26 | 8p | 4-OMe, 2-NO2 | 100.7 | 46.3 ± 1.6 | >100 | >0.99 | >2.11 |
| 27 | 10a | n-Propyl | 176.1 | 40.5 ± 2.7 | >100 | >0.57 | >1.44 |
| 28 | 10b | n-Nonyl | 38.5 ± 0.9 | 5.5 ± 0.5 | 62.67 | 1.63 | 6.83 |
| 29 | 10c | Cyclopropyl | 250.6 | 71.3 | >100 | >0.40 | >1.40 |
| 30 | 10d | 4-t-Butyl phenyl | 15.1 ± 1.0 | >500 | >100 | >6.37 | — |
| 31 | 11a | n-Propyl | 475.1 | 7.4 ± 0.9 | >100 | >0.21 | >3.09 |
| 32 | 11b | n-Nonyl | 62.0 ± 2.7 | 0.72 ± 0.57 | >100 | >1.63 | >6.12 |
| 33 | 11c | Cyclopropyl | >500 | 131.9 | >100 | >0.19 | >0.76 |
| 34 | 11d | Phenyl | >250 | >500 | >100 | >0.35 | — |
| 35 | 11e | 4-t-Butyl phenyl | 171.1 | >500 | >100 | >1.49 | >0.76 |
| 36 | Miltefosine | 12.6 ± 0.5 | — | ||||
| 37 | Melarsoprol | — | 0.05 ± 0.01 | ||||
Among the tested four series of thirty-five compounds, five compounds (8d, 8f, 8j, 10b, and 10d) showed significant inhibition on the growth of promastigote forms of L. major with IC50 values in the range of 15 to 47 μM. The most significantly active compound 10d showed comparable IC50 value (15.1 μM) as standard drug miltefosine (12.6 μM). Apart from this, around nine compounds (7d, 8c, 8e, 8h, 8k, 8l, 8n, 8o, and 11b) exhibited moderate activity against the tested strain of L. major with IC50 values in the range of 63 to 91 μM. The remaining compounds were found weakly active against the tested forms of the parasite in comparison to the standard drug. Finally, among the four tested series, the majority of 8a–p and 10a–d compounds possessed significant activity against the tested strain of promastigote of L. major and 7a–j, 11a–e series compounds exhibited weak activity against L. major. On the other side, among the tested thirty-five compounds, three compounds (10b, 11a, and 11b) showed significant activity against the T. brucei parasite with IC50 values of 5.5, 7.4, and 0.7 μM, respectively. The later compound 11b showed only a ten-fold high IC50 value than that of standard drug melarsoprol (0.05 μM). Apart from this, around twelve compounds (7j, 8c, 8d, 8f, 8h, 8i, 8j, 8l, 8n, 8o, 8p, 10a) exhibited moderate antitrypanosomal activity with IC50 values in the range of 23 to 53.9 μM. Around five compounds (7c, 7d, 8e, 8k, and 10c) possessed weak activity with IC50 values in the range of 61 to 95.9 μM against the tested T. brucei parasite in the in vitro assay. The remaining compounds were found largely inactive against the tested forms of the parasite in comparison to the standard drug. In summary, compound 10b showed significant, and five compounds (8c, 8h, 8l, 8n, and 8o) exhibited moderate activity against both the tested strains of L. major and T. brucei.
2.4 Structure–activity relationship (SAR) description
Electron withdrawing groups especially at the para position of the phenyl group attached to triazole nucleus (8d, 8f), enhanced the antileishmanial activity. In contrast, a majority of the compounds substituted with electron-withdrawing groups at the ortho, meta, and ortho, meta disubstitution (8h, 8k, 8l, 8n, 8o) showed moderate antileishmanial activity against the tested L. major. Concerning the antitrypanosomal activity, compounds substituted with bulky group/long chain at the triazole nucleus (10b and 11b) showed significant activity among all the tested series of compounds. Electron-withdrawing groups, especially at the para position of the phenyl group attached to the triazole nucleus (7c, 7d, 8e, 8k), resulted in a weak antitrypanosomal activity.2.5 In silico prediction of ADME and toxicity parameters
Lipinski's rule of five parameters (mol. weight, donor HB, acceptor HB, and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) of all the titled compounds are satisfied compared with the marketed approved drugs. The solvent-accessible surface area, aqueous solubility pattern, brain/blood partition coefficient, and the number of rotatable bonds of the titled compounds were found to be in the prescribed range as that of marketed drugs. The Caco-2 permeability of all the compounds except few compounds (7b, 7f, 7j, 8b, and 8g) was found to be moderate when compared to the marketed drugs. Percent of human oral absorption was 100% in the compounds 10b and 10d, while in compound 8p, it is 48%. For the rest of the final compounds, it lies between 84–55% (Table 2).
P) of all the titled compounds are satisfied compared with the marketed approved drugs. The solvent-accessible surface area, aqueous solubility pattern, brain/blood partition coefficient, and the number of rotatable bonds of the titled compounds were found to be in the prescribed range as that of marketed drugs. The Caco-2 permeability of all the compounds except few compounds (7b, 7f, 7j, 8b, and 8g) was found to be moderate when compared to the marketed drugs. Percent of human oral absorption was 100% in the compounds 10b and 10d, while in compound 8p, it is 48%. For the rest of the final compounds, it lies between 84–55% (Table 2).
| S. no. | Comp. code | Mol. wt. | SASA | Donor HB | Acceptor HB | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w Po/w |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
PCaco | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BB BB |
Rotor | % Human oral absorption |
|---|---|---|---|---|---|---|---|---|---|---|---|
a Comp. code – compound code; mol. wt. – molecular weight (130–725); SASA – solvent accessible surface area (300–1000); donor HB – hydrogen bond donor (0–6); acceptor HB – hydrogen bond acceptor (0–20); log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w – partition co-efficient (octanol/water) (−2.0 to 6.50); log Po/w – partition co-efficient (octanol/water) (−2.0 to 6.50); log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S – solubility compound (−6.5 to 0.5 mol dm−3); PCaco – Caco-2 cell permeability in nm s−1 (<25 poor, >500 good); log S – solubility compound (−6.5 to 0.5 mol dm−3); PCaco – Caco-2 cell permeability in nm s−1 (<25 poor, >500 good); log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BB – brain/blood partition coefficient (−3.0 to 1.2); rotor – number of rotatable bonds (0 to 15); % human oral absorption – percentage human oral intestional absorption (up to 100). BB – brain/blood partition coefficient (−3.0 to 1.2); rotor – number of rotatable bonds (0 to 15); % human oral absorption – percentage human oral intestional absorption (up to 100). |
|||||||||||
| 1 | 7a | 415.50 | 757.26 | 0.00 | 8.50 | 3.07 | −4.41 | 245.81 | −0.38 | 3.00 | 87.72 |
| 2 | 7b | 460.49 | 797.80 | 0.00 | 9.50 | 2.39 | −4.49 | 34.87 | −1.49 | 4.00 | 55.57 |
| 3 | 7c | 483.50 | 812.89 | 0.00 | 8.50 | 4.12 | −5.98 | 256.96 | −0.12 | 3.00 | 94.18 |
| 4 | 7d | 443.55 | 821.72 | 0.00 | 8.50 | 3.79 | −5.47 | 256.76 | −0.48 | 4.00 | 92.25 |
| 5 | 7e | 494.39 | 786.30 | 0.00 | 8.50 | 3.66 | −5.30 | 245.73 | −0.22 | 3.00 | 91.14 |
| 6 | 7f | 460.49 | 797.65 | 0.00 | 9.50 | 2.38 | −4.49 | 33.53 | −1.52 | 4.00 | 55.23 |
| 7 | 7g | 445.52 | 795.51 | 0.00 | 9.25 | 3.20 | −4.58 | 282.14 | −0.40 | 4.00 | 89.53 |
| 8 | 7h | 449.94 | 778.99 | 0.00 | 8.50 | 3.57 | −5.07 | 274.69 | −0.20 | 3.00 | 91.50 |
| 9 | 7i | 467.93 | 789.71 | 0.00 | 8.50 | 3.84 | −5.50 | 283.04 | −0.07 | 3.00 | 93.32 |
| 10 | 7j | 490.52 | 823.13 | 0.00 | 10.25 | 2.59 | −4.42 | 56.64 | −1.30 | 5.00 | 60.54 |
| 11 | 8a | 435.92 | 748.21 | 0.00 | 8.50 | 3.25 | −4.54 | 256.45 | −0.18 | 3.00 | 89.11 |
| 12 | 8b | 480.91 | 789.14 | 0.00 | 9.50 | 2.56 | −4.63 | 34.89 | −1.31 | 4.00 | 56.61 |
| 13 | 8c | 503.91 | 802.61 | 0.00 | 8.50 | 4.28 | −6.09 | 256.02 | 0.06 | 3.00 | 82.16 |
| 14 | 8d | 463.97 | 812.97 | 0.00 | 8.50 | 4.01 | −5.61 | 291.36 | −0.23 | 4.00 | 94.52 |
| 15 | 8e | 453.91 | 757.74 | 0.00 | 8.50 | 3.50 | −4.93 | 256.01 | −0.08 | 3.00 | 90.53 |
| 16 | 8f | 514.81 | 777.17 | 0.00 | 8.50 | 3.84 | −5.43 | 257.36 | −0.02 | 3.00 | 79.61 |
| 17 | 8g | 480.91 | 788.44 | 0.00 | 9.50 | 2.56 | −4.63 | 33.38 | −1.32 | 4.00 | 56.24 |
| 18 | 8h | 503.91 | 800.22 | 0.00 | 8.50 | 4.26 | −6.05 | 243.67 | 0.04 | 3.00 | 81.63 |
| 19 | 8i | 465.94 | 787.38 | 0.00 | 9.25 | 3.38 | −4.74 | 281.53 | −0.22 | 4.00 | 90.56 |
| 20 | 8j | 470.36 | 774.46 | 0.00 | 8.50 | 3.82 | −5.36 | 282.23 | 0.02 | 3.00 | 93.17 |
| 21 | 8k | 453.91 | 757.34 | 0.00 | 8.50 | 3.54 | −4.88 | 304.91 | −0.01 | 3.00 | 92.11 |
| 22 | 8l | 470.36 | 770.05 | 0.00 | 8.50 | 3.79 | −5.21 | 307.83 | 0.04 | 3.00 | 93.66 |
| 23 | 8m | 488.35 | 782.52 | 0.00 | 8.50 | 4.03 | −5.68 | 281.51 | 0.11 | 3.00 | 94.37 |
| 24 | 8n | 463.97 | 813.29 | 0.00 | 8.50 | 3.90 | −5.78 | 242.65 | −0.26 | 3.00 | 92.49 |
| 25 | 8o | 463.97 | 805.29 | 0.00 | 8.50 | 3.87 | −5.63 | 243.04 | −0.25 | 3.00 | 92.29 |
| 26 | 8p | 510.94 | 814.65 | 0.00 | 10.25 | 2.77 | −4.57 | 56.18 | −1.12 | 5.00 | 48.54 |
| 27 | 10a | 395.51 | 770.01 | 0.00 | 9.00 | 2.69 | −3.90 | 263.71 | −0.55 | 6.00 | 86.02 |
| 28 | 10b | 479.67 | 957.13 | 0.00 | 9.00 | 4.96 | −6.47 | 245.83 | −1.11 | 12.00 | 100.00 |
| 29 | 10c | 393.49 | 697.95 | 0.00 | 9.00 | 2.23 | −2.86 | 257.06 | −0.31 | 4.00 | 83.11 |
| 30 | 10d | 485.63 | 866.91 | 0.00 | 9.00 | 4.46 | −5.90 | 302.69 | −0.44 | 5.00 | 100.00 |
| 31 | 11a | 415.93 | 734.35 | 0.00 | 9.00 | 2.78 | −3.53 | 326.11 | −0.21 | 6.00 | 88.23 |
| 32 | 11b | 500.09 | 947.49 | 0.00 | 9.00 | 5.13 | −6.59 | 246.81 | −0.93 | 12.00 | 73.90 |
| 33 | 11c | 413.91 | 689.03 | 0.00 | 9.00 | 2.40 | −3.00 | 256.07 | −0.12 | 4.00 | 84.08 |
| 34 | 11d | 449.94 | 764.40 | 0.00 | 9.00 | 3.41 | −4.43 | 329.49 | −0.11 | 4.00 | 91.95 |
| 35 | 11e | 506.05 | 882.00 | 0.00 | 9.00 | 4.73 | −6.49 | 270.88 | −0.36 | 5.00 | 85.20 |
2.6 In silico predicted toxicity profile of the titled compounds
The results of anticipated toxicity studies of the final compounds (7a–j, 8a–p, 10a–d, and 11a–e) revealed that the tested compounds are non-toxic. Cancer-causing properties like mutagenicity and tumorigenicity of these agents were found to be nil. The rest of the features like reproductive effect and irritant effects were also found to be none. With these satisfactory results, we conclude that all the predicted ADME and toxicity parameters of the designed compounds displayed productive results. The results disclosed that these compounds might not face any problems during clinical trials.2.7 In silico molecular docking studies
To find the putative binding mode of the titled compounds, a docking study was performed, and the results were analyzed (Table 3). The validation of the docking protocol was done by checking the root mean square deviation of the X-ray native pose of the ligand with its docked pose of ligand (Fig. 8) and found to be 0.20 Å. This RMSD could be reliable to proceed for the docking study of the designed scaffolds.| Comp. code | Hydrogen bond with distances (Å) | Π–Π interactions with distances (Å) | Gide score (kcal mol−1) |
|---|---|---|---|
| 10d | SER-14 (2.26) | ASP-327 (2.78) | −6.74 |
| LYS-60 (2.24) | |||
| Co-crystal ligand (FAD) | SER-14 (2.26) | TYR-198 (5.45) | −16.24 |
| GLY-15 (2.14) | |||
| ASP-35 (1.86, 2.04) | |||
| THR-51 (1.82) | |||
| LYS-60 (2.11) | |||
| GLY-127 (2.09, 2.13) | |||
| ASP-327 (1.85, 2.36) | |||
| THR-335 (2.26) |
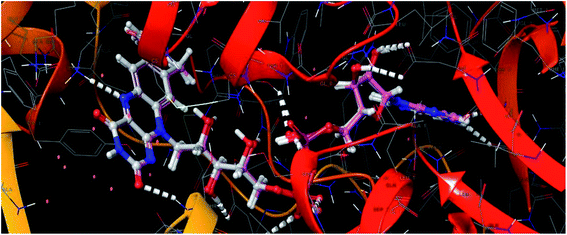 | ||
| Fig. 8 Superimposed view of the FAD molecule in the active site of the target PDB-2JK6, and its redocked pose in the same target (RMSD – 0.20 Å). | ||
From the docked pose view of extracted co-crystal ligand (Fig. 9), eight hydrogen bond interactions with the amino acid residues SER14, GLY15, ASP35, THR51, LYS60, GLY127, ASP327 and THR335 of the active site of the target protein and one Π–Π interaction with the amino acid residue TYR198 with a docking score value of −16.24 kcal mol−1 was observed. The docking score exhibited by the significantly active compound 10d is −6.74 kcal mol−1. Like co-crystal ligand, compound 10d also made two hydrogen bond interactions with the amino acid residues SER14, LYS60 and one Π–Π interaction with the amino acid residue ASP327 of the active site of the target protein (Fig. 10). Upon critical examination of the interactions formed by the co-crystal ligand to the target protein and the compound 10d to the active site amino acid residues of the target protein, it was found that the same amino acid residues (SER14, LYS60) are involved in the similar kind of hydrogen bond interactions with both co-crystal ligand and compound 10d.
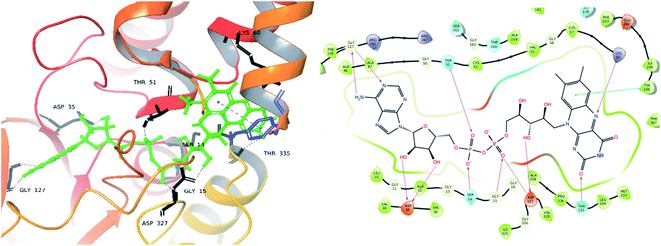 | ||
| Fig. 9 Exposed amino-acid residue interactions of the FAD molecule in the actives site of the target PDB-2JK6 (black lines – hydrogen bonds, blue – pi interaction in 3D, magenta – hydrogen bond, green – pi interaction in 2D). | ||
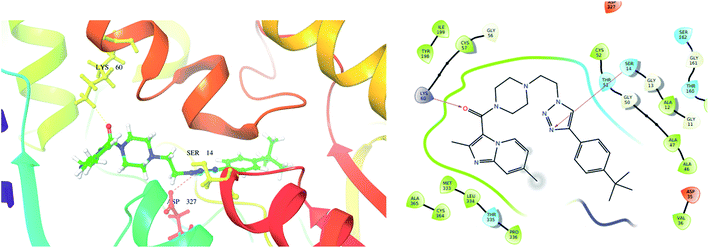 | ||
| Fig. 10 Exposed amino-acid residue interactions of the significantly active compound 10d in the actives site of the target PDB-2JK6 (yellow lines – hydrogen bonds, blue – pi interaction in 3D, magenta – hydrogen bond in 2D). | ||
2.8 Single crystal X-ray crystallographic structure of compound 8f
The suitable crystals of compound 8f for single-crystal X-ray diffraction (SCXRD) study were grown from the mixture of methanol and dichloromethane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). The SCXRD measurements were performed on the Rigaku XtaLAB P200 diffractometer using graphite monochromated Mo-Kα radiation (λ = 0.71073 Å). The data was collected and reduced using CrysAlisPro (Rigaku Oxford Diffraction) software. The data collection was carried out at 100 K, and the structures were solved using Olex2 with the ShelX structure solution program using Direct Methods and refined with the ShelXL refinement package using Least Squares minimization. The basic crystallographic data is given in a table in ESI.†
3). The SCXRD measurements were performed on the Rigaku XtaLAB P200 diffractometer using graphite monochromated Mo-Kα radiation (λ = 0.71073 Å). The data was collected and reduced using CrysAlisPro (Rigaku Oxford Diffraction) software. The data collection was carried out at 100 K, and the structures were solved using Olex2 with the ShelX structure solution program using Direct Methods and refined with the ShelXL refinement package using Least Squares minimization. The basic crystallographic data is given in a table in ESI.†
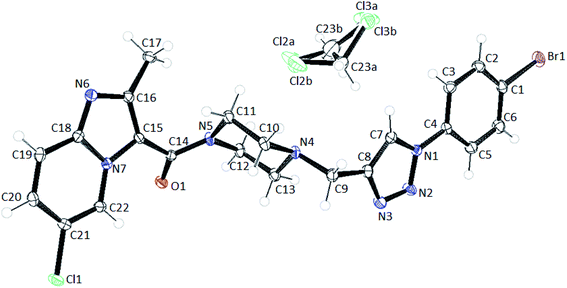
The compound 8f crystallizes in Pbca space group with one linker per asymmetric unit and one molecule of DCM which is disordered over two positions (Fig. 11). The ORTEP diagram of the structure shows that the ellipsoids of all the atoms are well behaved. Crystallographic Data Center and corresponding deposition number is CCDC 1991585.†
Upon closer look at the structure, it was found that the ligand packs in an interesting manner where the individual units stack antiparallel to each other to minimize the electronic repulsions. The structure show pi–pi stacking between the benzene units of one ligand and the imidazole pyridine units of the other ligand (Fig. 12). The pi–pi stacking distance is in the range of 3.6–3.9 Å.
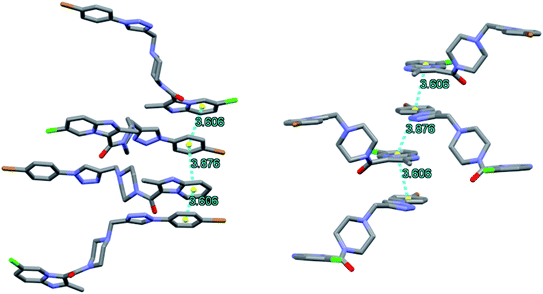 | ||
| Fig. 12 Packing diagram of 8f showing pi–pi stacking arrangement as observed along a (left) and c (right) axis. | ||
3. Conclusion
In conclusion, we have designed and synthesized thirty-five novel 1,2,3-triazole analogues of imidazo-[1,2-a]-pyridine-3-carboxamides using substituted pyridine, propargyl bromide, 2-azidoethyl-4-methyl benzenesulfonate and substituted acetylenes. The synthesized compounds were well characterized using 1H NMR, 13C NMR, LCMS and elemental analyses. These compounds were further screened in vitro for antileishmanial and antitrypanosomal activity against L. major and T. brucei parasites, respectively. Few compounds exhibited significant to moderate antileishmanial and antitrypanosomal activity in the tested assay system. Five compounds exhibited significant antileishmanial activity (8d, 8f, 8j, 10b and 10d), while three compounds (10b, 11a and 11b) showed significant activity against the T. brucei parasite. In silico ADME prediction studies also depicted that the titled compounds do not violate Lipinski's rule of five parameters in comparison with the marketed approved drugs. The predicted toxicity profile also confirmed that the compounds are non-toxic. Finally, molecular docking studies was also performed for significantly active compound (10d) in order to study its putative binding pattern at the active site of the selected leishmanial trypanothione reducatse target. The crystal structure developed for one of the significantly active antileishmanial compound 8f was found to occupy Pbca space group. Further, the crystal structure revealed pi–pi stacking between the benzene units of one ligand and the imidazole pyridine units of the other. We also observed that all the final synthesized compounds were non-toxic to HeLa cell line at 100 μM.4. Experimental section
4.1 Chemistry
The intermediates 2a, 2b, 3a, and 3b were prepared as per the earlier reported literature protocol.46 The procedures and the analytical data of the remaining intermediates and final compounds (7a–j, 8a–p, 10a–d, and 11a–e) are given in ESI.† The detailed procedures used for cytotoxicity, in vitro activity studies, in silico toxicity parameters, molecular docking studies, ligand and protein preparation methods, receptor grid generation, docking validation, and docking analysis are also given in ESI.†4.2 Analytical data of final compounds (7a–j, 8a–p, 10a–d, and 11a–e)
Conflicts of interest
Authors declare that there is no conflict of interest.Acknowledgements
KVGCS and SM thank DBT, New Delhi [BT/IN/Spain/39/SMl2017-18] for providing financial support. The financial assistance provided by DIST FIST grant (SR/FST/CSI-240/2012), New Delhi is gratefully acknowledged. SS thanks CSIR for providing SRF fellowship. Central analytical lab facilities of BITS Pilani Hyderabad Campus are gratefully acknowledged.References
- P. J. Hotez and N. C. Lo, Neglected Tropical Diseases: Public Health Control Programs and Mass Drug Administration, Elsevier Inc., 10th edn, 2020 Search PubMed.
- D. Steverding, Parasites Vectors, 2017, 10, 1–10 CrossRef.
- S. Kapil, P. K. Singh and O. Silakari, Eur. J. Med. Chem., 2018, 157, 339–367 CrossRef CAS.
- N. D. Karunaweera and M. U. Ferreira, Parasitology, 2018, 145, 425–429 CrossRef.
- K. Pandey, B. Pal, V. N. R. Das, K. Murti, C. S. Lal, N. Verma, S. Bimal, V. Ali, R. B. Verma, R. K. Topno, N. A. Siddiqi and P. Das, Br. J. Dermatol., 2017, 177, 557–559 CrossRef CAS.
- https://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness).
- N. Devi, D. Singh, R. K. Rawal, J. Bariwal and V. Singh, Curr. Top. Med. Chem., 2016, 16, 2963–2994 CrossRef CAS.
- S. Chitti, S. R. Singireddi, P. Santosh Kumar Reddy, P. Trivedi, Y. Bobde, C. Kumar, K. Rangan, B. Ghosh and K. V. G. C. Sekhar, Bioorg. Med. Chem. Lett., 2019, 29, 2551–2558 CrossRef CAS.
- Y. Yu, Y. Han, F. Zhang, Z. Gao, T. Zhu, S. Dong and M. Ma, J. Med. Chem., 2020, 63, 3028–3046 CrossRef.
- B. Zivkovic, E. Morel, D. Joly, G. Perrault, D. J. Sanger and K. G. Llyod, Pharmacopsychiatry, 1990, 23, 108–113 CrossRef.
- A. Dang, A. Garg and P. V. Rataboli, CNS Neurosci. Ther., 2011, 17, 387–397 CrossRef CAS.
- S. Mohana Roopan, S. M. Patil and J. Palaniraja, Res. Chem. Intermed., 2016, 42, 2749–2790 CrossRef CAS.
- W. M. Almirante, L. Polo, A. Mugnaini, E. Provinciali, P. Rugarli, A. Biancotti and A. Gamba, J. Med. Chem., 1965, 1125, 305–312 CrossRef.
- E. Esposito, E. Mazzon, I. Paterniti, D. Impellizzeri, P. Bramanti and S. Cuzzocrea, PLoS One, 2010, 5, 1–16 CrossRef.
- L. A. Sorbera, J. Castañer and P. A. Leeson, Drugs Future, 2002, 27, 935–941 CrossRef CAS.
- W. A. Simon, M. Herrmann, T. Klein, J. M. Shin, R. Huber, J. Senn-Bilfinger and S. Postius, J. Pharmacol. Exp. Ther., 2007, 321, 866–874 CrossRef CAS.
- S. Boggs, V. I. Elitzin, K. Gudmundsson, M. T. Martin and M. J. Sharp, Org. Process Res. Dev., 2009, 13, 781–785 CrossRef CAS.
- O. Kim, Y. Jeong, H. Lee, S. Hong and S. Hong, J. Med. Chem., 2011, 54, 2455–2466 CrossRef CAS.
- A. A. Trabanco, G. Tresadern, G. J. MacDonald, J. A. Vega, A. I. De Lucas, E. Matesanz, A. García, M. L. Linares, S. A. Alonso De Diego, J. M. Alonso, D. Oehlrich, A. Ahnaou, W. Drinkenburg, C. MacKie, J. I. Andrés, H. Lavreysen and J. M. Cid, J. Med. Chem., 2012, 55, 2688–2701 CrossRef CAS.
- J. Rether, G. Erkel, T. Anke, J. Bajtner and O. Sterner, Bioorg. Med. Chem., 2008, 16, 1236–1241 CrossRef CAS.
- C. Castera-Ducros, L. Paloque, P. Verhaeghe, M. Casanova, C. Cantelli, S. Hutter, F. Tanguy, M. Laget, V. Remusat, A. Cohen, M. D. Crozet, P. Rathelot, N. Azas and P. Vanelle, Bioorg. Med. Chem., 2013, 21, 7155–7164 CrossRef CAS.
- C. Fersing, L. Basmaciyan, C. Boudot, J. Pedron, S. Hutter, A. Cohen, C. Castera-Ducros, N. Primas, M. Laget, M. Casanova, S. Bourgeade-Delmas, M. Piednoel, A. Sournia-Saquet, V. Belle Mbou, B. Courtioux, É. Boutet-Robinet, M. Since, R. Milne, S. Wyllie, A. H. Fairlamb, A. Valentin, P. Rathelot, P. Verhaeghe, P. Vanelle and N. Azas, ACS Med. Chem. Lett., 2019, 10, 34–39 CrossRef CAS.
- J. J. Allocco, R. Donald, T. Zhong, A. Lee, Y. S. Tang, R. C. Hendrickson, P. Liberator and B. Nare, Int. J. Parasitol., 2006, 36, 1249–1259 CrossRef CAS.
- S. Marhadour, P. Marchand, F. Pagniez, M. A. Bazin, C. Picot, O. Lozach, S. Ruchaud, M. Antoine, L. Meijer, N. Rachidi and P. Le Pape, Eur. J. Med. Chem., 2012, 58, 543–556 CrossRef CAS.
- S. Haider, M. S. Alam and H. Hamid, Inflammation Cell Signaling, 2014, 1, e95 Search PubMed.
- D. Dheer, V. Singh and R. Shankar, Bioorg. Chem., 2017, 71, 30–54 CrossRef CAS.
- S. Vanaparthi, R. Bantu, N. Jain, S. Janardhan and L. Nagarapu, Bioorg. Med. Chem. Lett., 2020, 30, 127304 CrossRef CAS.
- S. Narsimha, S. K. Nukala, T. Savitha Jyostna, M. Ravinder, M. Srinivasa Rao and N. Vasudeva Reddy, J. Heterocycl. Chem., 2020, 57, 1655–1665 CrossRef CAS.
- I. Baccelli, J. Krosl, G. Boucher, I. Boivin, V. P. Lavallée, J. Hébert, S. Lemieux, A. Marinier and G. Sauvageau, Blood Cancer J., 2017, 7, e529 CrossRef CAS.
- E. A. Johnson, R. S. Marks, S. J. Mandrekar, S. L. Hillman, M. D. Hauge, M. D. Bauman, E. J. Wos, D. F. Moore, J. W. Kugler, H. E. Windschitl, D. L. Graham, A. M. Bernath, T. R. Fitch, G. S. Soori, J. R. Jett, A. A. Adjei and E. A. Perez, Lung Cancer, 2008, 60, 200–207 CrossRef.
- https://clinicaltrials.gov/ct2/show/NCT00003869Title.
- K. Das, J. D. Bauman, A. S. Rim, C. Dharia, A. D. Clark, M. J. Camarasa, J. Balzarini and E. Arnold, J. Med. Chem., 2011, 54, 2727–2737 CrossRef CAS.
- S. G. Agalave, S. R. Maujan and V. S. Pore, Chem.–Asian J., 2011, 6, 2696–2718 CrossRef CAS.
- W. H. Mudd and E. P. Stevens, Tetrahedron Lett., 2010, 51, 3229–3231 CrossRef CAS.
- T. B. Cassamale, E. C. Costa, D. B. Carvalho, N. S. Cassemiro, C. C. Tomazela, M. C. S. Marques, M. Ojeda, M. F. C. Matos, S. Albuquerque, C. C. P. Arruda and A. C. M. Baroni, J. Braz. Chem. Soc., 2016, 27, 1217–1228 CAS.
- P. Dwivedi, K. B. Mishra, B. B. Mishra, N. Singh, R. K. Singh and V. K. Tiwari, Glycoconjugate J., 2015, 32, 127–140 CrossRef CAS.
- A. Tahghighi, S. Razmi, M. Mahdavi, P. Foroumadi, S. K. Ardestani, S. Emami, F. Kobarfard, S. Dastmalchi, A. Shafiee and A. Foroumadi, Eur. J. Med. Chem., 2012, 50, 124–128 CrossRef CAS.
- T. T. Guimarães, M. D. C. F. R. Pinto, J. S. Lanza, M. N. Melo, R. L. Do Monte-Neto, I. M. M. De Melo, E. B. T. Diogo, V. F. Ferreira, C. A. Camara, W. O. Valença, R. N. De Oliveira, F. Frézard and E. N. Da Silva Júnior, Eur. J. Med. Chem., 2013, 63, 523–530 CrossRef.
- M. M. Masood, P. Hasan, S. Tabrez, M. B. Ahmad, U. Yadava, C. G. Daniliuc, Y. A. Sonawane, A. Azam, A. Rub and M. Abid, Bioorg. Med. Chem. Lett., 2017, 27, 1886–1891 CrossRef CAS.
- C. M. Horn, J. Aucamp, F. J. Smit, R. Seldon, A. Jordaan, D. F. Warner and D. D. N’Da, Med. Chem. Res., 2020, 29, 1387–1399 CrossRef CAS.
- M. Shaquiquzzaman, G. Verma, A. Marella, M. Akhter, W. Akhtar, M. F. Khan, S. Tasneem and M. M. Alam, Eur. J. Med. Chem., 2015, 102, 487–529 CrossRef CAS.
- F. Alireza, H. Adibi, S. K. Ardestani, S. Shirooie, A. Bozorgomid and A. Jafari, Iran. J. Pharm. Sci., 2017, 16, 904–909 Search PubMed.
- B. Carlos, A. Pabón, S. Galiano, S. Pérez-Silanes, G. Gonzalez, C. Deyssard, A. Monge, E. Deharo and I. Aldana, Molecules, 2012, 17, 9451–9461 CrossRef.
- B. M. Fardmoghadam, F. Poorrajab, S. K. Ardestani, S. Emami, A. Shafiee and A. Foroumadi, Bioorg. Med. Chem., 2008, 16, 4509–4515 CrossRef.
- C. Subhash, P. Ashok, R. M. Reguera, M. Y. Perez-Pertejo, R. Carbajo-Andres, R. Balana-Fouce, K. V. G. C. Sekhar and M. Sankaranarayanan, Exp. Parasitol., 2018, 189, 49–60 CrossRef.
- Z. Wu, Y. Lu, L. Li, R. Zhao, B. Wang, K. Lv, M. Liu and X. You, ACS Med. Chem. Lett., 2016, 7, 1130–1133 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1991585. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0ra07881f |
| This journal is © The Royal Society of Chemistry 2020 |