DOI:
10.1039/D0RA06565J
(Paper)
RSC Adv., 2020,
10, 36920-36929
Isolation and characterization of novel acetylcholinesterase inhibitors from Ficus benghalensis L. leaves†
Received
29th July 2020
, Accepted 19th September 2020
First published on 7th October 2020
Abstract
Metabolic profiling of the crude methanolic extract of Ficus benghalensis leaves has revealed the presence of different phenolic and nitrogenous compounds including cerebrosides and tetrapyrrole pigments. A phytochemical study of the ethyl acetate fraction resulted in the identification of three known compounds, namely carpachromene (1), alpha amyrine acetate (2), and mucusoside (3) together with one new fatty acid glycoside, named 2-O-α-L-rhamnopyranosyl-hexacosanoate-β-D-glucopyranosyl ester (4). The compounds were identified using 1D, 2D NMR, and HR-ESIMS techniques as well as via comparison to other literature. Studies on the acetylcholinesterase inhibition potential and antioxidant activity were carried out on the total methanolic leaf extract, ethyl acetate fraction, and the isolated compounds. The results revealed the potent acetylcholinesterase inhibition of mucusoside alongside a new compound. Docking studies were also performed to confirm the possible interaction between the isolated compounds and acetylcholinesterase accompanying Alzheimer's disease progress.
1 Introduction
Alzheimer's disease (AD) is a neurodegenerative disease that is characterized by slow progression, dysfunction, and loss of neurons in the central nervous system.1 Alzheimer's disease could be associated with oxidative stress,2 aggregated protein deposit,3 neuroinflammation,4 and cholinergic deficit.5 Although the pathogenesis of such neurodegenerative disease or selecting an appropriate treatment is not yet discovered,4 the management of AD may be by hitting one or more of those associated conditions.6
Most of the treatment therapies only relieve symptoms and reduce progress.7 Thus, seeking for alternative and better therapeutic options is still under research. Acetylcholine (ACh) was the first neurotransmitter to be identified.8 It is used by all cholinergic neurons as it has a very important role in the peripheral and central nervous systems. ACh is considered to play a role in the pathology of Alzheimer's disease.9 Acetylcholinesterase (AChE) is a biologically important enzyme that hydrolyzes acetylcholine (ACh).10 One of the most important approaches for the treatment of this disease involves the enhancement of the acetylcholine levels in the brain using AChE inhibitors.11 Several studies have reported anticholinesterase activity of the plant extracts and drugs.12,13
In spite of great advances in modern medicine, plants still make an important contribution to health care. Pharmaceutical companies are showing renewed interest in investigating higher plants as sources for new lead structures and also for the development of efficacious drugs. F. benghalensis, belonging to the Moraceae family, is commonly known as Banyan tree (English), and is native to a wide area of Asia. The tree is cultivated in botanical gardens and parks throughout the tropical regions of the world.14 The chemical and pharmacological properties of the bark of F. benghalensis were extensively studied; however, only a few studies refer to its leaves that are involved in both diet and medicine.15 It exhibits antimicrobial,16 antidiabetic17,18 anti-inflammatory and antitumor activities.19 It is also useful for the treatment of gastric ulcers, painful skin diseases, and burning sensation.19 The F. benghalensis methanolic extract also exhibits acetylcholinesterase inhibition.20 These studies provoked us to delve deeper into studying the acetylcholinesterase inhibition activity of the isolated compounds. Metabolomics, a valued analytical tool, is used in combination with pattern differentiation approaches and bioinformatics to explore metabolites in a given metabolome.21 It encompasses several techniques of analysis, including fingerprinting of each metabolite that measures a subset of the whole profile. This gives the opportunity of differentiation and quantitation of metabolites.22 Metabolomic profiling was performed on the methanolic extract of leaves of F. benghalensis and resulted in the identification of 13 compounds. Concomitantly, the phytochemical study was performed on the ethyl acetate fraction. Then, the neuroprotective potentials of the total extract, the ethyl acetate fraction, and the identified compounds were evaluated by assessing the antioxidant and acetylcholinesterase inhibitory activities.
A docking study was also performed to investigate the possible interaction between the identified compounds and acetylcholinesterase.
2 Experimental
2.1. Plant material
Leaves of F. benghalensis were collected from the Experimental Station of Ornamental Plants, Faculty of Agriculture, Minia University, Egypt, in September 2016. Authentication of the plant species was established by Prof. Nasser Barakat, Professor of Botany, (Faculty of Science, Minia University, Egypt). A voucher specimen was deposited in the herbarium section of the Pharmacognosy Department, Faculty of Pharmacy, Minia University, Minia, Egypt under the registration number Mn-Ph-Cog-039.
2.2. Chemicals and reagents
Solvents, including petroleum ether, dichloromethane, ethyl acetate, methanol, and dimethylsulfoxide (DMSO), used in this study were of analytical grade and distilled before use. They were obtained from El-Nasr Company for Pharmaceuticals and Chemicals, Egypt. Distilled water was also used.
2.3. Extraction, fractionation and isolation
Fresh leaves of F. benghalensis (250 g) were air-dried and then mechanically ground to powder and extracted by maceration with 90% methanol till exhaustion. The methanolic extract was filtered and concentrated using a rotary evaporator to a syrupy residue (5.0 g). The residue was first suspended in water, defatted with petroleum ether, and then fractionated with ethyl acetate to afford 1.5 g of an ethyl acetate fraction. The EtOAc fraction was then fractionated by a VLC to give six subfractions, DCM (100%) 1, DCM–MeOH (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) 2, (80
10) 2, (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) 3, (70
20) 3, (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) 4, (50
30) 4, (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) 5, MeOH (100%) 6. Subfraction 1 was subjected to a silica gel column using petroleum ether–EtOAc (100% to 50
50) 5, MeOH (100%) 6. Subfraction 1 was subjected to a silica gel column using petroleum ether–EtOAc (100% to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) to afford four compounds. Compound 1 (7 mg) was obtained as crystals. The petroleum ether–EtOAc (80
50) to afford four compounds. Compound 1 (7 mg) was obtained as crystals. The petroleum ether–EtOAc (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) fraction was further purified using a Sephadex column DCM–MeOH (1
20) fraction was further purified using a Sephadex column DCM–MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give compound 2 (8 mg). Subfraction 4 was subjected to a silica column DCM–MeOH (90
1) to give compound 2 (8 mg). Subfraction 4 was subjected to a silica column DCM–MeOH (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to 70
10 to 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30), and then its repetitive purification with the Sephadex column DCM–MeOH (1
30), and then its repetitive purification with the Sephadex column DCM–MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) afforded compounds 3 (1.5 mg) and 4 (0.9 mg).
1) afforded compounds 3 (1.5 mg) and 4 (0.9 mg).
2.4. Metabolic profiling
Metabolic profiling of the methanolic extract of F. benghalensis was carried out according to Abdelmohsen et al.23 using an Acquity Ultra Performance Liquid Chromatography system combined with a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, USA). Chromatographic separation was executed using a reversed-phase BEH C18 column (2.1 × 100 mm, 1.7 μm particle size; Waters, Milford, USA) connected to a guard column (2.1 × 5 mm, 1.7 μm particle size). The mobile phase used during the separation was composed of purified water (A) and acetonitrile (B) with 0.1% formic acid added in each solvent. The elution was started in a gradient manner at a flow rate of 300 μl min−1 with 10% B linearly increased to 100% B within 30 min and kept isocratic for an additional 5 min before linearly returning to 10% B for the next 1 min. The injection volume was 2 μl and the column temperature was adjusted at 40 °C. The MSConvert software was utilized to switch the raw data into separate positive and negative ionization files. These files were finally exported to the data mining software MZmine 2.10 for peak picking, deconvolution, deisotoping, alignment, and formula prediction. The dereplication of compounds was executed by comparing with the Dictionary of Natural Products (DNP)24 and Metlin databases 2015.
2.5. Acetylcholinesterase inhibitory activity
The acetylcholinesterase inhibition activity was carried out according to the Ellman method.25 Briefly, the cholinesterase enzyme hydrolyzes the substrate acetyl thiocholine and the thiocholine product reacts with the Ellman's reagent (DTNB) to produce 2-nitrobenzoic-5-mercaptothiocholine (thiocholine-thionitrobenzoate disulfide) and 5-thio-2-nitrobenzoic acid (thionitrobenzoate), which can be detected at 405 nm. The total volume of the reaction mixture was 1 ml. In a 1 ml cuvette, 50 μl of 3.5 mmol L−1 acetyl thio choline iodide (ATCI) in a buffer was added to 920 μl of 0.125 mmol L−1 DTNB/buffer mixture, and then 20 μl choline esterase enzyme was added and the absorbance was measured at 405 nm every 30 s for 3 times to determine the total activity of the enzyme before inhibition. Then, 10 μl of the sample was added and the absorbance was read again at the same wavelength 3 times with 30 s intervals to determine the percentage of inhibition after the addition of the extract. After adding the sample, the inhibition percentage of the enzyme activity was calculated. IC50 values (concentration at which there is 50% enzyme inhibition) of selected samples were calculated using the Graph Pad Prism-7 software. Donepezil was used as a standard reference.
2.6. Total antioxidant capacity
The determination of the antioxidative capacity was performed by the reaction with a defined amount of exogenously provided hydrogen peroxide. The antioxidants in the sample eliminate a certain amount of the provided hydrogen peroxide. The residual hydrogen peroxide was determined colorimetrically. Briefly, 500 μl of hydrogen peroxide was added to 20 μl of the sample and incubated for 10 min at 37 °C. Later, 500 μl of the enzyme/3,5dichloro-2- hydroxyl benzensulphonate mixture was added and incubated for 10 min at 37 °C. The intensity of the colored product was measured colorimetrically at 505 nm, and the percent inhibition that represents the total scavenging activity of the sample was calculated relative to the blank. IC50 of each sample was calculated after performing the assay at two different concentrations. IC50 was calculated using the Graph pad prism 7 software. Ascorbic acid was used as a standard reference.
2.7. Statistical analysis
IC50 was calculated using the Graphpad prism 7 software. All results were expressed as mean ± standard deviation. Statistical comparison was conducted using the Student's t-test using the GraphPad Prism 7 statistical software (GraphPad, La Jolla, CA, USA). The results were considered to be significant when the probability values (p) were less than 0.05.
2.8. Molecular docking
The target compounds were constructed into a 3D model. After checking their structures and the formal charges on atoms by the 2D depiction, the following steps were carried out: the target compounds were subjected to a conformational search. All conformers were subjected to energy minimization. All the minimizations were performed until a RMSD gradient of 0.01 kcal mole−1 and RMS distance of 0.1 Å with MMFF94X force-field and the partial charges were automatically calculated. The obtained database was then saved as an MDB file to be used in the docking calculations. The X-ray crystallographic structure of donepezil in the complex with AChE was retrieved from the Protein Data Bank through the internet (http://www.rcsb.org/, PDB code 1EVE).26 The enzyme was prepared for docking studies using the following steps: hydrogen atoms were added to the system with their standard geometry. The atom connection and type were checked for any errors with automatic correction. The selection of the receptor and its atoms potential were fixed. Site Finder was used for the active site search in the enzyme structure using all default items. Dummy atoms were created from the site finder of the pocket. The docking of the conformation database of the target compounds was done. The following methodology was generally applied: The enzyme active site file was loaded, and the Dock tool was initiated. The program specifications were adjusted to: dummy atoms as the docking site, and alpha triangle as the placement methodology to be used. London dG as a scoring methodology to be used and was adjusted to its default values. The MDB file of the ligand to be docked was loaded and dock calculations were run automatically. The obtained poses were studied, and the poses showing the best ligand–enzyme interactions were selected and stored for energy calculations. After the completion of docking, the pose having the lowest energy was chosen to visualize the ligand–protein interaction in Discovery Studio 2017.27–29
3 Results and discussion
3.1. Structure elucidation
The total methanolic extract of F. benghalensis leaves was subjected to different chromatographic processes to afford different classes of compounds including three known compounds and one new fatty acid ester (Fig. 1). The known compounds were identified as, carpachromene (1) (S1 and S2†),30 α-amyrin acetate (2) (S3 and S4†),31 mucusoside (3) (S5),32 based on comparisons of their NMR and MS data with that previously reported in the literature. Compound 4 (S6–10†) was isolated as a brown powder. The 1H-NMR and 13C-NMR spectral data of compound 4 showed signals at δC 175.01 and 71.78 with δH 3.93 (1H, dd, J = 17.4, 3.6), indicating the presence of a carbonyl of an ester group and an oxygenated methine carbon attached to a methylene group. The signals at δC 14.0 with δH 0.89 (3H, t, J = 7.2) could be assigned to a terminal methyl group (C26) of a longchain fatty acid. The 1HNMR and 13C NMR spectral data of this compound revealed the presence of two sugar moieties, the chemical shift value at δH 4.25 (1H, d, J = 7.2) with δC 104.72 suggested the presence of the β-glucopyranosyl unit and the signals at δH 4.89 (1H, d, J = 3.6) with δC 100.02 and a methyl group with δH 0.95 (3H, d, J = 6.7) and δC 19.26 revealed the presence of an α-rhamnopyranosyl unit. The attachment of glucose to form an ester linkage was confirmed from HMBC as there is a noticeable correlation between anomeric proton and carbonyl group (H-1, δH 4.25 and C-1, 175.01). In addition, the presence of HMBC correlation between the anomeric proton of rhamnose δH 4.89 (1H, d, J = 3.6) and C-2 δC 71.78 confirmed the attachment of rhamnose at C-2. A characteristic mass ion peak at m/z 413.2652 was suggested for the molecular formula C26H52O3 [M+ − (glucose + rhamnose)]. Accordingly, from the previously mentioned physical, chromatographic properties, spectral data, the structure of compound 4 was suggested to be 2-O-α-L-rhamnopyranosyl-hexacosanoate-β-D-glucopyranosyl ester.
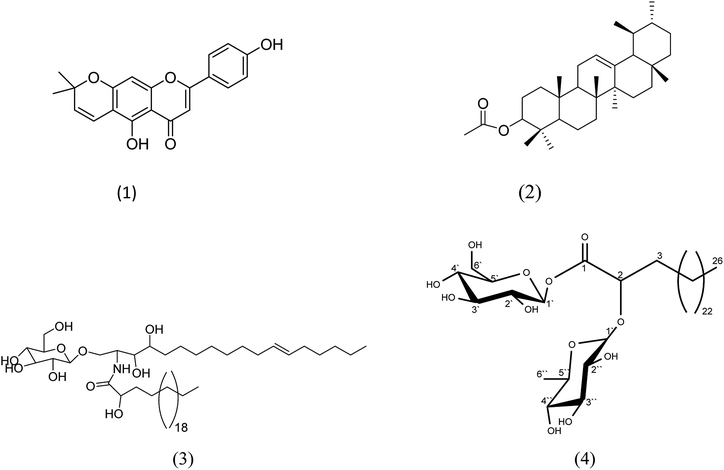 |
| | Fig. 1 Structures of the compounds isolated from the ethyl acetate fraction of F. benghalensis. | |
3.2. Metabolic profiling
Metabolic profiling of the leaves of F. benghalensis using LC-HR-ESI-MS for dereplication processes has led to the characterization of a number of secondary metabolites, including phenolic compounds, which were noticed to prevail. The identification of the detected compounds (Fig. 2) was processed by coupling MZmine with some databases such as Metlin and DNP. The results (Table S1†) showed a mass ion peak at m/z 275.092 for the predicted molecular formula C15H14O5, which was dereplicated as ficuformodiol A; (S)-form (5), ficuformodiol B; (S)-form (6). They were previously isolated from Ficus formosana.33 Another mass ion peak at m/z 287.055 suggested for the molecular formula C15H12O6 gave hits of steppogenin (7), which belongs to flavonoids. It was previously obtained from Ficus formosana.33
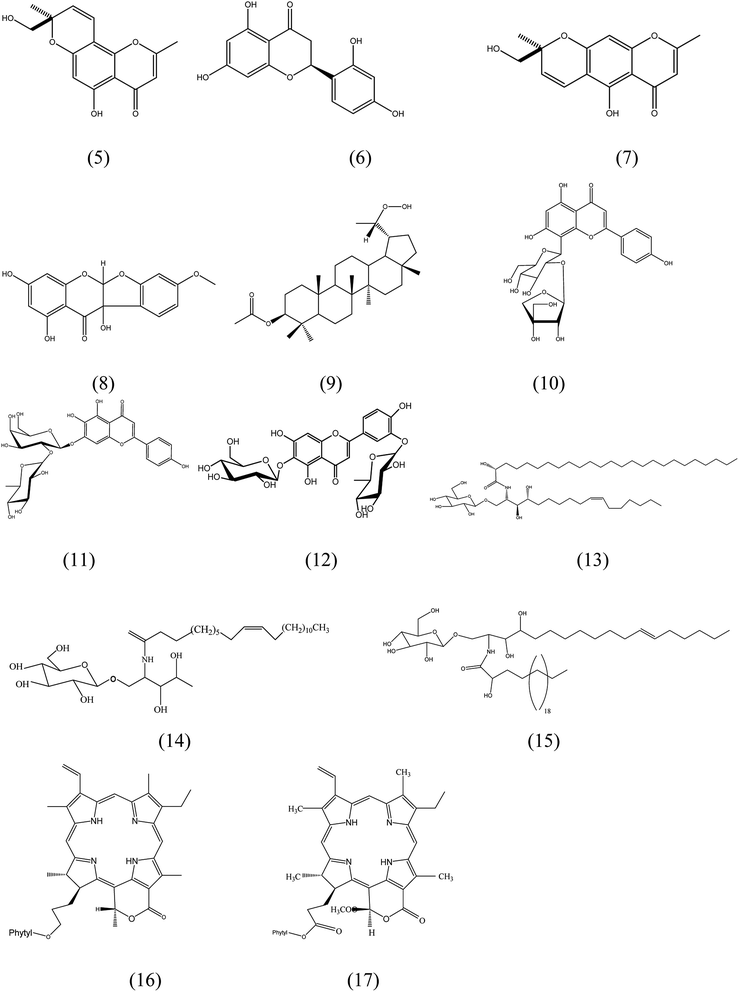 |
| | Fig. 2 Structures of the compounds dereplicated from the methanolic extract of leaves of F. benghalensis. | |
An isoflavonoid with a mass ion peak at m/z 317.066, corresponding to the molecular formula C16H12O7 had been detected. It was identified as 5a,10b-dihydro-1,3,8,10b-tetrahydroxy-11H-benzofuro[2,3-b][1]benzopyran-11-one; 4′-Me ether (8), which was formerly isolated from Ficus benjamina.34 Furthermore, the mass ion peak at m/z 487.378 belongs to C31H52O4 was also dereplicated as 30-nor-3,20-lupanediol; (3β,20S)-form, 20-hydroperoxide, 3-Ac (9), a pentacyclic triterpene, which was isolated earlier from F. microcarpa.35 In addition, a mass ion peak at m/z 563.139 in accordance with C26H28O14. It gave hits of a hydroxyflavone named ficuflavoside (10). It was previously purified from F. microcarpa.36 Likewise, a hydroxyflavone named 4′,5,6,7-tetrahydroxyflavone 6-O-[α-L-rhamnopyranosyl-(1→2)-β-D-galactopyranoside] (11), was dereplicated from the extract. It fits the molecular formula C27H30O15 as it has a mass ion peak at m/z 595.164. It was reported from F. infectoria in 1994.37 3′,4′,5,6,7-Pentahydroxyflavone; 3′-O-α-L-rhamnopyranoside, 6-O-β-D-glucopyranoside (12), another hydroxyflavone has the mass ion peak at m/z 609.145, consistent with the molecular formula C27H30O16. It was also obtained from F. infectoria.38 Among the dereplicated compounds, a mass ion peak at m/z 843.681 fits the molecular formula C48H93NO10 that could be characterized as ficusoside (13), mucusoside (14), 2-amino-10-heptadecene-1,3,4-triol; (2S,3S,4R,10Z)-form, N-(2R-hydroxypentacosanoyl), 1-O-β-D-glucopyranoside (15). These compounds were already reported from F. elastica, F. mucuso, and F. glumosa, respectively.39,32,40 In the same context, lactone chlorines with the molecular formula C55H74N4O6 were characterized as ficuschlorine A (16) and B (17) that was previously detected in F. microcarpa.41
3.3. Acetylcholinesterase inhibition
The acetylcholinesterase inhibitory activity of several medicinal plants has been reported in the literature.13,42,43 F. benghalensis has been considered as a prolific source of variable secondary metabolites, including phenolic compounds, cerebrosides, and terpenoids, which were endorsed as the major contributors to its biological potentials. The assessment of the in vitro acetylcholine esterase inhibition of the total methanolic extract, the ethyl acetate fraction and the isolated compounds of F. benghalensis showed that the total methanolic extract was the most potent acetylcholine esterase inhibitor with IC50 = 194.6 ± 7.961 μg ml−1 that is close to that of donepezil (IC50 = 186.1 ± 7.1), as illustrated in Table 1, Fig. 3, followed by compound 3 and compound 1 with IC50 = 390.5 ± 2.829 and 449.4 ± 8.924 μg ml−1, respectively. When comparing the results of the ethyl acetate fraction and the tested compounds with that of the standard, there were significant differences. Only the total extract did not show any significance, which means that it has a strong acetylcholinesterase inhibitory activity.
Table 1 Acetylcholinesterase inhibition (IC50) of the methanolic extract, ethyl acetate fraction and isolated compounds of F. benghalensis
| Sample |
IC50 |
| Methanolic extract |
194.6 ± 7.961 μg ml−1 |
| Ethyl acetate fraction |
604.6 ± 5.492 μg ml−1 |
| Compound 1 |
449.4 ± 8.924 μg ml−1 |
| Compound 2 |
NIL |
| Compound 3 |
390.5 ± 2.829 μg ml−1 |
| Compound 4 |
777.5 ± 5.603 μg ml−1 |
| Donepezil |
186.1 ± 7.1 μg ml−1 |
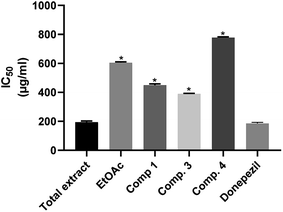 |
| | Fig. 3 Acetylcholinesterase inhibition activity of the total extract, ethyl acetate fraction and the isolated compounds of leaves of F. benghalensis. | |
However, the acetylcholinesterase inhibitory activity of methanolic extract of F. benghalensis was previously reported with IC50 = 228.3 μg ml−1.20 These results suggested the hypothesis that there is a synergistic effect between the compounds either of the ethyl acetate fraction or the methanolic extract. To the best of our knowledge, this is the first report of acetylcholinesterase inhibition for the compounds or the ethyl acetate fraction.
3.4. Total antioxidant activity
Oxidative stress is a substantial risk factor in the pathogenesis of miscellaneous chronic diseases. Free radicals are major causative agents of sicknesses such as diabetes, inflammatory arthropathies, asthma, Parkinson's, and Alzheimer's diseases.44,45 The presence of high doses of antioxidants in AD patients could lead to a slower rate of cognitive deterioration.46 Total antioxidant capacities of the total methanolic extract, ethyl acetate fraction, and four isolated compounds of F. benghalensis were investigated based on the in vitro antioxidant assay.47 The results listed in Table 2, Fig. 4 showed that total methanolic extract displayed the most potent antioxidant potential with IC50 = 178.2 ± 1.750 μg ml−1. This may be attributed to the phenolic, flavonoid content in addition to the diverse nitrogenous compounds of the extract.48 It is noteworthy that the antioxidant capacity of the total extract is comparable to that of ascorbic acid that had IC50 = 174.8 ± 12.3. The radical scavenging capacity of the compounds could be arranged in the descending order to show that compound 2 had higher activity than compound 1, which is followed by compound 3 and then compound 4 with IC50 286.9 ± 1.353, 358 ± 2.454, 430 ± 4.157 and 691.7 ± 4.038 μg ml−1, respectively. The values of the tested samples were significantly different as compared with the standard except for the total extract. This leads to the assumption that the total extract is a potent antioxidant.
Table 2 Total anti-oxidant activity of the methanolic extract, ethyl acetate fraction and isolated compounds of F. benghalensis
| Sample |
IC50 |
| Methanolic extract |
178.2 ± 1.750 μg ml−1 |
| Ethyl acetate fraction |
603.1 ± 6.573 μg ml−1 |
| Compound 1 |
430 ± 4.157 μg ml−1 |
| Compound 2 |
286.9 ± 1.353 μg ml−1 |
| Compound 3 |
358 ± 2.454 μg ml−1 |
| Compound 4 |
691.7 ± 4.038 μg ml−1 |
| Ascorbic acid |
174.8 ± 12.3 μg ml−1 |
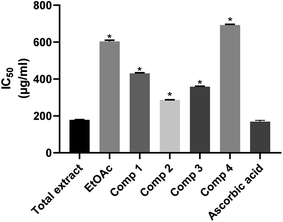 |
| | Fig. 4 Antioxidant activity of the total extract, ethyl acetate fraction and the isolated compounds of leaves of F. benghalensis. | |
3.5. Molecular docking
Based on the biological evaluation results and to explore the possible binding mode of the isolated compounds with AChE (PDB code 1EVE),26 docking studies were performed using the Molecular Operating Environment (MOE) package version 2014.10 (Chemical Computing Group, Inc. Molecular Operating Environment (MOE). CCG, Montreal, Canada.2014. http://www.chemcomp.com, accessed in February 2012) was used for performing docking studies. In the first step, the validation of the used protocol was done by redocking donepezil into the binding site of the crystal structures of AChE (PDB code 1EVE resolved at 2.5 Å). The crystal structure of AChE in complex with inhibitors revealed that there are two binding sites, a peripheral anionic site (PAS) and catalytic active site (CAS). The obtained RMSD (0.98) less than 2, which indicates the validity of the used protocol.
Moreover, as shown in Table 3 Fig. 5, donepezil exhibited two binding modes with AChE. In the bottom of the gorge, donepezil interacts with Trp84 and Trp279 via the π–π stacking interactions, carbon–carbon hydrogen bonds with Tyr70 and Ser286, two attractive charges with Asp72 and Phe330, and Pi-sigma interaction with Trp279 and Tyr334. In addition, the oxygen atom of the donepezil carbonyl group creates a hydrogen bond with the hydroxyl group of Phe288. The isolated compounds (1, 2, 3, and 4) bind nicely into the binding site of the crystal structures of AChE (PDB 1EVE). However, the most active compound 3 showed the best binding mode as it incorporated in five hydrogen bonds with the amino acid residues Tyr70 (3 hydrogen bonds), Glu73 and Gln74, in addition to two hydrophobic interactions with bTrp84 and Trp279 (Fig. 6).
Table 3 Docking scores of the compounds isolated from the ethyl acetate fraction of F. benghalensis leavesa
| Compound |
Molecular formula |
Molecular weight |
BE (kcal mol−1) |
NHBD |
NHBA |
| NHBD: number of hydrogen bond donor, NHBA: number of hydrogen bond acceptor. |
| 1 |
C20H16O5 |
336.34 |
−5.852 |
2 |
5 |
| 2 |
C32H52O2 |
468.75 |
5.724 |
Zero |
2 |
| 3 |
C48H93NO10 |
844.25 |
−10.913 |
8 |
10 |
| 4 |
C38H72O12 |
720.97 |
−8.781 |
7 |
12 |
| Donepezil |
C24H29NO3 |
379.49 |
−10.886 |
Zero |
4 |
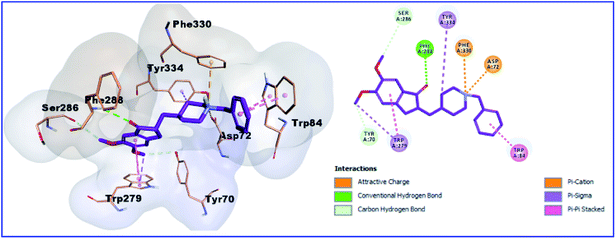 |
| | Fig. 5 3D and 2D binding pattern of donepezil into the active site of the AChE receptor (PDB entry: 1EVE) (pink). | |
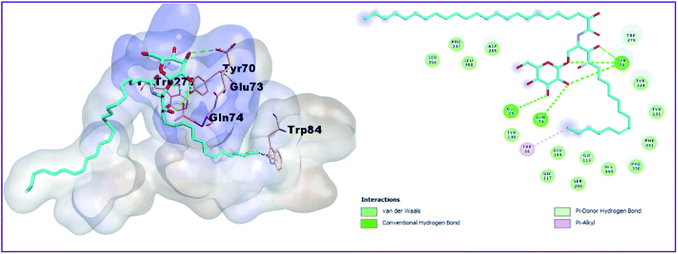 |
| | Fig. 6 3D and 2D binding pattern of compound 3 into the active site of the AChE receptor (PDB entry: 1EVE) (cyan). | |
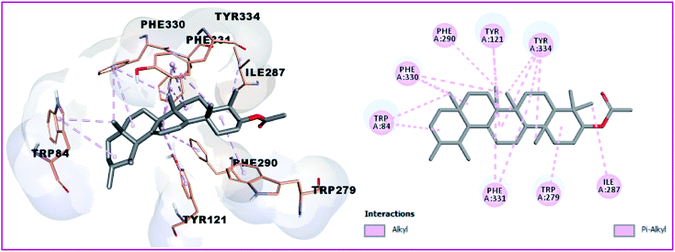
Compound 1 and 4 both of them engaged with two hydrogen bonds and many hydrophobic interactions. For instance, compound 1 creates two hydrogen bonds with Asp72 and Gly118, in addition to hydrophobic interactions with Tyr121, Tyr334, and Ile287 (Fig. 7). Compound 4 (Fig. 8) also showed two hydrogen bonds with Tyr70 and Gly333 in addition to hydrophobic interactions with Leu127, Tyr130, and Trp279. Finally, the least active compound 2 (Fig. 9), incorporated in hydrophobic interactions only without engagement in any hydrogen bonds. It showed a positive binding energy value, which means that the binding is energy consuming and it occurs only if the required energy was available. The complexes with high energies are less stable. Therefore, the negative Gibbs free energy is preferable for better binding.49
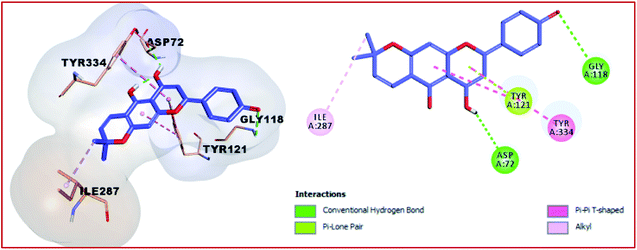 |
| | Fig. 7 3D and 2D binding pattern of compound 1 into the active site of the AChE receptor (PDB entry: 1EVE) (moov). | |
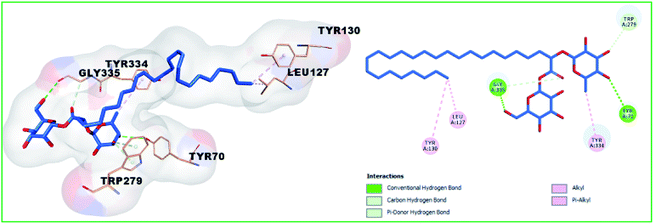 |
| | Fig. 8 3D and 2D binding pattern of compound 4 into the active site of the AChE receptor (PDB entry: 1EVE) (blue). | |
 |
| | Fig. 9 3D and 2D binding pattern of compound 2 into the active site of the AChE receptor (PDB entry: 1EVE) (grey). | |
To sum up, the docking results were in agreement with the biological activity and it can be proposed that the isolated compounds particularly compound 1, 3, and 4 can interact both with CAS and PAS of AChE.
4 Conclusion
Metabolic profiling of the secondary metabolites of F. benghalensis has resulted in diverse classes of compounds, particularly phenolics and cerebrosides. The phytochemical study of the ethyl acetate fraction showed four compounds, one of them (mucusoside) was detected in metabolomics. When testing the acetylcholinesterase inhibition and antioxidant potentials, the total extract exhibited promising results together with the isolated compounds. Accordingly, the potential inhibition of acetylcholinesterase and antioxidant results in this study reflects their usefulness in the management of AD. We also correlated the phytochemical content with the observed activities of the plant. In addition, the aforementioned results were correlated with docking studies that confirmed our approaches. Further investigations are therefore strongly recommended in order to exploit such active metabolites as possible candidates of new chemotherapeutic regimens. Moreover, a study concerning the synergism between the compounds will be conducted.
Author contributions
Conceptualization, M. S. K., U. R. A., K. M. M. and S. Y. D; methodology, H. A. H., D. H. A., M. F. A. M.; data curation, M. S. K., K. M. M., U. R. A., A. M. H., S. Y. D, M. A. E., A. E. A., H. A. H and K. S.; original draft preparation, H. A. H.; writing, review and editing, all authors.
Conflicts of interest
The authors declare there is no conflict of interest.
Acknowledgements
We thank Markus Krischke and M Müller for LC-MS measurement (University of Würzburg).
References
- S. Amor, F. Puentes, D. Baker and P. Van Der Valk, Immunology, 2010, 129, 154–169 CrossRef CAS.
- D. A. Butterfield, A. M. Swomley and R. Sultana, Antioxid. Redox Signaling, 2013, 19, 823–835 CrossRef CAS.
- M. Takalo, A. Salminen, H. Soininen, M. Hiltunen and A. Haapasalo, American Journal of Neurodegenerative Disease, 2013, 2, 1 Search PubMed.
- W. W. Chen, X. Zhang and W. J. Huang, Mol. Med. Rep., 2016, 13, 3391–3396 CrossRef CAS.
- R. Schliebs and T. Arendt, Behav. Brain Res., 2011, 221, 555–563 CrossRef CAS.
- T. O. Elufioye, C. G. Chinaka and A. O. Oyedeji, Antioxidants, 2019, 8, 400 CrossRef CAS.
- A. Kumar and A. Singh, Pharmacol. Rep., 2015, 67, 195–203 CrossRef CAS.
- T. H. Ferreira-Vieira, I. M. Guimaraes, F. R. Silva and F. M. Ribeiro, Curr. Neuropharmacol., 2016, 14, 101–115 CrossRef CAS.
- H. A. Lester, Sci. Am., 1977, 236, 106–120 CrossRef CAS.
- L. E. Hebert, P. A. Scherr, L. A. Beckett, M. S. Albert, D. M. Pilgrim, M. J. Chown, H. H. Funkenstein and D. A. Evans, JAMA, J. Am. Med. Assoc., 1995, 273, 1354–1359 CrossRef CAS.
- G. M. Bores, F. P. Huger, W. Petko, A. E. Mutlib, F. Camacho, D. K. Rush, D. E. Selk, V. Wolf, R. Kosley and L. Davis, J. Pharmacol. Exp. Ther., 1996, 277, 728–738 CAS.
- F. Ahmed and A. Urooj, Pharmacogn. Mag., 2010, 6, 142–144 CrossRef.
- J. d. C. Rojas-Vera, A. A. Buitrago-Díaz, L. M. Possamai, L. F. Timmers, L. R. Tallini and J. Bastida, S. Afr. J. Bot., 2020 DOI:10.1016/j.sajb.2020.06.025.
- R. L. Riffle, The tropical look: An encyclopedia of dramatic landscape plants, Portland, US, Timber Press, 1998, p. 1998 Search PubMed.
- Y. Shi, A. M. Mon, Y. Zhang, Y. Fu, C. Wang, X. Yang and Y. Wang, J. Ethnopharmacol., 2018, 226, 185–196 CrossRef.
- H. A. Hassan, S. F. Abdelwahab, S. Y. Desoukey, K. M. Mohamed and M. S. Kamel, Indian J. Public Health Res. Dev., 2019, 10, 1938–1943 CrossRef.
- P. Khanal and B. Patil, Asian Pac. J. Trop. Biomed., 2019, 9, 263 CrossRef CAS.
- M. Gayathri and K. Kannabiran, Indian J. Clin. Biochem., 2008, 23, 394–400 CrossRef.
- Z. Etratkhah, S. E. S. Ebrahimi, N. K. Dehaghi and Y. Seifalizadeh, J. Rep. Pharm. Sci., 2019, 8, 24 CrossRef.
- M. Vignesh, V. V. Priya and R. Gayathri, Drug Invent. Today, 2019, 12(6), 1281–1283 Search PubMed.
- D. J. Raheem, A. F. Tawfike, U. R. Abdelmohsen, R. Edrada-Ebel and V. Fitzsimmons-Thoss, Sci. Rep., 2019, 9, 2547 CrossRef.
- N. F. Tawfik, Ů. Tawfike, R. Abdo, G. Abbott, U. Abdelmohsen, R. Edrada-Ebel and E. Haggag, J. Adv. Pharm. Educ. Res., 2017, 1, 100–109 CrossRef.
- U. Abdelmohsen, C. Cheng, C. Viegelmann, T. Zhang, T. Grkovic, S. Ahmed, R. Quinn, U. Hentschel and R. Edrada-Ebel, Mar. Drugs, 2014, 12, 1220–1244 CrossRef CAS.
- J. Buckingham, J. Acoust. Soc. Am., 2017, 2273–2287 Search PubMed.
- G. L. Ellman, K. D. Courtney, V. Andres Jr and R. M. Featherstone, Biochem. Pharmacol., 1961, 7, 88–95 CrossRef CAS.
- Z. Soyer, S. Uysal, S. Parlar, A. H. Tarikogullari Dogan and V. Alptuzun, J. Enzyme Inhib. Med. Chem., 2017, 32, 13–19 CrossRef CAS.
- H. A. Abou-Zied, B. G. Youssif, M. F. Mohamed, A. M. Hayallah and M. Abdel-Aziz, Bioorg. Chem., 2019, 89, 102997 CrossRef CAS.
- M. M. Al-Sanea, L. Gotina, M. F. Mohamed, D. G. T. Parambi, H. A. Gomaa, B. Mathew, B. G. Youssif, K. S. Alharbi, Z. M. Elsayed and M. A. Abdelgawad, Drug Des., Dev. Ther., 2020, 14, 497 CrossRef CAS.
- B. G. Youssif, M. F. Mohamed, M. M. Al-Sanea, A. H. Moustafa, A. A. Abdelhamid and H. A. Gomaa, Bioorg. Chem., 2019, 85, 577–584 CrossRef CAS.
- Z. P. Zheng, K. W. Cheng, J. T. K. To, H. Li and M. Wang, Mol. Nutr. Food Res., 2008, 52, 1530–1538 CrossRef CAS.
- N. N. Okoye, D. L. Ajaghaku, H. N. Okeke, E. E. Ilodigwe, C. S. Nworu and F. B. C. Okoye, Pharm. Biol., 2014, 52, 1478–1486 CrossRef CAS.
- J. J. K. Bankeu, S. A. A. Mustafa, A. S. Gojayev, B. D. Lenta, D. T. Noungoue, S. A. Ngouela, K. Asaad, M. I. Choudhary, S. Prigge and A. A. Guliyev, Chem. Pharm. Bull., 2010, 58, 1661–1665 CrossRef CAS.
- Y.-W. Sheu, L.-C. Chiang, J.-J. Chen, Y.-C. Chen and I.-L. Tsai, Planta Med., 2006, 71, 1165–1167 CrossRef.
- J. Dai, D. Shen, W. Y. Yoshida, S. M. Parrish and P. G. Williams, Planta Med., 2012, 78, 1357–1362 CrossRef CAS.
- Y.-M. Chiang and Y.-H. Kuo, J. Nat. Prod., 2001, 64, 436–439 CrossRef CAS.
- P. Van Kiem, N. X. Cuong, N. X. Nhiem, V. K. Thu, N. K. Ban, C. Van Minh, B. H. Tai, T. N. Hai, S. H. Lee and H. D. Jang, Bioorg. Med. Chem. Lett., 2011, 21, 633–637 CrossRef.
- J. Neeru and R. Yadava, Fitoterapia, 1994, 65 Search PubMed.
- E. P. Lansky and H. M. Paavilainen, Figs: the genus Ficus, CRC Press, 2010 Search PubMed.
- E. J. T. Mbosso, J. C. A. Nguedia, F. Meyer, B. N. Lenta, S. Ngouela, B. Lallemand, V. Mathieu, P. V. Antwerpen, A. L. Njunda, D. Adiogo, E. Tsamo, Y. Looze, R. Kiss and R. Wintjens, Phytochemistry, 2012, 83, 95–103 CrossRef CAS.
- F. Nana, L. P. Sandjo, F. Keumedjio, P. Ambassa, R. Malik, V. Kuete, V. Rincheval, M. I. Choudhary and B. T. Ngadjui, J. Braz. Chem. Soc., 2012, 23, 482–487 CrossRef CAS.
- H. Y. Lin, H. L. Chiu, Y. H. Lan, C. Y. Tzeng, T. H. Lee, C. K. Lee, Y. Y. Shao, C. R. Chen, C. I. Chang and Y. H. Kuo, Chem. Biodiversity, 2011, 8, 1701–1708 CrossRef CAS.
- S. P. Samaradivakara, R. Samarasekera, S. M. Handunnetti and O. V. D. S. J. Weerasena, Ind. Crops Prod., 2016, 83, 227–234 CrossRef CAS.
- D. Sheeja Malar, R. Beema Shafreen, S. Karutha Pandian and K. Pandima Devi, Pharm. Biol., 2017, 55, 381–393 CrossRef CAS.
- J. R. Kanwar, R. K. Kanwar, H. Burrow and S. Baratchi, Curr. Med. Chem., 2009, 16, 2373–2394 CrossRef CAS.
- V. Chiavaroli, C. Giannini, S. De Marco, F. Chiarelli and A. Mohn, Redox Rep., 2011, 16, 101–107 CrossRef CAS.
- P. Mecocci and M. C. Polidori, Biochim. Biophys. Acta, Mol. Basis Dis., 2012, 1822, 631–638 CrossRef CAS.
- D. Koracevic, G. Koracevic, V. Djordjevic, S. Andrejevic and V. Cosic, J. Clin. Pathol., 2001, 54, 356–361 CrossRef CAS.
- B. Kaurinovic and D. Vastag, in Antioxidants, IntechOpen, 2019 Search PubMed.
- X. Du, Y. Li, Y.-L. Xia, S.-M. Ai, J. Liang, P. Sang, X.-L. Ji and S.-Q. Liu, Int. J. Mol. Sci., 2016, 17, 144 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06565j |
|
| This journal is © The Royal Society of Chemistry 2020 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *b,
Dalia H. Abu-Baihc,
Mamdouh F. A. Mohamed
*b,
Dalia H. Abu-Baihc,
Mamdouh F. A. Mohamed d,
Usama Ramadan Abdelmohsen
d,
Usama Ramadan Abdelmohsen ae,
Kuniyoshi Shimizuf,
Samar Y. Desoukeye,
Alaa M. Hayallahgh,
Mahmoud A. Elrehanyc,
Khaled M. Mohamedi and
Mohamed S. Kamelae
ae,
Kuniyoshi Shimizuf,
Samar Y. Desoukeye,
Alaa M. Hayallahgh,
Mahmoud A. Elrehanyc,
Khaled M. Mohamedi and
Mohamed S. Kamelae
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) 2, (80
10) 2, (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) 3, (70
20) 3, (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) 4, (50
30) 4, (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) 5, MeOH (100%) 6. Subfraction 1 was subjected to a silica gel column using petroleum ether–EtOAc (100% to 50
50) 5, MeOH (100%) 6. Subfraction 1 was subjected to a silica gel column using petroleum ether–EtOAc (100% to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) to afford four compounds. Compound 1 (7 mg) was obtained as crystals. The petroleum ether–EtOAc (80
50) to afford four compounds. Compound 1 (7 mg) was obtained as crystals. The petroleum ether–EtOAc (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) fraction was further purified using a Sephadex column DCM–MeOH (1
20) fraction was further purified using a Sephadex column DCM–MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give compound 2 (8 mg). Subfraction 4 was subjected to a silica column DCM–MeOH (90
1) to give compound 2 (8 mg). Subfraction 4 was subjected to a silica column DCM–MeOH (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to 70
10 to 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30), and then its repetitive purification with the Sephadex column DCM–MeOH (1
30), and then its repetitive purification with the Sephadex column DCM–MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) afforded compounds 3 (1.5 mg) and 4 (0.9 mg).
1) afforded compounds 3 (1.5 mg) and 4 (0.9 mg).









