Open Access Article
Open Access ArticleNetwork pharmacology-based study of the mechanisms of action of anti-diabetic triterpenoids from Cyclocarya paliurus†
Zixin Linab,
Yingpeng Tongbc,
Na Lib,
Ziping Zhub and
Junmin Li *bc
*bc
aSchool of Life Science, Shanghai Normal University, Shanghai 200234, China
bZhejiang Provincial Key Laboratory of Evolutionary Ecology and Conservation, Taizhou University, Taizhou 318000, China. E-mail: lijmtzc@126.com
cSchool of Advanced Study, Taizhou University, Taizhou 318000, China
First published on 7th October 2020
Abstract
Diabetes is a complex illness requiring long-term therapy. Cyclocarya paliurus, a recently confirmed new food resource, shows significant hypoglycemic and hypolipidemic effects in type II diabetes. Triterpenoid saponins are considered as the effective medicinal components of C. paliurus and are useful for the treatment of diabetes mellitus. However, little is known regarding their specific mechanism of actions. In this study, we used active ingredient screening and target prediction techniques to determine the components of C. paliurus responsible for its anti-diabetic effects as well as their targets. In addition, we used bioinformatics technology and molecular docking analysis to determine the mechanisms underlying their anti-diabetic effects. A total of 39 triterpenes were identified through a literature search and 1 triterpene compound by experiments. In all, 33 potential target proteins associated with 36 pathways were predicted to be related to diabetes. Finally, 7 compounds, 15 target proteins, and 15 signaling pathways were found to play important roles in the therapeutic effects of C. paliurus against diabetes. These results provide a theoretical framework for the use of C. paliurus against diabetes. Moreover, molecular docking verification showed that more than 90% of the active ingredients had binding activity when tested against key target proteins, and a literature search showed that the active ingredients identified had anti-diabetic effects, indicating that the results were highly reliable.
1. Introduction
Diabetes is a disorder of glucose metabolism that leads to high blood sugar levels and to a series of complications which reduce the quality of life of patients, increasing their mortality.1 Based on its pathogenesis, it can be divided into type 1 diabetes (T1DM) and type 2 diabetes (T2DM), with T2DM accounting for more than 90% of cases.2 At present, diabetes is treated with drugs, such as sulfonylureas, DPP-4 inhibitors, oral antidiabetic agents, sodium-glucose co-transport inhibitors, and glucokinase activators,3–7 but many drugs have side effects.8 Therefore, it is urgent to find new, safe and effective early intervention medicinal compounds for diabetes.9Traditional botanical and herbal medicines have been widely used for the treatment of diabetes,10,11 e.g. Lobelia chinensis Lour.,7 Hydrangea macrophylla var. thunbergii,12 and Cyclocarya paliurus (Batal.) Iljinskaja.13 C. paliurus is a medicinal plant native to southern China. Its leaves have been traditionally used in the form of herbal tea for their beneficial effects against diabetes. Various bioactive compounds have been extracted from the leaves of C. paliurus, including flavonoids, triterpenoids, polysaccharides, and organic acids, which may contribute to its antihyperglycemic, antihyperlipidemic, and antihypertensive effects.13–24 Although the mechanisms underlying the anti-diabetic effects of the ethanol and aqueous extracts of C. paliurus leaves have been explored,18 the specific bioactive constituents of C. paliurus and their targets are still unknown. Due to their safety and efficacy, plant-derived triterpenoids have attracted attention for the treatment of diabetes.25–27 For example, new cucurbitane-type triterpenoids have shown potential for the prevention and management of diabetes by improving insulin sensitivity and glucose homeostasis.28 Triterpenoids, such as cyclocaric acid B and cyclocarioside H, extracted from C. paliurus leaves, promote glucose uptake in the absence of insulin, and ameliorate inflammation by inhibiting the insulin receptor substrate 1 (IRS-1)/phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) pathway.29 However, no C. paliurus triterpenoids with anti-diabetic effects or their specific targets have been described.
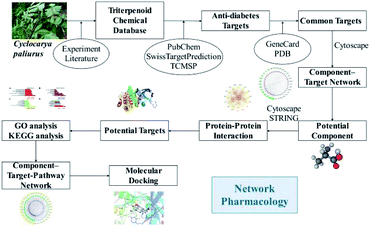
Network pharmacology is a new systematic method to study target-drug interactions and their influence on disease. Combined with pharmacology and pharmacodynamics, network pharmacology has been successfully applied to explore the mechanisms of action of traditional botanical or herbal medicines at the molecular level,30 to dissect the relationship between medicines and diseases, and to identify signaling pathways. This approach makes it feasible to explore the therapeutic mechanisms of multi-component and multi-target traditional botanical or herbal medicines.13 Investigated the beneficial effects of C. paliurus against diabetic dyslipidemia and its mechanisms of action using lipidomics, serum pharmacochemistry, and network pharmacology approaches.13 Based on network pharmacology analysis, they identified the predicted targets of C. paliurus' s active ingredients (ALOX12, APP, BCL2, CYP2C9, PTPN1) and their linked lipidome targets [PLD2, PLA2G(s), and PI3K(s) families], which could be responsible for the effects of C. paliurus leaf extracts against diabetic dyslipidemia.13 In this study, we used network pharmacology to explore the effects of C. paliurus triterpenoids on diabetes and elucidate their mechanisms of action. The detailed procedures are shown in Fig. 1.
2. Materials and methods
2.1 Plant species
C. paliurus is commonly called “tian sha Chun” in China, meaning “sweat tree”, due to the flavor of its leaves. This tree can reach 30 m and grows in moist forests on mountains at altitudes between 400 and 2500 m. It is distributed in many regions of China with temperatures between 7.8 °C and 19.9 °C and with precipitation between 825.9 mm and 2394.5 mm,31,32 including Anhui, Fujian, Guangdong, Guangxi, Guizhou, Hainan, Hubei, Hunan, Jiangsu, Jiangxi, Sichuan, Taiwan, Yunnan, and Zhejiang Provinces. The leaves of this plant are used in traditional Chinese medicine (TCM) for treating disorders like hypertension, high cholesterol, and diabetes, as well as for improving the function of the immune system.15,33 The first health tea from China approved by the U. S. Food and Drug Administration (FDA) was made with the leaves of C. paliurus.312.2 Chemical ingredients database
Leaves of C. paliurus obtained from Taizhou University in October 2018 were air-dried (total weight, 1.6 kg). Dried leaves of C. paliurus (1.6 kg) were pulverized and extracted three times with 90% ethanol (8.0 L) at room temperature. After filtration, the solvents were removed under vacuum to obtain a crude extract (1.1 kg), which was resuspended in H2O (1.0 L) and then partitioned successively with petroleum ether (1.0 L), ethyl acetate (EtOAc, 1.0 L) and butyl alcohol (1.0 L) three times. After removal of solvent, the entire EtOAc extract was fractionated by silica gel column chromatography using a gradient of petroleum ether (PE)/EtOAc (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 0
1 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and then methanol to obtain a pure component.
1, v/v) and then methanol to obtain a pure component.
With the exception of the previous triterpenoids, other triterpenoids compounds from C. paliurus were identified by reviewing previously published studies (ESI Table 1†) and the Traditional Chinese Medicine Systems Pharmacology database (TCMSP, http://lsp.nwu.edu.cn/tcmsp.php).34 The specific structures were obtained from Pubchem databases (https://pubchem.ncbi.nlm.nih.gov/). If the specific structures could not be found in the databases, the original research articles were reviewed. In addition, two natural products were isolated from the C. paliurus extract. Then, all triterpenes were subjected to network pharmacologic prediction. ChemDraw 14.0 software (https://www.chemdraw.com.cn/xiazai.html) was used to draw the structures, and the files were saved in mol and mol2 formats.
2.3 Target protein database
Proteins involved in diabetes were selected from the literature and from the following public databases: PubChem (https://pubchem.ncbi.nlm.nih.gov/), which provides information about chemical substances and their biological activities as a public repository; Genecards database (https://www.genecards.org/), which is a unique bioinformatics and chemical informatics resource; Potential Drug Target Database (http://dddc.ac.cn/pdtd/index.php), which is a protein database for drug target recognition; and RCSB Database (http://www.rcsb.org/pdb/home/home.do), which provides information about therapeutic proteins, nucleic acid targets, targeted diseases, pathways, and the corresponding drugs for each of these targets. Based on the determination of the protein–ligand complex and crystal structures, protein targets were obtained. The crystal structures of the target protein complexes with the original ligands were downloaded from the Protein Data Bank (PDB) database, which is a free database containing structural data of biological macromolecules. These proteins were used as receptors in molecular docking experiments. The structures of the compounds were imported into the Prediction Target of Swiss Target Prediction network database as a *.mol format35 (http://www.swisstargetprediction.ch/), setting the species to “Homo sapiens” to predict active targets. This platform automatically conducts a docking simulation between the compound and the background target database, selecting 15 potential active targets with the highest affinity. Predicted targets showing high probability were selected and duplicates were deleted. Diabetes mellitus was searched in the GeneCards online database (http://www.genecards.org/) to identify genes related to diabetes. Common targets between the database and C. paliurus were identified by using excel. The results were imported into Cytoscape 3.6.1 software and the component-target interaction network was constructed and analyzed.2.4 Component-target network
To elucidate the relationship between C. paliurus chemical constituents and diabetes targets, a “component-target-disease” network was analyzed by linking the selected chemical constituents, component-related targets, diabetes-related targets, and the corresponding targets of interactive proteins. The networks were visually analyzed and screened using Cytoscape 3.6.1 software. The screening criteria were as follows: potential targets for the treatment of diabetes with C. paliurus had a node value (degree) and BC (betweenness centrality) greater than the median values of all points, and the closeness centrality was greater than the median values of all points. The “degree” and “betweenness” were calculated using the analysis function of Cytoscape 3.6.1 to evaluate its topological features.362.5 Protein–protein interaction (PPI) network
To explain interactions between target proteins of related C. paliurus compounds, these were uploaded to the STRING database (https://string-db.org/). The reliability of protein interactions analyzed with the STRING database is divided into multiple levels, with interaction scores between 0.15 and 0.4 considered of low reliability, between 0.4 and 0.7 of medium reliability, and greater than 0.7 of high reliability.37 Targets showing interactions greater than medium reliability were selected to construct the interaction network of C. paliurus anti-diabetic targets. The node interaction relationship parameters were uploaded into Cytoscape 3.6.1 software to draw the interaction network, and the internet was used to obtain the final protein interaction diagram.2.6 Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis of target proteins
To elucidate the role played by target proteins interacting with the active ingredients of C. paliurus in terms of gene function and signaling pathways, the Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.ncifcrf.gov/) 6.8 was used to analyze GO function and KEGG pathway enrichment. The biological process (BP), molecular function (MF) and cell components (CC) of the total targets of C. paliurus were analyzed by GO enrichment, and the column graphs were drawn according to the P value. The KEGG pathway was also analyzed using DAVID6.8, setting P ≤ 0.05 and introducing Gene ID from the total targets of C. paliurus. According to the enrichment degree and P value, the column graphs of the KEGG pathway were drawn. The main signaling pathways of diabetes mellitus were selected and imported into Cytoscape 3.6.1 software for the construction and visualization of the “component-target-pathway” interaction network.2.7 Component-target-pathway network
After obtaining all the interaction information of the active ingredient compounds, the target proteins, and the pathways, the “Component-Target-Pathway” network was constructed using Cystoscope 3.6.1 software. In this network, nodes represent components, targets, and pathways, and edges represent interactions between each other. A hypothetical schematic diagram of the target proteins involved in the pathways was drawn. Based on the network model map, a pathway involving active components and target proteins for this disease is initially explored to provide a preliminary theoretical basis for the design of targeted drugs.2.8 Molecular docking
Target proteins were modified using Pymol software, including hydrogenation, water content, optimization, and repair of amino acid residues. The target proteins' docking pockets were constructed to test the ligands. Interaction between active ingredients and targets was evaluated based on binding energy values by means of AutoDock. The smaller the docking binding energy, the better the molecule will bind to the target protein. When the binding energy is less than 0, the ligand and receptor can bind spontaneously. It is generally believed that energy values below 0 indicate there is a certain binding activity between the target and the ligand; energy values below −5.0 kJ mol−1 indicate that the molecule has a good binding activity for the target, and values below −10.0 kJ mol−1 indicate a strong binding activity.38 The 3D structures of the prepared compounds were used as ligands for molecular docking, and the receptors were the target proteins.39–44 A size of CASP3′ grid box was set as x, y, z center coordinate i.e. 41.391, 14.109, 77.896 and x, y, z size coordinate i.e. 15, 15, 15. A size of MAPK14′ grid box was set as x, y, z center coordinate i.e. 20.647, 13.217, 31.550 and x, y, z size coordinate i.e. 15, 15, 15.3. Results
3.1 Screening of potential targets
Based on an extensive survey of the scientific literature, a total of 39 triterpenoids were identified; this includes one compound isolated and identified in our study and 38 other reported compounds. These were used to create a library of compounds for the network analysis (Table 1). The ADME data for the compounds is shown in ESI Table 2.† We used SwissTargetPrediction to predict the targets of the active compounds, and a total of 585 targets proteins were identified. A total of 33 potential target proteins of C. paliurus anti-diabetes compounds were identified, as shown in Table 2.| MOL ID | Compound | Degree | Betweenness centrality | Molecular formula | CAS |
|---|---|---|---|---|---|
| a * means the core active ingredients. | |||||
| M1 | 2α,3α,23-Trihydroxyurs-12-en-28-oic acid | 5 | 0.0017618 | C30H48O5 | 103974-74-9 |
| M2 | 3β-Hydroxy-urs-11-en-28,13-lactone | 5 | 0.0017618 | C30H46O3 | 35959-05-8 |
| M3 | 2α-Hydroxyursolic acid | 5 | 0.0017618 | C30H48O4 | 72881-13-1 |
| M4 | 2α,3α,23-Trihydroxyurs-12,20(30)-dien-28-oic acid | 5 | 0.0017618 | C30H46O5 | 143839-01-4 |
| M5 | 2α,3β,23-Trihydroxy-12-ene-28-ursolic acid | 5 | 0.0017618 | C30H48O5 | 464-92-6 |
| M6 | 3β-O-trans-Caffeoyl-morolic acid | 4 | 0.00594168 | C39H54O6 | 97534-10-6 |
| M7 | Actinidic acid | 4 | 0.00111799 | C30H46O5 | 341971-45-7 |
| M8 | Arjunolic acid | 4 | 0.00111799 | C30H48O5 | 465-00-9 |
| M9 | Corosolic acid | 5 | 0.00508509 | C30H48O4 | 4547-24-4 |
| M10 | Cyclocaric acid B | 5 | 0.0017618 | C30H46O5 | 182315-46-4 |
| M11 | Daucosterol | 5 | 0.00979396 | C35H60O6 | 474-58-8 |
| M12 | Hederagenin | 5 | 0.0017618 | C30H48O4 | 465-99-6 |
| M13 | Maslinic acid | 5 | 0.0017618 | C30H48O4 | 4373-41-5 |
| M14 | Olean-12-en-28-oic acid | 5 | 0.0017618 | C30H48O2 | 17990-43-1 |
| M15 | Oleanolic acid | 5 | 0.0017618 | C30H48O3 | 508-02-1 |
| M16 | Taraxerol | 5 | 0.00598173 | C30H50O0 | 127-22-0 |
| M17 | β-Amyrin | 5 | 0.00590412 | C30H50O | 559-70-6 |
| M18 | β-Amyrone | 4 | 0.00397423 | C30H48O | 638-97-1 |
| M19 | β-Sitosterol | 5 | 0.00270752 | C29H50O | 83-46-5 |
| M20 | Cyclocariosides I | 6 | 0.01121781 | C35H56O8 | 1644624-82-7 |
| M21 | Cyclocarioside N | 2 | 0.00019562 | C44H74O13 | 2093058-24-1 |
| M22* | 3β,23-Dihydroxy-1,12-dioxo-olean-28-oic acid | 6 | 0.01175987 | C30H46O6 | 2093058-20-7 |
| M23* | 3β,23,27-Trihydroxy-1-oxo-olean-12-ene-28-oic acid | 6 | 0.01175987 | C30H46O6 | 2093058-21-8 |
| M24* | 2α,3β,23-Trihydroxyurs-11-oxo-12-ene-28-oic acid | 6 | 0.00987916 | C30H46O6 | 107302-99-8 |
| M25 | Cyclocarioside II | 3 | 0.00464055 | C35H36O8 | 173294-76-3 |
| M26* | Arjunglucoside II | 7 | 0.01868779 | C36H58O10 | 62369-72-6 |
| M27 | Quadranoside IV | 6 | 0.00588117 | C36H58O10 | 267001-55-8 |
| M28* | α-Boswellic acid | 7 | 0.0127971 | C30H48O3 | 471-66-9 |
| M29 | 3α,4β,18α-3-Hydroxyurs-12-en-23-oic acid | 6 | 0.00860482 | C30H48O3 | 2243454-82-0 |
| M30 | β-Boswellic acid | 6 | 0.00860482 | C30H48O3 | 631-69-6 |
| M31 | Cyclocarioside H | 2 | 0.0001863 | C43H72O13 | 1403937-87-0 |
| M32 | Cyclocarioside J | 2 | 0.0001863 | C35H58O9 | 1644624-86-1 |
| M33* | 2α,3β,23-Trihydroxyoleana-11,13(18)-dien-28-oic acid | 5 | 0.01046971 | C30H46O5 | 6790-76-7 |
| M34 | Cyclocaric acid A | 5 | 0.00786033 | C30H46O3 | 21754-17-6 |
| M35 | (+)-Betulinic acid | 4 | 0.00368899 | C30H48O3 | 472-15-1 |
| M36* | Cyclocarioside K | 5 | 0.01444275 | C36H58O8 | 1644624-87-2 |
| M37 | Cyclocarioside III | 2 | 0.00041134 | C36H60O9 | 173294-77-4 |
| M38 | Cyclocarioside L | 2 | 0.00041134 | C38H62O12 | 2093058-22-9 |
| M39 | Cyclocarioside M | 1 | 0 | C40H64O13 | 2093058-23-0 |
| UniProt IDs | Protein name | Gene name | Betweenness centrality | Degree |
|---|---|---|---|---|
| P24666 | Low molecular weight phosphotyrosine protein phosphatase | ACP1 | 0.04820819 | 16 |
| P15121 | Aldose reductase | AKR1B1 | 0.00271300 | 2 |
| P10275 | Androgen receptor | AR | 0.01510033 | 6 |
| P42574 | Caspase-3 | CASP3 | 0.00164939 | 2 |
| P21554 | Cannabinoid receptor 1 (by homology) | CNR1 | 0.01369327 | 2 |
| P11511 | Cytochrome P450 19A1 | CYP19A1 | 0.02352865 | 8 |
| P03372 | Estrogen receptor alpha | ESR1 | 0.01221787 | 5 |
| P00734 | Thrombin | F2 | 0.00971275 | 5 |
| P12104 | Fatty acid binding protein intestinal | FABP2 | 0.00305104 | 3 |
| P15090 | Fatty acid binding protein adipocyte | FABP4 | 0.00487387 | 4 |
| P09038 | Basic fibroblast growth factor | FGF2 | 0.02471924 | 3 |
| P04035 | HMG-CoA reductase | HMGCR | 0.01564161 | 6 |
| P28845 | 11-Beta-hydroxysteroid dehydrogenase 1 | HSD11B1 | 0.14538756 | 24 |
| P80365 | 11-Beta-hydroxysteroid dehydrogenase 2 | HSD11B2 | 0.00419333 | 3 |
| O14920 | Inhibitor of nuclear factor kappa B kinase beta subunit | IKBKB | 0.00271300 | 2 |
| P60568 | Interleukin-2 | IL2 | 0.01315293 | 6 |
| P35968 | Vascular endothelial growth factor receptor 2 | KDR | 0.00265707 | 2 |
| Q16539 | MAP kinase p38 alpha | MAPK14 | 0.01369327 | 2 |
| P35228 | Nitric oxide synthase, inducible | NOS2 | 0.03106597 | 13 |
| Q96RI1 | Bile acid receptor FXR | NR1H4 | 0.00625265 | 4 |
| P04150 | Glucocorticoid receptor | NR3C1 | 0.00661281 | 4 |
| P42336 | PI3-kinase p110-alpha subunit | PIK3CA | 0.00164939 | 2 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | PPARA | 0.00487387 | 4 |
| Q03181 | Peroxisome proliferator-activated receptor delta | PPARD | 0.00487387 | 4 |
| P37231 | Peroxisome proliferator-activated receptor gamma | PPARG | 0.00487387 | 4 |
| P35354 | Cyclooxygenase-2 | PTGS2 | 0.00176559 | 2 |
| P18031 | Protein-tyrosine phosphatase 1B | PTPN1 | 0.20514580 | 28 |
| P10586 | Receptor-type tyrosine-protein phosphatase F (LAR) | PTPRF | 0.03748757 | 15 |
| O00767 | Acyl-CoA desaturase | SCD | 0.00139742 | 2 |
| P04278 | Testis-specific androgen-binding protein | SHBG | 0.01774612 | 7 |
| P40763 | Signal transducer and activator of transcription 3 | STAT3 | 0.04628878 | 9 |
| P11473 | Vitamin D receptor | VDR | 0.07454926 | 10 |
| P15692 | Vascular endothelial growth factor A | VEGFA | 0.03145277 | 4 |
3.2 Analysis of interactions between active components and target proteins
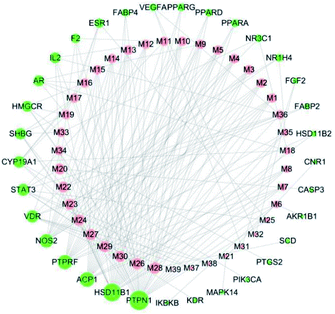
Cytoscape was used to visualize the component-target interactions. The component-target network consists of a total of 73 nodes and 213 edges; 33 of these nodes represent target proteins and 39 nodes active ingredients directly related to the anti-diabetic effects of C. paliurus (Fig. 2). The network shows that the anti-diabetic effects of C. paliurus may be the result of multi-component and multi-target protein effects. The top 7 components based on degree values were arjunglucoside II (M26), α-boswellic acid (M28), 3β,23-trihydroxyurs-11-oxo-12-ene-28-oic acid (M24), 3β,23,27-trihydroxy-1-oxo-olean-12-ene-ene-28-oic acid (M24), dihydroxy-1,12-dioxo-olean-28-oic acid (M22), 23-3β,23,27-trihydroxy-1-oxo-olean-12-ene-28-oic acid (M23), 2α,3β,23-trihydroxyoleana-11,13(18)-dien-28-oic acid (M33) and cyclocarioside K (M36). Of these, four compounds, M23, M24, M25 and M34, complied with the “Lipinski's Rule of Five”. These components are the key active ingredients underlying the antipyretic effects of C. paliurus and their structural formulas are shown in ESI Fig. 2.† The component-targets network revealed that several target proteins could interact with multiple compounds. Network values for active components and target proteins are listed in Tables 1 and 2.3.3 Analysis of target protein PPI network
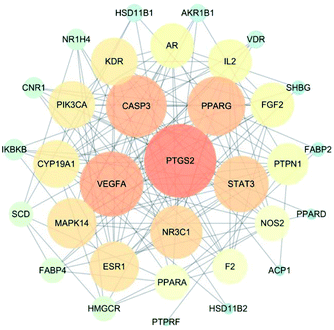
The interaction network for the anti-diabetic target proteins of C. paliurus, including 33 nodes and 153 edges, was constructed based on an interaction reliability greater than mid-range (Fig. 3). In protein interaction networks, the position of node proteins is not the same. The most important nodes are key nodes. The network node degree parameter is a common method for evaluating key nodes of the network. The PPI network showed that the median value of betweenness centrality and closeness centrality was greater than the median with high degree for a total of 15 target proteins. These 15 target proteins (Table 3) can be considered as core target proteins for the treatment of diabetes and may play an important role in the treatment of diabetes. Among them, PTGS2, VEGFA, CASP3, PPARG, STAT3, and NR3C1 are centrally located in the PPI network (Fig. 3), indicating that these proteins are involved in the pathogenesis of diabetes mellitus.| Gene symbol | Name | Betweenness centrality | Closeness centrality | Degree |
|---|---|---|---|---|
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 0.15607784 | 0.76190476 | 22 |
| VEGFA | Vascular endothelial growth factor A | 0.05924204 | 0.71111111 | 19 |
| CASP3 | Caspase 3 | 0.04400629 | 0.68085106 | 18 |
| PPARG | Peroxisome proliferator activated receptor gamma | 0.14458119 | 0.68085106 | 17 |
| STAT3 | Signal transducer and activator of transcription 3 | 0.02315961 | 0.66666667 | 16 |
| NR3C1 | Nuclear receptor subfamily 3 group C member 1 | 0.07062907 | 0.62745098 | 15 |
| MAPK14 | Mitogen-activated protein kinase 14 | 0.01666107 | 0.64000000 | 14 |
| ESR1 | Estrogen receptor 1 | 0.04629238 | 0.62745098 | 14 |
| KDR | Kinase insert domain receptor | 0.04561908 | 0.60377358 | 13 |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha | 0.01214958 | 0.60377358 | 13 |
| CYP19A1 | Cytochrome P450 family 19 subfamily a member 1 | 0.07381888 | 0.60377358 | 13 |
| IL2 | Interleukin 2 | 0.01306559 | 0.57142857 | 12 |
| AR | Androgen receptor | 0.01695036 | 0.58181818 | 12 |
| PTPN1 | Protein tyrosine phosphatase non-receptor type 1 | 0.09420179 | 0.59259259 | 11 |
| PPARA | Peroxisome proliferator activated receptor alpha | 0.03537093 | 0.54237288 | 9 |
3.4 GO analysis of target proteins
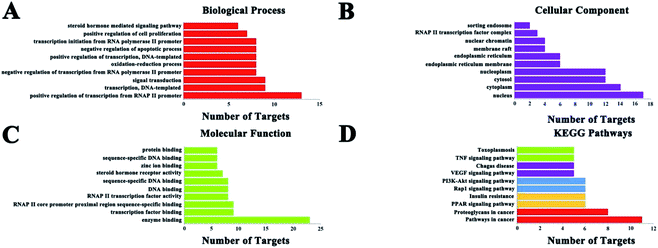
GO enrichment analysis results (Fig. 4) showed that the numbers of target proteins involved in the BP, MF, and CC categories were 116 (70.7%), 37 (22.5%), and 11 (6.7%), respectively. In the BP category, the target proteins were mainly involved in positive regulation of transcription from RNA polymerase II promoter (13, 39.4%), signal transduction (9, 29.3%), and transcription, DNA-templated (9, 29.3%). In the MF category, the target proteins were mainly involved in protein binding (23, 69.7%), zinc ion binding (9, 27.3%) and transcription factor activity, sequence-specific DNA binding (9, 27.3%). In the CC category, target proteins were mainly associated with the nucleus (17, 51.5%), cytoplasm (14, 42.4%), nucleoplasm (12, 36.4%) and cytosol (12, 36.4%). These results demonstrate that C. paliurus acts on diabetes probably by engaging the above mentioned pathways.3.5 KEGG classification of target proteins
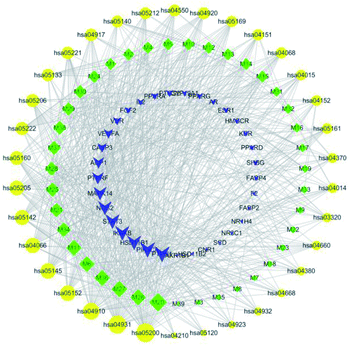
KEGG pathway annotation showed that 27 of the 33 (81.8%) potential target proteins were enriched and involved in 41 pathways; of these, 35 pathways were significantly correlated with the target proteins (P ≤ 0.05). The following pathways included the largest number of proteins: cancer pathways (11, 40.7%), proteoglycans in cancer (8, 29.6%), insulin resistance (6, 22.2%), acute myeloid leukemia (6, 22.2%), PI3K-Akt signaling pathway (6, 22.2%), PPAR signaling pathway (6, 22.2%), and so on. The top 10 pathways with the largest number of proteins involved are shown in Fig. 4, and the complete KEGG classification results are shown in Table 4. The target proteins involved in cancer pathways included| Term | Count | Percent% | P value | Genes |
|---|---|---|---|---|
| a Bold text means the important pathways reported and involved in diabetes. | ||||
| hsa05200:Pathways in cancer | 11 | 33.33 | 0.00 | AR, CASP3, PPARD, PTGS2, VEGFA, PPARG, PIK3CA, NOS2, IKBKB, FGF2, STAT3 |
| hsa05205:Proteoglycans in cancer | 8 | 24.24 | 0.00 | CASP3, MAPK14, VEGFA, ESR1, PIK3CA, FGF2, STAT3, KDR |
| hsa03320:PPAR signaling pathway | 6 | 18.18 | 0.00 | PPARA, PPARD, SCD, PPARG, FABP4, FABP2 |
| hsa04931:Insulin resistance | 6 | 18.18 | 0.00 | PPARA, PTPRF, PIK3CA, PTPN1, IKBKB, STAT3 |
| hsa04015:Rap1 signaling pathway | 6 | 18.18 | 0.00 | MAPK14, CNR1, VEGFA, PIK3CA, FGF2, KDR |
| hsa04151:PI3K-Akt signaling pathway | 6 | 18.18 | 0.02 | VEGFA, PIK3CA, IKBKB, FGF2, KDR, IL2 |
| hsa04370:VEGF signaling pathway | 5 | 15.15 | 0.00 | PTGS2, MAPK14, VEGFA, PIK3CA, KDR |
| hsa05142:Chagas disease | 5 | 15.15 | 0.00 | MAPK14, PIK3CA, NOS2, IKBKB, IL2 |
| hsa04668:TNF signaling pathway | 5 | 15.15 | 0.00 | CASP3, PTGS2, MAPK14, PIK3CA, IKBKB |
| hsa05145:Toxoplasmosis | 5 | 15.15 | 0.00 | CASP3, MAPK14, NOS2, IKBKB, STAT3 |
| hsa05160:Hepatitis C | 5 | 15.15 | 0.00 | PPARA, MAPK14, PIK3CA, IKBKB, STAT3 |
| hsa04014:Ras signaling pathway | 5 | 15.15 | 0.02 | VEGFA, PIK3CA, IKBKB, FGF2, KDR |
| hsa05206:MicroRNAs in cancer | 5 | 15.15 | 0.03 | CASP3, PTGS2, VEGFA, IKBKB, STAT3 |
| hsa05221:Acute myeloid leukemia | 4 | 12.12 | 0.00 | PPARD, PIK3CA, IKBKB, STAT3 |
| hsa05212:Pancreatic cancer | 4 | 12.12 | 0.00 | VEGFA, PIK3CA, IKBKB, STAT3 |
| hsa04917:Prolactin signaling pathway | 4 | 12.12 | 0.00 | MAPK14, ESR1, PIK3CA, STAT3 |
| hsa05222:Small cell lung cancer | 4 | 12.12 | 0.01 | PTGS2, PIK3CA, NOS2, IKBKB |
| hsa04066:HIF-1 signaling pathway | 4 | 12.12 | 0.01 | VEGFA, PIK3CA, NOS2, STAT3 |
| hsa04660:T cell receptor signaling pathway | 4 | 12.12 | 0.01 | MAPK14, PIK3CA, IKBKB, IL2 |
| hsa05169:Epstein-Barr virus infection | 4 | 12.12 | 0.02 | MAPK14, PIK3CA, IKBKB, STAT3 |
| hsa04152:AMPK signaling pathway | 4 | 12.12 | 0.02 | HMGCR, SCD, PPARG, PIK3CA |
| hsa04380:Osteoclast differentiation | 4 | 12.12 | 0.02 | MAPK14, PPARG, PIK3CA, IKBKB |
| hsa04068:FoxO signaling pathway | 4 | 12.12 | 0.02 | MAPK14, PIK3CA, IKBKB, STAT3 |
| hsa04910:Insulin signaling pathway | 4 | 12.12 | 0.02 | PTPRF, PIK3CA, PTPN1, IKBKB |
| hsa04550:Signaling pathways regulating pluripotency of stem cells | 4 | 12.12 | 0.02 | MAPK14, PIK3CA, FGF2, STAT3 |
| hsa05161:Hepatitis B | 4 | 12.12 | 0.02 | CASP3, PIK3CA, IKBKB, STAT3 |
| hsa04932:Non-alcoholic fatty liver disease (NAFLD) | 4 | 12.12 | 0.03 | PPARA, CASP3, PIK3CA, IKBKB |
| hsa05152:Tuberculosis | 4 | 12.12 | 0.04 | VDR, CASP3, MAPK14, NOS2 |
| hsa04923:Regulation of lipolysis in adipocytes | 3 | 9.09 | 0.02 | PTGS2, PIK3CA, FABP4 |
| hsa00140:Steroid hormone biosynthesis | 3 | 9.09 | 0.03 | HSD11B1, HSD11B2, CYP19A1 |
| hsa04210:Apoptosis | 3 | 9.09 | 0.03 | CASP3, PIK3CA, IKBKB |
| hsa05120:Epithelial cell signaling in Helicobacter pylori infection | 3 | 9.09 | 0.03 | CASP3, MAPK14, IKBKB |
| hsa04920:Adipocytokine signaling pathway | 3 | 9.09 | 0.04 | PPARA, IKBKB, STAT3 |
| hsa05140:Leishmaniasis | 3 | 9.09 | 0.04 | PTGS2, MAPK14, NOS2 |
| hsa05133:Pertussis | 3 | 9.09 | 0.04 | CASP3, MAPK14, NOS2 |
AR, CASP3, PPARD, PTGS2, VEGFA, PPARG, PIK3CA, NOS2, IKBKB, FGF2 and STAT3; the target proteins involved in insulin resistance included PPARA, PTPRF, PIK3CA, PTPN1, IKBKB and STAT3; and the target proteins involved in the PI3K-Akt signaling pathway included VEGFA, PIK3CA, IKBKB, FGF2, KDR and IL2. As can be seen, there are multiple target proteins in a pathway and the same target protein exists in multiple pathways. These results suggest that C. paliurus may exert its effects on diabetes by regulating these pathways via core target proteins.
3.6 Component-target-pathway network
The component-target-pathway network was constructed to visualize all interactions between target proteins and the anti-diabetic-related pathways. Based on the previous KEGG enrichment analysis, a component-target-pathway network was generated by connecting compounds, targets and pathways (Fig. 5). This network included 106 nodes (39 active compound nodes, 33 composite target protein nodes and 34 pathways nodes) and 745 edges. The component-target-pathway network results are shown in ESI Table 2 and ESI Fig. 1.†The network analysis showed that the median value of betweenness centrality and closeness centrality was greater than the median with high degree for a total of 15 pathways, including cancer (hsa05200), insulin resistance (hsa04931), HIF-1 signaling pathway (hsa04066), PI3K-Akt signaling pathway (hsa04151) etc. These 15 pathways with several diabetes-associated target proteins can be considered as core pathways for the treatment of diabetes. Among these pathways, insulin resistance, HIF-1 signaling, and PI3K-Akt signaling pathways have been shown to have a clear link with the occurrence of diabetes. As shown in Fig. 6, the active anti-inflammatory and anti-diabetic components present in C. paliurus can synergize with multiple target proteins within these pathways to form a multi-component-multi-target-multi-pathway mechanism.
3.7 Docking analysis
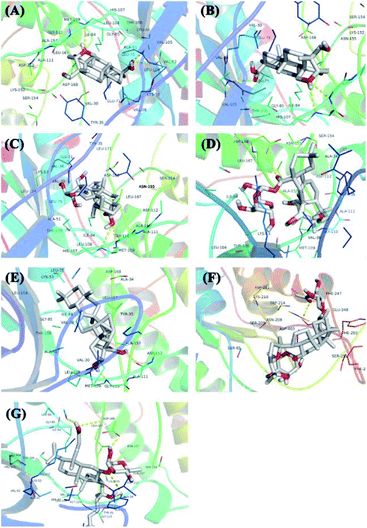
The protein structure was set to a rigid macromolecule, and the algorithm to local search parameters. Then, the object of molecular docking visualization was selected according to the lowest binding energy for docking. The core compounds and diabetes target proteins were ranked according to the docking binding energy. The top 10 core target proteins in the previous ranking were docked with 7 key medicinal components (Table 5). The docking results showed that the lowest binding energy for the 7 core medicinal components and CASP3 were all below −5.0 kJ mol−1, indicating that the 7 key medicinal components bound better to the CASP3 target protein. With the exception of 2α,3β,23-trihydroxyoleana-11,13(18)-dien-28-oic acid, the other 6 components showed the lowest docking binding energies with MAPK14 target protein when compared with the other 9 target proteins. Interactions between the 7 core components of C. paliurus and the previously mentioned 10 core target proteins with the lowest binding energy and the smallest binding energy were visualized using Pymol (Fig. 7 and 8).| No. | Compound name | PTGS2 | VEGFA | CASP3 | PPARG | STAT3 | NR3C1 | MAPK14 | ESR1 | KDR | PIK3CA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a Ligand means a substance that has the ability to recognize the receptor and can bind to it. The docking method is rigid docking. Algorithm is all local search parameters. | |||||||||||
| 1 | 3β,23-Dihydroxy-1,12-dioxo-olean-28-oic acid | −5.2 | −4.5 | −6.1 | 2.9 | −5.8 | −2.4 | −6.7 | −4.7 | −5 | −5.3 |
| 2 | 3β,23,27-Trihydroxy-1-oxo-olean-12-ene-28-oic acid | −5.2 | −4.5 | −6.1 | 3 | −5.8 | −2.4 | −6.7 | −4.7 | −4.9 | −5.3 |
| 3 | 2α,3β,23-Trihydroxyurs-11-oxo-12-ene-28-oic acid | −4.9 | −4.6 | −5.2 | 0 | −5.2 | −3.6 | −8.7 | −2.9 | −5 | −6.1 |
| 4 | Arjunglucoside II | −4.8 | −4.6 | −5.5 | 2.3 | −5.4 | 8.2 | −6.7 | −4.4 | −4.6 | −5.1 |
| 5 | α-Boswellic acid | −5.7 | −5.3 | −6.1 | −1.7 | −6 | −6.3 | −9.1 | −6.2 | −5.2 | −5.4 |
| 6 | 2α,3β,23-Trihydroxyoleana-11,13(18)-dien-28-oic acid | −4.5 | −4 | −6 | 1.7 | −5.3 | 16.5 | −0.3 | 3.1 | −4.7 | −2.4 |
| 7 | Cyclocarioside K | −4.3 | −4.9 | −4.9 | −2.9 | −5.7 | 4.5 | −8 | −5.4 | −4.1 | −6.7 |
| 8 | Ligand | −3.1 | −3.2 | −3.4 | −5.5 | −6.1 | −11.9 | −12.7 | −11.4 | −3 | −7.9 |
4. Discussion
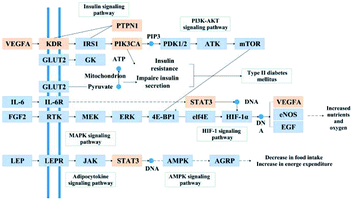
Used as a health tea, C. paliurus has a long history in the treatment of diabetes. In this study, 39 triterpenoids were selected to create a library of compounds, and network pharmacology technology was employed to explore the multiple pharmacological effects of triterpenoids from C. paliurus in the treatment of diabetes. We found that seven core ingredients found in the leaves of C. paliurus, including arjunglucoside II, α-boswellic acid, 3β,23-dihydroxy-1,12-dioxo-olean-28-oic acid, 23-trihydroxyurs-11-oxo-12-ene-28-oic acid, 3β,23,27-trihydroxy-1-oxo-olean-12-ene-28-oic acid, 2α,3β,23-trihydroxyoleana-11,13(18)-dien-28-oic acid and cyclocarioside K, showed potential for the treatment of diabetes.Among them, α-boswellic acid has been shown to exert some anti-diabetic effects by suppressing the expression of proinflammatory cytokines.45 Cyclocarioside K is a new epoxydammarane triterpenoid saponin isolated from the ethanol leaf extracts of C. paliurus.46 Arjunglucoside II is a common glucoside derivative of arjunic acid, found in Terminalia arjuna.47 The DPP-IV inhibitory activity of arjunic acid and its derivatives may have demonstrable therapeutic benefits in diabetes patients with cardiovascular comorbidities.48 3β,23-Dihydroxy-1,12-dioxo-olean-28-oicacid and 3β,23,27-trihydroxy-1-oxo-olean-12-ene-28-oic acid are new triterpenoid saponins isolated from the CH3Cl-soluble extract of C. paliurus leaves, whereas 23-trihydroxyurs-11-oxo-12-ene-28-oic acid and 2α,3β,23-trihydroxyoleana-11,13(18)-dien-28-oic acid are also known triterpenoid saponins isolated from the CH3Cl-soluble extract of the leaves of C. paliurus.49 These triterpenoid saponins may have an effect on diabetes by inhibiting apolipoprotein B48 overproduction.49,50 Based on the interaction network of C. paliurus's anti-diabetes target proteins, we found that STAT3, PTPN1, PTGS2, VEGFA, CASP3, etc. were the key target proteins. These targets may play an important role in C. paliurus's anti-diabetes effects. According to the results of the component-target and PPI networks, STAT3, CASP3, ACP1, PTPN1, HSD11B1 and other targets were regulated by multiple components, such as arjunglucoside II, cyclocarioside K, quadranoside IV, corosolic acid, cyclocariosides I and so on. Some of these ingredients have been proven to have strong anti-diabetic effects. For example, corosolic acid can reduce glucose levels in human hepatocellular carcinoma cells, as well as in zebrafish and in rats.51 Molecular docking is one of the most widely used methods to investigate binding of a compound to a target protein.39–42 We used AutoDock software to analyze molecular docking. Compounds with docking energies below −5 kJ mol−1 were considered to bind well to their targets. In the PPI network diagram, betweenness is defined as the number of shortest paths between pairs of nodes that run through nodes, and degree centrality is the most direct measure of the importance of a network node. The larger the node degree value, the more important the node is in the network. We used two indicators (betweenness centrality and closeness centrality) as the criteria for screening targets. Only target proteins with betweenness centrality and closeness centrality values greater than the median for all target proteins were selected. Then, all target proteins were sorted according to the degree value. Based on the docking results between the top ten target proteins and core compounds, we observed that although the docking energy of some core compounds to the target proteins was very small and the docking results were good, there were also poor docking results. Only STAT3 showed docking energies for all compounds below −5.0 kJ mol−1. STAT3 is an important signal transduction factor that participates in the signal transduction process of various cytokines, such as interferon, interleukins and growth factors, and it forms part of the JAK2/STAT3 signaling pathway.52,53 Many studies have shown that the JAK2/STAT3 signaling pathway is an important pathway that mediates the signal transduction process of various cytokines and growth factors, and can regulate cell growth, differentiation, migration, apoptosis, autophagy, immunity, and metabolism.54,55 It is considered to play an important role in the development of diabetes.56,57 STAT3 may regulate diabetes by acting on SOCS3 and thereby affecting the activity of insulin receptor substrate 1 (IRS-1).58 SOCS3 can mediate central leptin resistance in obese patients.59 Leptin signal transduction in the hypothalamus can regulate liver glucose and lipid metabolism60 as well as liver gluconeogenesis.61 It seems evident that STAT3 plays an important role regulating disorders of glucose metabolism. In network pharmacology analysis, component-target networks are mainly used to screen core compounds and targets. Betweenness centrality and closeness centrality values greater than the median are also the main criteria for screening core target proteins. All eligible target proteins were sorted according to the degree value, and the one with the highest value was PTPN1. In component-target networks, the higher the degree of a target protein, the more the compounds that regulate it. The more active compounds bind to a protein target, the greater the possibility of producing a drug effect, and the higher the possibility of becoming the key node. PTPN1 is a non-transmembrane protein tyrosine phosphatase which functions mainly by regulating the levels of protein tyrosine phosphorylation in cells, and is involved in the regulation of multiple cell signaling pathways modulating cell proliferation, growth and migration.62 Many studies have found correlations between DNA sequence variations in the PTPN1 gene, SNP and T2DM.63 PTPN1 can regulate insulin signaling by modulating the expression of the PTP1B enzyme. PTP1B is a key factor that regulates various metabolic processes in the body and is closely associated with the occurrence and development of diseases, especially type 2 diabetes.64 Studies have shown that inhibiting the expression of the PTP1B enzyme can regulate insulin sensitivity in diabetic mice and improve insulin resistance, producing a therapeutic effect in diabetes.65 Therefore, we can study the potential pharmacological effects of C. paliurus on diabetes by analyzing the relationship between active compounds and core targets. According to the results of the component-target-protein network analysis, the insulin resistance pathway, the PI3K-Akt signaling pathway and the HIF-1 pathway may play an important role in the anti-diabetes effects of C. paliurus. As shown in Fig. 5, the active anti-diabetes ingredients of C. paliurus can synergize with various target proteins in these pathways, resulting in a multi-component, multi-target and multi-pathway mechanism of action. Insulin resistance, a condition in which cells are resistant to the action of insulin, is often found in obese and diabetic patients and is closely related with the occurrence of diabetes.66 It is associated with increased activity of phosphatases, including PTPs, PTEN, and PP2A, and decreased activation of signaling molecules, such as PI3K/AKT, resulting in reduced GLUT4 translocation, glucose uptake and glycogen synthesis in skeletal muscle, as well as increased hepatic gluconeogenesis and decreased glycogen synthesis in the liver.67,68 Many studies have shown that this pathway is activated by changes in the levels of enzymes such as IL-6, Akt and IRS-1, or other abnormalities.69,70
HIF-1, a transcription factor which functions as a major regulator of oxygen homeostasis,71 is associated with diabetic nephropathy. HIF-1 expression indirectly affects renal oxygen metabolism.72,73 In hypoxic conditions, it can reduce damage to the body by regulating the expression of target genes coding for downstream factors related with the hypoxic response, and by modulating cell energy metabolism, glucose metabolism and apoptosis.74 Studies have shown that oxidative stress and microcirculatory disorders are important mechanisms underlying the development of diabetic nephropathy and other diseases.75 Antioxidant therapy can increase the expression of medullary HIF in diabetic kidneys and improve renal oxidative damage.76 HIF activation can attenuate changes in renal oxygen metabolism and mitochondrial function caused by diabetes, thereby reducing proteinuria and improving renal tubular interstitial damage.77 The PI3K-Akt signaling pathway plays an important role maintaining insulin stability.75 This signaling pathway is activated by many types of cellular stimuli or toxic insults and regulates fundamental cellular functions, such as transcription, translation, proliferation, growth, and survival.76 At present, there are studies showing that selective activation of the PI3K/AKT signaling pathway can modulate neuroprotection, angiogenesis, and islet β cell survival in diabetic rats.75 Many studies indicate that after insulin binds to its receptor, it will self-phosphorylate and phosphorylate IRS-2 tyrosine sites.77 Phosphorylated IRS-2 binds to the p85 subunit of PI3K to further activate PI3K and to activate Akt after phosphorylation.78 Akt2 is a subtype of Akt, a serine/threonine kinase and an important signaling molecule located downstream of PI3K79,80 which can be regulated by modulating Gsk-3β, GLUT-4, etc. A series of downstream molecules promote glycogen synthesis, glucose transport, and other pathways to regulate glucose metabolism, thereby regulating diabetes.70
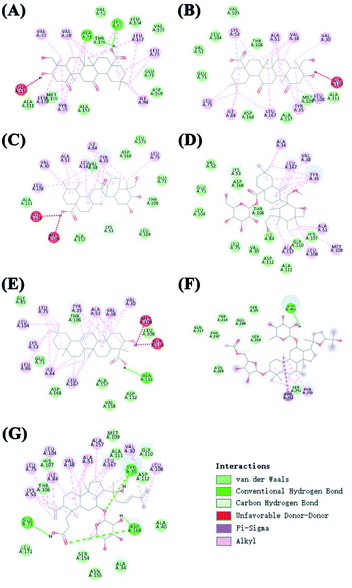
Receptor theory is the basic theory of pharmacodynamics. It postulates that the combination of drugs and target proteins is the basis of pharmacodynamics. Molecular docking is a method to evaluate binding of active drug ingredients to target proteins. In network pharmacology research, molecular docking is commonly used to study interactions between small molecules and key target proteins of the network, and to validate key target proteins identified in the network based on the interaction between active ingredients and target proteins.81 As shown in Fig. 7 and 8, molecular docking analysis showed that hydrogen bonds can form spontaneously between the 7 core compounds and the target proteins. These studies showed that the number of hydrogen bonds between the 4 core compounds and the target proteins surpassed 4, indicating that the core components bound well to the key target proteins of the network. Although the molecular docking results support the key node network findings, more in vivo or in vitro experiments are needed to verify the effects of ingredients on target proteins.
5. Conclusions
In summary, in this study we applied the network pharmacological approach to investigate the main target proteins of C. paliurus compounds with anti-diabetes activity by constructing a target interaction network, and used molecular docking methods to validate the key findings. Our results indicate that seven compounds, including arjunglucoside II, α-boswellic acid, 3β,23-dihydroxy-1,12-dioxo-olean-28-oic acid, 23-trihydroxyurs-11-oxo-12-ene-28-oic acid, 3β,23,27-trihydroxy-1-oxo-olean-12-ene-28-oic acid, 2α,3β,23-trihydroxyoleana-11,13(18)-dien-28-oic acid and cyclocarioside K show potential for the treatment of diabetes. These triterpene compounds were predicted to interact with PTGS2, VEGFA, CASP3, and other enzymes. These results provide valuable insights into the synergistic mechanism of action of natural medicines. In addition, biological process and pathway enrichment analysis of the target proteins of the active ingredients of C. paliurus enhanced our understanding of its mechanism of action in diabetes. Our study provides scientific evidence supporting the use of C. paliurus for the treatment of diabetes and confirms that modern technologies can be used to explore the therapeutic value of natural medicines.Data availabIlity statement
All datasets generated for this study are included in the article/ESI.†Author contributions statement
Z. L., Y. T., N. L., Z. Z., and J. L. conceived the experiment. Z. L. collected data, conducted the analytical part, wrote the first version of the manuscript. Y. T., N. L., Z. Z., and J. L. revised the manuscript. J. L. finalized the manuscript. All authors read and approved the final manuscript.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This research work was financially supported by Zhejiang Provincial Key Research and Development Program (2018C02021) and the Ten Thousand Talent Program of Zhejiang Province (2019).Notes and references
- J. M. Baena-Díez, J. Peñafiel, I. Subirana, R. Ramos, R. Elosua, A. Marín-Ibañez, M. J. Guembe, F. Rigo, M. J. Tormo-Díaz, C. Moreno-Iribas, J. J. Cabré, A. Segura, M. García-Lareo, A. Gómez De La Cámara, J. Lapetra, M. Quesada, J. Marrugat, M. J. Medrano, J. Berjón, G. Frontera, D. Gavrila, A. Barricarte, J. Basora, J. M. García, N. C. Pavone, D. Lora-Pablos, E. Mayoral, J. Franch, M. Mata, C. Castell, A. Frances and M. Grau, Risk of Cause-Specific Death in Individuals with Diabetes: A Competing Risks Analysis, Diabetes Care, 2016, 39, 1987–1995 CrossRef.
- N. M. Maruthur, M. O. Gribble, W. L. Bennett, S. Bolen, L. M. Wilson, P. Balakrishnan, A. Sahu, E. Bass, W. H. L. Kao and J. M. Clark, The Pharmacogenetics of Type 2 Diabetes: A Systematic Review, Diabetes Care, 2014, 37, 876 CrossRef CAS.
- C. I. Ezema, E. Omeh, O. Onyeso, C. C. Anyachukwu, M. J. Nwankwo, A. Amaeze, J. U. Ugwulebor, E. O. Nna, E. O. Ohotu and I. Ugwuanyi, The Effect of an Aerobic Exercise Programme On Blood Glucose Level, Cardiovascular Parameters, Peripheral Oxygen Saturation, and Body Mass Index Among Southern Nigerians with Type 2 Diabetes Mellitus, Undergoing Concurrent Sulfonylurea and Metformin Treatment, Malays. J. Med. Sci., 2019, 26, 88–97 Search PubMed.
- S. H. Havale and M. Pal, Medicinal Chemistry Approaches to the Inhibition of Dipeptidyl Peptidase-4 for the Treatment of Type 2 Diabetes, Bioorg. Med. Chem., 2009, 17, 1783–1802 CrossRef CAS.
- A. J. Krentz and C. J. Bailey, Oral Antidiabetic Agents, Drugs, 2005, 65, 385–411 CrossRef CAS.
- J. J. Neumiller, J. R. White and R. K. Campbell, Sodium-Glucose Co-Transport Inhibitors, Drugs, 2010, 70, 377–385 CrossRef CAS.
- P. Kaur, A. Mittal, S. Nayak, M. Vyas, V. Mishra and G. Khatik, Current Strategies and Drug Targets in the Management of Type 2 Diabetes Mellitus, Curr. Drug Targets, 2018, 19, 1738–1766 CrossRef CAS.
- Z. Zhang, Y. Cao, Y. J. Tao, E. Meng, J. H. Tang, Y. C. Liu and F. P. Li, Sulfonylurea and Fracture Risk in Patients with Type 2 Diabetes Mellitus: A Meta-Analysis, Diabetes Res. Clin. Pract., 2019, 159, S168–S8227 Search PubMed.
- L. Wang, P. Gao, M. Zhang, Z. Huang, D. Zhang, Q. Deng, Y. Li, Z. Zhao, X. Qin, D. Jin, M. Zhou, X. Tang, Y. Hu and L. Wang, Prevalence and Ethnic Pattern of Diabetes and Prediabetes in China in 2013, JAMA, 2017, 317, 2515–2523 CrossRef.
- S. S. Kolhe and P. R. Rachh, Review On Potent Anti-Diabetic Plants Or Herbs From Traditional Medicine, J. Drug Deliv. Therapeut., 2018, 8, 92–98 CrossRef CAS.
- Q. Ge, L. Chen, Y. Yuan, L. Liu, F. Feng, P. Lv, S. Ma, K. Chen and Q. Yao, Network Pharmacology-Based Dissection of the Anti-Diabetic Mechanism of Lobelia Chinensis, Front. Pharmacol., 2020, 11, 347 CrossRef CAS.
- H. Zhang, H. Matsuda, A. Kumahara, Y. Ito, S. Nakamura and M. Yoshikawa, New Type of Anti-Diabetic Compounds From the Processed Leaves of Hydrangea Macrophylla Var. Thunbergii (Hydrangeae Dulcis Folium), Bioorg. Med. Chem. Lett., 2007, 17, 4972–4976 CrossRef CAS.
- L. Zhai, Z. Ning, T. Huang, B. Wen, C. Liao, C. Lin, L. Zhao, H. Xiao and Z. Bian, Cyclocarya paliurus Leaves Tea Improves Dyslipidemia in Diabetic Mice: A Lipidomics-Based Network Pharmacology Study, Front. Pharmacol., 2018, 9, 973 CrossRef.
- Z. Yang, K. Ouyang, J. Zhao, H. Chen, L. Xiong and W. Wang, Structural Characterization and Hypolipidemic Effect of Cyclocarya Paliurus Polysaccharide in Rat, Int. J. Biol. Macromol., 2016, 91, 1073–1080 CrossRef CAS.
- H. Kurihara, H. Fukami, A. Kusumoto, Y. Toyoda, H. Shibata, Y. Matsui, S. Asami and T. Tanaka, Hypoglycemic Action of Cyclocarya paliurus (Batal.) Iljinskaja in Normal and Diabetic Mice, Biosci., Biotechnol., Biochem., 2003, 67, 877–880 CrossRef CAS.
- S. Li, J. Li, X. Guan, J. Li, S. Deng, L. Li, M. Tang, J. Huang, Z. Chen and R. Yang, Hypoglycemic Effects and Constituents of the Barks of Cyclocarya Paliurus and their Inhibiting Activities to Glucosidase and Glycogen Phosphorylase, Fitoterapia, 2011, 82, 1081–1085 CrossRef CAS.
- Y. Ma, C. Jiang, N. Yao, Y. Li and Q. Wang, Antihyperlipidemic Effect of Cyclocarya Paliurus (Batal.) Iljinskaja Extract and Inhibition of Apolipoprotein B48 Overproduction in Hyperlipidemic Mice, J. Ethnopharmacol., 2015, 166, 286–296 CrossRef.
- Q. Wang, C. Jiang, S. Fang, J. Wang, Y. Ji, X. Shang, Y. Ni, Z. Yin and J. Zhang, Antihyperglycemic, Antihyperlipidemic and Antioxidant Effects of Ethanol and Aqueous Extracts of Cyclocarya Paliurus Leaves in Type 2 Diabetic Rats, J. Ethnopharmacol., 2013, 150, 1119–1127 CrossRef.
- J. Xie, D. Cai-jun, N. Shao-Ping, L. Feng, W. Zhi-Jun, S. Ming-Yue and M. Xie, Extraction, Chemical Composition and Antioxidant Activity of Flavonoids From Cyclocarya Paliurus (Batal.) Iljinskaja Leaves, Food Chem., 2015, 186, 186–197 Search PubMed.
- J. Xie, M. Xie, S. Nie, M. Shen, Y. Wang and C. Li, Isolation, Chemical Composition and Antioxidant Activities of a Water-Soluble Polysaccharide From Cyclocarya Paliurus (Batal.) Iljinskaja, Food Chem., 2010, 119, 1626–1632 CrossRef CAS.
- H. Yoshitomi, R. Tsuru, L. Li, J. Zhou, M. Kudo, T. Liu and M. Gao, Cyclocarya Paliurus Extract Activates Insulin Signaling Via Sirtuin1 in C2C12 Myotubes and Decreases Blood Glucose Level in Mice with Impaired Insulin Secretion, PLoS One, 2017, 12, e183988 CrossRef.
- C. Jiang, Q. Wang, Y. Wei, N. Yao, Z. Wu, Y. Ma, Z. Lin, M. Zhao, C. Che, X. Yao, J. Zhang and Z. Yin, Cholesterol-Lowering Effects and Potential Mechanisms of Different Polar Extracts From Cyclocarya Paliurus Leave in Hyperlipidemic Mice, J. Ethnopharmacol., 2015, 176, 17–26 CrossRef CAS.
- W. Lin, Y. Li, Q. Lu, H. Lu and J. Li, Combined Analysis of the Metabolome and Transcriptome Identified Candidate Genes Involved in Phenolic Acid Biosynthesis in the Leaves of Cyclocarya Paliurus, Int. J. Mol. Sci., 2020, 21, 1337 CrossRef CAS.
- C. Sun, X. Shang, H. Ding, Y. Cao and S. Fang, Natural Variations in Flavonoids and Triterpenoids of Cyclocarya Paliurus Leaves, J. For. Res., 2020, 31 DOI:10.1007/s11676-020-01139-1.
- J. G. A. W. Du, M. Zhang and Li Chen, Compound K, a Final Intestinal Metabolite of Ginsenosides, Enhances Insulin Secretion in MIN6 Pancreatic β-Cells by Upregulation of GLUT2, Fitoterapia, 2013, 87, 84–88 CrossRef.
- A. E. Ardiles, Á. González-Rodríguez, M. J. N. Ez, N. R. Perestelo, V. Pardo, I. A. Jiménez, Á. M. Valverde and I. L. Bazzocchi, Studies of Naturally Occurring Friedelane Triterpenoids as Insulin Sensitizers in the Treatment Type 2 Diabetes Mellitus, Phytochemistry, 2012, 84, 116–124 CrossRef CAS.
- D. K. Burdi, S. N. Qureshi and A. B. Ghanghro, An Overview of Available Hypoglycemic Triterpenoids and Saponins to Cure Diabetes Mellitus, Adv. Life Sci., 2014, 1, 119–128 Search PubMed.
- J. Han, N. Tuan, M. Park, K. Quan, J. Oh, K. Heo, M. Na and C. Myung, Cucurbitane Triterpenoids From the Fruits of Momordica Charantia Improve Insulin Sensitivity and Glucose Homeostasis in Streptozotocin-Induced Diabetic Mice, Mol. Nutr. Food Res., 2018, 62, 1700769 CrossRef.
- K. N. Zhu, C. H. Jiang, Y. S. Tian, N. Xiao, Z. F. Wu, Y. L. Ma, Z. Lin, S. Z. Fang, X. L. Shang, K. Liu, J. Zhang, B. L. Liu and Z. Q. Yin, Two Triterpeniods From Cyclocarya Paliurus (Batal) Iljinsk (Juglandaceae) Promote Glucose Uptake in 3T3-L1 Adipocytes: The Relationship to AMPK Activation, Phytomedicine, 2015, 22, 837–846 CrossRef CAS.
- S. Yue, L. Xin, Y. Fan, S. Li, Y. Tang, J. Duan, H. Guan and C. Wang, Herb Pair Danggui-Honghua: Mechanisms Underlying Blood Stasis Syndrome by System Pharmacology Approach, Sci. Rep., 2017, 7, 40318 CrossRef CAS.
- M. U. Kakar, M. Naveed, M. Saeed, S. Zhao, M. Rasheed, S. Firdoos, R. Manzoor, Y. Deng and R. Dai, A Review On Structure, Extraction, and Biological Activities of Polysaccharides Isolated From Cyclocarya Paliurus (Batalin) Iljinskaja, Int. J. Biol. Macromol., 2020, 156, 420–429 CrossRef CAS.
- Z. Wu and T. Ding, Seed Plants of China, Kunming: Yunnan Science and Technology Press., 1999 Search PubMed.
- Z. Li, X. Ma, D. Wang, Y. Li, C. Wang and X. Jin, Evolution of Plastid Genomes of Holcoglossum (Orchidaceae) with Recent Radiation, BMC Evol. Biol., 2019, 19, 63 CrossRef.
- J. Ru, P. Li, J. Wang, W. Zhou, B. Li, C. Huang, P. Li, Z. Guo, W. Tao, Y. Yang, X. Xu, Y. Li, Y. Wang and L. Yang, TCMSP: A Database of Systems Pharmacology for Drug Discovery From Herbal Medicines, J. Cheminf., 2014, 6, 13 Search PubMed.
- D. Gfeller, O. Michielin and V. Zoete, Shaping the Interaction Landscape of Bioactive Molecules, Bioinformatics, 2013, 29, 3073–3079 CrossRef CAS.
- X. Liu, J. Wu, D. Zhang, K. Wang, X. Duan and X. Zhang, A Network Pharmacology Approach to Uncover the Multiple Mechanisms of Hedyotis Diffusa Willd. On Colorectal Cancer, J. Cheminf., 2018, 2018, 6517034 Search PubMed.
- C. von Mering, L. J. Jensen, B. Snel, S. D. Hooper, M. Krupp, M. Foglierini, N. Jouffre, M. A. Huynen and P. Bork, STRING: Known and Predicted Protein-Protein Associations, Integrated and Transferred Across Organisms, Nucleic Acids Res., 2004, 33, D433–D437 CrossRef.
- R. Yu, L. Chen, R. Lan, R. Shen and P. Li, Computational Screening of Antagonists Against the SARS-CoV-2 (COVID-19) Coronavirus by Molecular Docking, Int. J. Antimicrob. Agents, 2020, 56, 106012 CrossRef CAS.
- X. G. Liu, M. C. Lv, M. Y. Huang, Y. Q. Sun, P. Y. Gao and D. Q. Li, A Network Pharmacology Study On the Triterpene Saponins from Medicago Sativa L. For the Treatment of Neurodegenerative Diseases, J. Food Biochem., 2019, 43, e12955 Search PubMed.
- J. Li, X. Ma, C. Liu, H. Li, J. Zhuang, C. Gao, C. Zhou, L. Liu, K. Wang and C. Sun, Exploring the Mechanism of Danshen against Myelofibrosis by Network Pharmacology and Molecular Docking, Evid. base Compl. Alternative Med., 2018, 2018, 1–11 Search PubMed.
- R. Liu, C. Su, Y. Xu, K. Shang, K. Sun, C. Li and J. Lu, Identifying Potential Active Components of Walnut Leaf that Action Diabetes Mellitus through Integration of UHPLC-Q-Orbitrap HRMS and Network Pharmacology Analysis, J. Ethnopharmacol., 2020, 253, 112659 CrossRef CAS.
- Q. Luo, X. Shi, J. Ding, Z. Ma, X. Chen, Y. Leng, X. Zhang and Y. Liu, Network Pharmacology Integrated Molecular Docking Reveals the Antiosteosarcoma Mechanism of Biochanin A, Evid. base Compl. Alternative Med., 2019, 2019, 1–10 Search PubMed.
- Y. Xiong, Y. Yang, W. Xiong, Y. Yao, H. Wu and M. Zhang, Network Pharmacology-Based Research On the Active Component and Mechanism of the Antihepatoma Effect of Rubia Cordifolia L., J. Cell. Biochem., 2019, 120, 12461–12472 CrossRef CAS.
- C. Mu, Y. Sheng, Q. Wang, A. Amin and Y. Xie, Potential Compound From Herbal Food of Rhizoma Polygonati for Treatment of COVID-19 Analyzed by Network Pharmacology and Molecular Docking Technology, J. Funct. Foods, 2020, 104149 CrossRef.
- H. P. T. Ammon, Boswellic Extracts and 11-Keto-β-Boswellic Acids Prevent Type 1 and Type 2 Diabetes Mellitus by Suppressing the Expression of Proinflammatory Cytokines, Phytomedicine, 2019, 63, 153002 CrossRef CAS.
- B. Cui and S. Li, New Triterpenoid Saponins From the Leaves of Cyclocarya Paliurus, Chin. Chem. Lett., 2015, 26, 585–589 CrossRef CAS.
- T. Honda, T. Murae, T. Tsuyuki, T. Takahashi and M. Sawai, Arjungenin, Arjunglucoside I, and Arjunglucoside II. A New Triterpene and New Triterpene Glucosides From Terminalia Arjuna, Bull. Chem. Soc. Jpn., 1976, 49, 3213–3218 CrossRef CAS.
- I. R. Mohanty, M. Borde, S. Kumar C and U. Maheshwari, Dipeptidyl Peptidase IV Inhibitory Activity of Terminalia Arjuna Attributes to its Cardioprotective Effects in Experimental Diabetes: In Silico, in Vitro and in Vivo Analyses, Phytomedicine, 2019, 57, 158–165 CrossRef CAS.
- Z. Wu, F. Meng, L. Cao, C. Jiang, M. Zhao, X. Shang, S. Fang, W. Ye, Q. Zhang, J. Zhang and Z. Yin, Triterpenoids From Cyclocarya Paliurus and their Inhibitory Effect On the Secretion of Apoliprotein B48 in Caco-2 Cells, Phytochemistry, 2017, 142, 76–84 CrossRef CAS.
- C. Phillips, G. Murugasu, D. Owens, P. Collins, A. Johnson and G. H. Tomkin, Improved Metabolic Control Reduces the Number of Postprandial Apolipoprotein B-48-containing Particles in Type 2 Diabetes, Atherosclerosis, 2000, 148, 283–291 CrossRef CAS.
- S. Xu, G. Wang, W. Peng, Y. Xu, Y. Zhang, Y. Ge, Y. Jing and Z. Gong, Corosolic Acid Isolated From Eriobotrya Japonica Leaves Reduces Glucose Level in Human Hepatocellular Carcinoma Cells, Zebrafish and Rats, Sci. Rep., 2019, 9, 4388 CrossRef.
- J. Jiang, C. Yu, X. Guo, H. Zhang, S. Tian, K. Cai, Z. He and C. Sun, G Protein-Coupled Receptor GPR87 Promotes the Expansion of PDA Stem Cells through Activating JAK2/STAT3, Mol. Ther.--Oncolytics, 2020, 17, 384–393 CrossRef CAS.
- S. J. Thomas, J. A. Snowden, M. P. Zeidler and S. J. Danson, The Role of JAK/STAT Signalling in the Pathogenesis, Prognosis and Treatment of Solid Tumours, Br. J. Cancer, 2015, 113, 365–371 CrossRef CAS.
- J. Zhou, Y. Jiang, J. Zhao, H. Zhang, J. Fu, P. Luo, Y. Ma, D. Zou, H. Gao, J. Hu, Y. Zhang and Z. Jing, Dp44mT, an Iron Chelator, Suppresses Growth and Induces Apoptosis Via RORA-mediated NDRG2-IL6/JAK2/STAT3 Signaling in Glioma, Cell. Oncol., 2020, 43, 461–475 CrossRef CAS.
- G. L. Zhao, L. M. Yu, W. L. Gao, W. X. Duan, B. Jiang, X. D. Liu, B. Zhang, Z. H. Liu, M. E. Zhai, Z. X. Jin, S. Q. Yu and Y. Wang, Berberine Protects Rat Heart From Ischemia/Reperfusion Injury Via Activating JAK2/STAT3 Signaling and Attenuating Endoplasmic Reticulum Stress, Acta Pharmacol. Sin., 2016, 37, 354–367 CrossRef CAS.
- C. M. Taniguchi, B. Emanuelli and C. R. Kahn, Critical Nodes in Signalling Pathways: Insights Into Insulin Action, Nat. Rev. Mol. Cell Biol., 2006, 7, 85–96 CrossRef CAS.
- Y. Gao, M. Zhang, R. Zhang, L. You, T. Li and R. H. Liu, Whole Grain Brown Rice Extrudate Ameliorates the Symptoms of Diabetes by Activating the IRS1/PI3K/AKT Insulin Pathway in Db/Db Mice, J. Agric. Food Chem., 2019, 67, 11657–11664 CrossRef CAS.
- L. Rui, M. Yuan, D. Frantz, S. Shoelson and M. F. White, SOCS-1 and SOCS-3 Block Insulin Signaling by Ubiquitin-Mediated Degradation of IRS1 and IRS2, J. Biol. Chem., 2002, 277, 42394–42398 CrossRef CAS.
- A. L. Rodrigues, E. G. de Moura, M. C. F. Passos, I. H. Trevenzoli, E. P. S. da Coceição, I. T. Bonono, J. F. N. Neto and P. C. Lisboa, Postnatal Early Overfeeding Induces Hypothalamic Higher SOCS3 Expression and Lower STAT3 Activity in Adult Rats, J. Nutr. Biochem., 2011, 22, 109–117 CrossRef CAS.
- G. J. Morton and M. W. Schwartz, Leptin and the Central Nervous System Control of Glucose Metabolism, Physiol. Rev., 2011, 91, 389–411 CrossRef CAS.
- L. Cheng, Y. Yu, A. Szabo, Y. Wu, H. Wang, D. Camer and X. Huang, Palmitic Acid Induces Central Leptin Resistance and Impairs Hepatic Glucose and Lipid Metabolism in Male Mice, J. Nutr. Biochem., 2015, 26, 541–548 CrossRef CAS.
- C. E. Nunes-Xavier, O. Aurtenetxe, L. Zaldumbide, R. López-Almaraz, A. Erramuzpe, J. M. Cortés, J. I. López and R. Pulido, Protein Tyrosine Phosphatase PTPN1 Modulates Cell Growth and Associates with Poor Outcome in Human Neuroblastoma, Diagn. Pathol., 2019, 14, 134 CrossRef CAS.
- J. Xu, L. Li, Z. Qian, J. Hong, S. Shen and W. Huang, Reduction of PTP1B by RNAi Upregulates the Activity of Insulin Controlled Fatty Acid Synthase Promoter, Biochem. Biophys. Res. Commun., 2005, 329, 538–543 CrossRef CAS.
- S. Legeay, P. Fautrat, J. B. Norman, G. Antonova, S. Kennard, T. Bruder-Nascimento, V. S. Patel, S. Faure and E. J. B. de Chantemèle, Selective Deficiency in Endothelial PTP1B Protects From Diabetes and Endoplasmic Reticulum Stress-Associated Endothelial Dysfunction Via Preventing Endothelial Cell Apoptosis, Biomed. Pharmacother., 2020, 127, 110200 CrossRef CAS.
- C. M. Rondinone, J. M. Trevillyan, J. Clampit, R. J. Gum, C. Berg, P. Kroeger, L. Frost, B. A. Zinker, R. Reilly, R. Ulrich, M. Butler, B. P. Monia, M. R. Jirousek and J. F. Waring, Protein Tyrosine Phosphatase 1B Reduction Regulates Adiposity and Expression of Genes Involved in Lipogenesis, Diabetes, 2002, 51, 2405–2411 CrossRef CAS.
- A. Janus, E. Szahidewicz-Krupska, G. Mazur and A. Doroszko, Insulin Resistance and Endothelial Dysfunction Constitute a Common Therapeutic Target in Cardiometabolic Disorders, Mediators Inflammation, 2016, 2016, 3634948 CAS.
- D. Leto and A. R. Saltiel, Regulation of Glucose Transport by Insulin: Traffic Control of GLUT4, Nat. Rev. Mol. Cell Biol., 2012, 13, 383–396 CrossRef CAS.
- P. R. Shepherd and B. B. Kahn, Glucose Transporters and Insulin Action-Implications for Insulin Resistance and Diabetes Mellitus, N. Engl. J. Med., 1999, 341, 248–257 CrossRef CAS.
- S. Polsky and S. L. Ellis, Obesity, Insulin Resistance, and Type 1 Diabetes Mellitus, Curr. Opin. Endocrinol. Diabetes Obes., 2015, 22, 277 CrossRef CAS.
- Y. Gao, M. Zhang, R. Zhang, L. You, T. Li and R. H. Liu, Whole Grain Brown Rice Extrudate Ameliorates the Symptoms of Diabetes by Activating the IRS1/PI3K/AKT Insulin Pathway in Db/Db Mice, J. Agric. Food Chem., 2019, 67, 11657–11664 CrossRef CAS.
- E. S. Popravka, N. S. Linkova, S. V. Trofimova and V. K. Khavinson, HIF-1 as a Marker of Age-Related Diseases Associated with Tissue Hypoxia, Biol. Bull., 2018, 8, 497–508 CrossRef.
- M. T. Coughlan, A. L. Mibus and J. M. Forbes, Oxidative Stress and Advanced Glycation in Diabetic Nephropathy, Ann. N. Y. Acad. Sci., 2008, 1126, 190–193 CrossRef CAS.
- L. Nordquist, M. Friederich-Persson, A. Fasching, P. Liss, K. Shoji, M. Nangaku, P. Hansell and F. Palm, Activation of Hypoxia-Inducible Factors Prevents Diabetic Nephropathy, J. Am. Soc. Nephrol., 2015, 26, 328 CrossRef.
- C. Rosenberger, M. Khamaisi, Z. Abassi, V. Shilo, S. Zangen, M. Goldfarb, A. Shina, F. Zibertrest, K. Eckardt, S. Rosen and S. Heyman, Adaptation to Hypoxia in the Diabetic Rat Kidney, Kidney Int., 2008, 73, 34–42 CrossRef CAS.
- R. U. Ostrovskaya, S. V. Ivanov, T. A. Gudasheva and S. B. Seredenin, A Novel Dipeptide NGF Mimetic GK-2 Selectively Activating the PI3K/AKT Signaling Pathway Promotes the Survival of Pancreatic β-Cells in a Rat Model of Diabetes, Acta Naturae, 2019, 11, 48–57 CrossRef CAS.
- X. Li, H. Liu, J. Wang, J. Qin, Z. Bai, B. Chi, W. Yan and X. Chen, Curcumol Induces Cell Cycle Arrest and Apoptosis by Inhibiting IGF-1R/PI3K/Akt Signaling Pathway in Human Nasopharyngeal Carcinoma CNE-2 Cells, Phytother Res., 2018, 32, 2214–2225 CrossRef CAS.
- J. F. Wojtaszewski, B. F. Hansen, B. Kiens and E. A. Richter, Insulin Signaling in Human Skeletal Muscle: Time Course and Effect of Exercise, Diabetes, 1997, 46, 1775–1781 CrossRef CAS.
- L. R. Pearce, D. Komander and D. R. Alessi, The Nuts and Bolts of AGC Protein Kinases, Nat. Rev. Mol. Cell Biol., 2010, 11, 9–22 CrossRef CAS.
- G. Xi, X. Shen, C. Wai, M. White and D. Clemmons, Hyperglycemia Induces Vascular Smooth Muscle Cell Dedifferentiation by Suppressing Insulin Receptor Substrate-1-Mediated p53/KLF4 Complex Stabilization, J. Biol. Chem., 2018, 294, A118–A5398 Search PubMed.
- C. Kim, J. Lee, M. Kim and J. Hwang, Hypoglycemic Effect of Whole Grain Diet in C57BL/KsJ-db/db Mice by Activating PI3K/Akt and AMPK Pathways, J. Food Sci. Biotechnol., 2019, 28, 895–905 CrossRef CAS.
- W. Feng, H. Ao, S. Yue and C. Peng, Systems Pharmacology Reveals the Unique Mechanism Features of Shenzhu Capsule for Treatment of Ulcerative Colitis in Comparison with Synthetic Drugs, Sci. Rep., 2018, 8, 16160 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06846b |
| This journal is © The Royal Society of Chemistry 2020 |