Open Access Article
Open Access ArticleA review on biological activities of Schiff base, hydrazone, and oxime derivatives of curcumin
Sakineh Omidi
 * and
Ali Kakanejadifard
* and
Ali Kakanejadifard

Department of Chemistry, Faculty of Science, Lorestan University, Khorramabad, Iran. E-mail: sakineh.omidi@yahoo.com; Tel: +98-9181438542
First published on 17th August 2020
Abstract
Curcumin (1,7-bis[4-hydroxy-3-methoxyphenyl]-1,6-heptadiene-3,5-dione) is the main pigment present in the turmeric rhizome and shows various biological properties. The synthesis of different derivatives is an effective way to improve the medicinal and biological properties of curcumin. Many researchers have chosen the carbonyl group of curcumin for modification and preparation of new analogues. This review critically surveys a general overview of the literature and summarizes the synthesis and biological activities of Schiff base, hydrazone and oxime derivatives of curcumin over the last decade. These compounds and also their metal complexes possess higher potency in biological activity.
1. Introduction
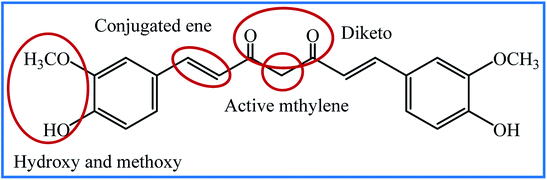
Curcumin is a natural yellow pigment isolated from the rhizomes of Curcuma longa and is broadly used as an additive in food. Curcumin has been used for several centuries in herbal medicines in India, China, and Asian countries. Curcumin has attracted much attention due to its various medicinal properties against expanding and dangerous diseases, such as different types of cancer, and diabetes.1 Various biological activities, including antioxidant,2 anti-inflammatory,3 antiviral,4 antibacterial,5 neuroprotective,6 antifungal,7 wound healing,8 and many more have been reported for curcumin. Also, it has been found that curcumin shows pharmacological activity against many human diseases, including multiple sclerosis, rheumatoid arthritis, Parkinson's and Alzheimer's disease, and atherosclerosis.9–11Despite all the outstanding properties of curcumin, this molecule has insignificant absorption and bioavailability due to limitations such as rapid metabolism, and low water solubility and stability. Many researchers have tried to overcome these problems by modifying the structure, synthesizing the different derivatives and designing the smart drug delivery systems. The chemical structure of curcumin and its active groups is shown in Fig. 1. From the synthetic point of view, there are four active sites in the structure of curcumin for use in the synthesis of new curcumin derivatives. Many researchers have chosen the hydroxyl group to modify the curcumin structure.12,13 Nucleophilic addition to the enone moiety is another way to synthesize new derivatives.14,15 The methylene group is an active site that causes the rapid metabolism of curcumin and removes it from the body. Hence, the modification of curcumin through the condensation reaction on the methylene site has attracted a lot of attention.16,17 Curcumin is a rare example of naturally ligands having the β-diketone group. This group can contribute to condensation reactions, including Schiff base reaction. Although several review articles have been published on medicinal and biological activities of curcumin analogues,18–22 to the best of our knowledge, Schiff base, hydrazone, and oxime analogues of curcumin have not been reviewed. In this review, we provide an account of the synthesized curcumin-based derivatives using Schiff base reaction on the β-diketone group, which results in the formation of Schiff base, hydrazone, oxime, and some oxime ether compounds. The most detailed discussion will be focused on the medicinal and biological information of these compounds.
2. Schiff base derivatives of curcumin
2.1. Schiff bases
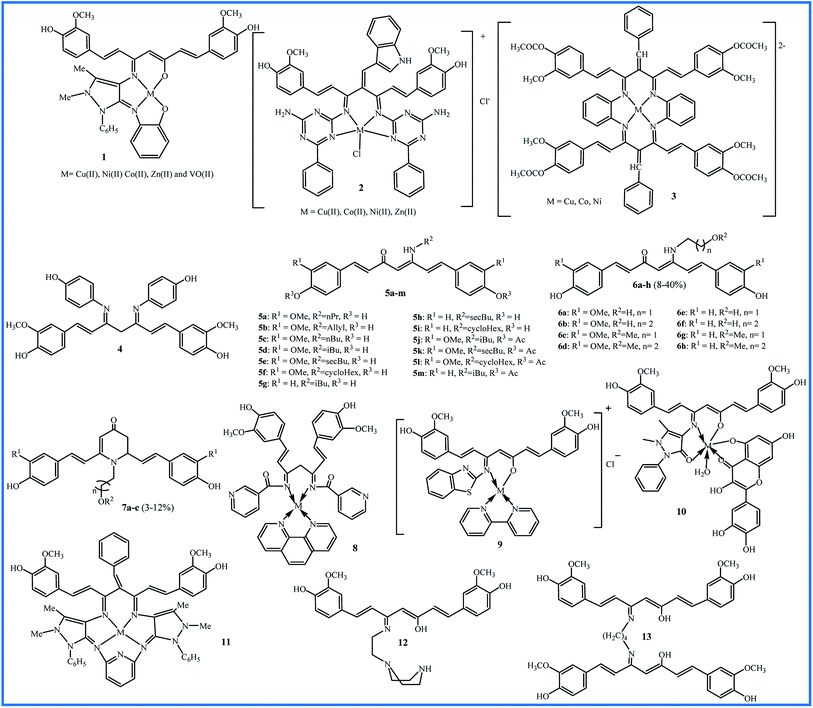
The reaction between the substances containing amino groups (NH2, NH2OH, NH2–NH2, etc.) with carbonyl groups (aldehydes or some ketones) in honor of the German chemist, Hugo Schiff, is called the Schiff base reaction. In 2011, Kulandaisamy reported the synthesis of tetradentate chelate 1 and its Cu(II), Ni(II), Co(II), VO(II), and Zn(II) complexes (Fig. 2). The antibacterial and antifungal activities of the ligand and their complexes were evaluated against Escherichia coli (E. coli), Salmonella typhimurium (S. typhi), Staphylococcus aureus (S. aureus), Streptococci, and Pseudomonas aeruginosa (P. aeruginosa) bacteria and Candida albicans (C. albicans) fungi by disc diffusion method. All the metal complexes showed better biological properties than the free ligand, and Zn(II) complex exhibited the highest antimicrobial activity (Zone of inhibition in mm: 9–14).23The Knoevenagel condensation can lock the curcumin in the keto tautomer, which leads to an effective reaction with amines to form the Schiff bases. With this strategy, Tharmaraj and co-workers synthesized a non-enolizable diketone of curcumin by indole-3-aldehyde. The reaction of curcumin diketimine and 2,4-diamino-6-phenyl-1,3,5-triazine formed the Schiff base 2. The synthesis process was followed with forming the metal(II) complexes. The authors evaluated the nonlinear properties and pharmacological activities of compounds. Nonlinear optical (NLO) materials play significant roles in a broad range of photonic applications. Curcumin Schiff base 2 with a conjugated electron structure can have great nonlinear polarizabilities. The compounds were screened against Streptococcus pyogenes (S. pyogenes) and S. aureus bacteria and Aspergillus flavus (A. flavus) and Penicillium digitatum (P. digitatum) fungi. All metal(II) complexes showed improved activities compared to compound 2. The copper complex exhibited higher antibacterial activity (zone of inhibition in mm: 23) than the amikacin (zone of inhibition in mm: 18) against S. aureus.24
In 2014, Haneefa et al. synthesized a Schiff base ligand by a simple condensation of curcumin and aniline with improved biological activities compared the curcumin.25 Later, they utilized the obtained curcuminaniline to functionalization of copper oxide nanoparticles in a green synthesis.26 The bio-functionalized copper oxide nanoparticles indicated potent antimicrobial activity, particularly against Bacillus subtilis (B. subtilis) (zone of inhibition in mm: 19) and Aspergillus niger (A. niger) (zone of inhibition in mm: 20) species.
Using tetraaza 14-membered 3, a series of [M(LL)]2+ complexes metals were synthesized and evaluated for interaction with Calf thymus (CT)-DNA using of UV-visible, emission and circular dichroism (CD) spectroscopic methods. This structure can be a mimicking of natural tetraaza macrocycles, such as porphyrins. All complexes showed the interactions with DNA by a groove binding mode and copper complex exhibited a higher degree of interactions with binding constant (Kb) = 1.4 × 105 (ref. 27). In 2019, a supported catalyst was organized by immobilization of the copper complex 3 on silica gel and used as the catalyst for the synthesis of some tetrazole compounds.28
Revathi and co-workers modified the curcumin in methylene and keto (4 in Fig. 2) groups and evaluated their antioxidant activities using 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay. The obtained Knoevenagel condensates bearing N,N-dimethylamino benzaldehyde and 4-hydroxy benzaldehyde were slightly more active than curcumin (IC50 values about 50 μM). Substitution on the keto site reduced the antioxidant activity, and IC50 value for compound 4 was not attained even to 250 μM for compound 4.29
Camp and D'hooghe and co-workers considered the labile β-diketo structure responsible for the low bioavailability of curcumin. Therefore, they synthesized thirteen new derivatives bearing a β-enaminone using the microwave irradiation (5 in Fig. 2). The antioxidant activities were examined by DPPH and the ferric reducing ability of plasma (FRAP) assays. Compounds 5a–f exhibited high antioxidant activities by both tests (0.08–0.13% inhibition per μM by DPPH assay and 0.83–1.29 Trolox equiv. per μM by FRAP assay) which was comparable to the activity of curcumin (0.15% inhibition per μM by DPPH assay and 1 Trolox equiv. per μM by FRAP assay). Compounds 5a–f, like curcumin, contain the –OCH3 group in the ortho position of the phenol group on the aromatic rings. Compounds without an ortho methoxy group (5g–i) showed only insignificant antioxidant activities by both assays. For compounds 5j–m that phenolic groups were protected through acetylation, no antioxidant activity was observed. Cell growth inhibition (IC50 values in μM) of curcumin derivative was evaluated against several cell lines included HT-29, Caco-2 (under undifferentiated and differentiated conditions), EA.hy962, and Chinese hamster ovarian CHO-K1 by determining two parameters of mitochondrial activity and protein content of cells. The results showed that none of the derivatives caused cytotoxic effects on differentiated Caco-2 cells. These cells are similar to in vivo enterocytes lining the intestinal tract. It can be concluded that analogues do not disturb the intestinal barrier by cell damage. Also, β-enaminone analogues 5g–i were cytotoxic for all undifferentiated cell lines, and 5i displayed the most significant cytotoxic (IC50 for CHO, Caco-2 undiff, and HT-29 were 9.2, 7.8, and 4.0 μM, respectively). Reactive oxygen species (ROS) assay did not show a relationship between ROS induction and decreased cell viability. Therefore, these derivatives did not exert the anticancer properties through ROS induction.30
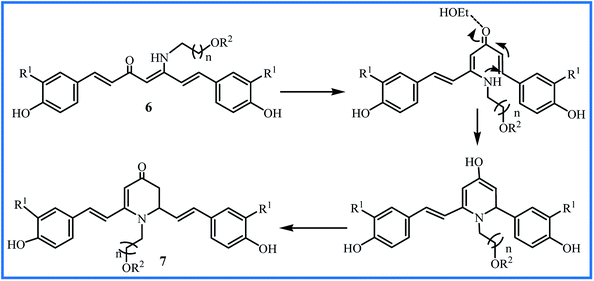
The water solubility of 5a–m derivatives was similar to curcumin. Therefore, following this work, Camp and D'hooghe research team extended the β-enaminone compound using more polar amines to enhance the water solubility without reducing the biological activity of the obtained curcuminoids. The reaction of the curcumin and hydroxyalkyl amines or methoxyalkyl amines produced eight new β-enaminone analogues (6a–h) with extra polar hydroxyl and methoxy groups. Three cyclic dihydropyridinone 7a–c side-products were also formed in low yields through the suggested mechanism in Fig. 3. The water solubility of 6a–h and 7a–c were enhanced 8- to 37-fold and 16- to 38-fold compared to curcumin, respectively. The new enaminone analogues 6a–d and 7a exhibited antioxidant activities comparable to curcumin and much better than bisdemethoxycurcumin derivatives 6e–h and 7b–c. In addition, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showed that cytotoxic activity of 6e–h (IC50 24–66 μM) was better than 6a–d (IC50 > 54 μM) in Caco-2, HepG2, HT-29 and CHO-K1 cells. The cyclic dihydropyridinone 7a–c did not show notably anti-proliferative activity.31
An impressive synthesis of the new curcumin-based ligand has been reported by Abdul Kareem et al. They showed that treatment of curcumin and nicotinamide produce a bidentate ligand that can form the Co(II), Ni(II) and Cu(II) complexes with 1,10-phenanthroline compound (8 in Fig. 2). The highest antibacterial activity against E. coli, S. aureus, Klebsiella pneumonia (K. pneumonia), S. pyogenes and P. aeruginosa were recorded for Cu(II) complexes (bacterial growth inhibition about 80–88%), slightly lower than the standard drug ciprofloxacin (bacterial growth inhibition about 90–98%). The Cu(II) complex also exhibited higher anthelmintic activity against the earthworms Pheretima posthuma than other metal complexes (shorter paralysis and mortality times of worms). The authors believed that the more lipophilic nature of the metal atom leads to efficient permeation of the compound through the lipid layer of cell membranes.32
In 2017, Raman et al. described the synthesis of a Schiff base ligand having 2-aminobenzothizole moiety and its metal complexes by 2,2′-bipyridine as co-ligand (9 in Fig. 2). Absorption spectral titration experiments showed that complexes have a significant binding affinity with CT DNA (Kb values 1.1–4.3 × 104 M−1). DNA cleavage study using pBR322 circular plasmid DNA in the presence of oxidant H2O2 showed that all complexes are the most potent nucleases. The oxidative damage to DNA over the formation of active oxygen species may result from the oxidation of metal ion from +2 to +3 through Fenton-type reactions. The antibacterial test against S. aureus, B. subtilis, E. coli, P. aeruginosa and S. typhi showed MIC values 8.1–14.4 × 104 μM. The antifungal test against A. niger, Fusarium solani (F. solani), Curvularia lunata (C. lunata), Rhizoctonia bataticola (R. bataticola), and C. albicans showed MIC values 8.4–14.8 × 104 μM. The most significant activities were observed for copper complex due to its high polarizing nature and lipophilicity.33 In the same year, Raman reported another Schiff base ligand having 4-aminoantipyrine moiety and its metal complexes by quercetin as co-ligand (10 in Fig. 2). Interactions of these metal complexes with DNA (Kb = 1.2–2.75 × 104 M−1) were slightly weaker than complexes of ligand 9. MIC values of antibacterial and antifungal activities of complexes against the same microbes were about 4.0–6.8 × 104 μM and 12.1–17.5 × 104 μM, respectively that indicated complexes of 10 are potent antibacterial agents.34
The evaluation of antibacterial and antifungal activities of 14-membered macrocyclic pentaza 11 and its Cu(II), Ni(II), Co(II), V(II) and Zn(II) complexes showed that all metal compounds have lower MIC values. The copper and zinc complexes exhibited the most potent antimicrobial activities against E. coli and C. albicans with MIC values 14 and 21 mg L−1. The antibacterial activity of the vanadium complex did not show notable differences with the free ligand. The copper complex was also showed higher cytotoxicity against breast cancer cell line MCF-7 (IC50 value 47.98 μM) compared to ligand 11 (IC50 value 75.70 μM).35
In 2018, Schiff base ligand containing ethylene piperazine 12 and their Co(II), Ni(II), Cu(II) and Zn(II) complexes was reported and studied for their antioxidant activity by DPPH assay and cytotoxic potential against MDA-MB-231, KCL22 and HeLa human cancer cell lines by MTT assay. The most significant antioxidant activity was observed for copper and zinc complexes with % scavenging activity 41.2 ± 1.75 and 40.5 ± 1.39 at 15 μM concentrations. The highest cytotoxicity was also observed for copper complex against MDA-MB-231 and KCL22 cells (IC50 values of 9.13 ± 1.18 and 9.87 ± 1.95 μM) and zinc complex against HeLa cells (IC50 value of 12.19 ± 1.52 μM). All metal complexes revealed the enhancement in the biological activities compared to the free ligand. It can be attributed to the existence of azomethine bond in the chelate ring and lipophilic nature of the metal ions, which improve their penetration via the lipid layer of the cell membrane.36
Recently, a new Schiff base of curcumin (13 in Fig. 2) was synthesized and used as a self-stabilizing and reducing agent in green synthesis of silver nanoparticles in water due to its high solubility in aqueous solutions. Compared to curcumin, compound 13 exhibited higher activity against Gram-negative bacteria (E. coli and P. aeruginosa) and lower activity against Gram-positive bacteria (S. aureus and B. subtilis).37
The synthetic methodology, yields, and distinct biological activity of 1–13 are summarized in Table 1.
| Compound | Condition of synthesis | Yield (%) | Activity | Outcome | Reference |
|---|---|---|---|---|---|
| 1 | 6 h reflux of precursors in EtOH | 80 | Antibacterial | Zn(II) complex showed the highest activity but was not compared with curcumin | 23 |
| 2 | 6 h stirrer of precursors in EtOH at room temperature and in presence of piperidine | 72 | Antibacterial and antifungal | Cu(II) complex showed the highest activities but was not compared with curcumin | 24 |
| 3 | 24 h reflux of precursors in MeOH | No reported | DNA cleavage | Cu(II) complex showed the highest Kb | 28 |
| 4 | 6 h stirrer of precursors in MeOH at room temperature and in presence of piperidine | 56.9 | Antioxidant | Lower antioxidant activity compared to curcumin | 29 |
| 5a–i | Heating of precursors in 2-methyl-THF or CHCl3 under microwave irradiation for 1–1.75 h at 80 °C in presence of MK10 clay and acetic acid | 11–58 | Anticancer and antioxidant | 5a–f showed comparable antioxidant activity to curcumin, 5i was strongest anticancer agent | 30 |
| 5j–m | Heating of 5d–g and acetic anhydride in 2-methyl-THF in presence of pyridine | 42–67 | Anticancer and antioxidant | No antioxidant activity and low anticancer activity | 30 |
| 6a–h | Heating of precursors in 2-methyl-THF or EtOH or DMF under microwave irradiation for 1–1.5 h at 70–100 °C in presence of MK10 clay and acetic acid | 8–40 | Anticancer and antioxidant | Enhanced water-solubility and anticancer activity and comparable antioxidant activity (6a–d) to curcumin | 31 |
| 7a–c | Heating of precursors in EtOH under microwave irradiation for 1–1.5 h at 70–100 °C in presence of MK10 clay and acetic acid | 3–12 | Anticancer and antioxidant | Enhanced water-solubility compared to curcumin | 31 |
| 8 | 12 h reflux of precursors in MeOH | 74 | Antibacterial and anthelmintic | Cu(II) complex showed the highest activities but was not compared with curcumin | 32 |
| 9 | 4 h reflux of precursors in EtOH | 66 | Antibacterial/antifungal and DNA cleavage | Cu(II) complex showed the highest activities but was not compared with curcumin | 33 |
| 10 | 3 h reflux of precursors in EtOH | 75 | Antibacterial/antifungal and DNA cleavage | Cu(II) complex showed the highest activities but was not compared with curcumin | 34 |
| 11 | 24 h reflux of precursors in EtOH in presence of anhydrous K2CO3 | No reported | Antibacterial/antifungal and anticancer | Cu(II) complex showed the highest activities but was not compared with curcumin | 35 |
| 12 | 6 h reflux of precursors in MeOH | 74 | Anticancer and antioxidant | Cu(II) and Zn(II) complexes showed the highest activities but was not compared with curcumin | 36 |
| 13 | 6 h reflux of precursors in EtOH in presence of acetic acid | 85 | Antibacterial | Higher activity against Gram-positive and lower activity against Gram-negative bacteria compared to curcumin | 37 |
2.2. Schiff base derivatives with other functional groups
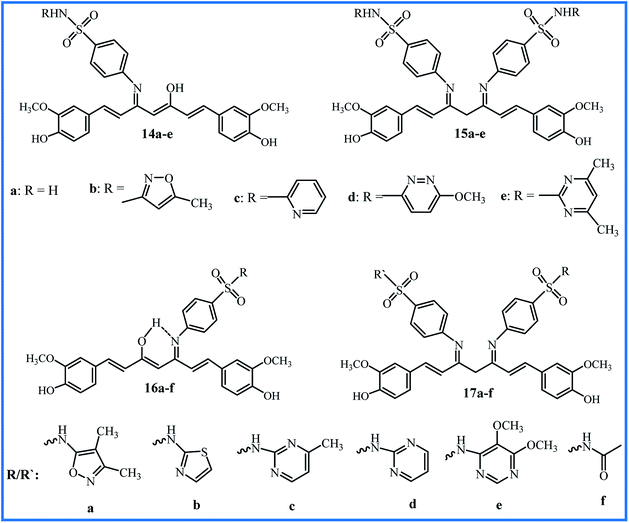
Ahmed et al. reported the synthesis of a series of curcumin–Schiff bases bearing one or two benzenes sulfonamides moieties (16a–f and 17a–f in Fig. 4). The compounds were investigated for antibacterial activity against Salmonella enterica (S. enterica), E. coli, P. aerugenosa, K. pneumonia, S. aureus, and Methicillin-resistant S. aureus (MRSA) bacteria, antifungal activity against A. niger, A. flavus, F. solani, Fusarium oxysporum (F. oxysporum), Alternaria alternate (A. alternate) and P. digitatum fungi, anti-inflammatory activity (using carrageenan-induced hind paw edema technique), and anti-nociceptive activity (by acetic acid-induced method). Compound 16a with isoxazole moiety was the most potent compound in anti-inflammatory (% inhibition 82.0), anti-nociceptive (% inhibition 66.8), and antibacterial (MIC 15.63–31.25 μg mL−1) activities; compound 16b with 4-methylpyrimidin moiety exhibited the highest antifungal activity (MIC 31.25–125 μg mL−1) against F. solani, F. oxysporum, and A. alternate. Furthermore, the combinations of 16a with standard antibacterial drug ciprofloxacin or standard antifungal drug nystatin indicated the notable synergic effects. The combination of ciprofloxacin (0.12 μg mL−1) with compound 16a (7.8 μg mL−1) was 8-fold more potent than ciprofloxacin, and inhibited the growth of MRSA.39 This research group also conducted in vitro urease inhibition studies, and indicated these compounds are moderate to good urease enzyme inhibitors.40
The synthetic methodology, yields, and distinct biological activity of 14–17 are summarized in Table 2.
| Compound | Condition of synthesis | Yield (%) | Activity | Outcome | Reference |
|---|---|---|---|---|---|
| 14a–e | 3–4 h reflux of precursors in EtOH | 87–91 | Antibacterial/antifungal and anti-inflammatory | 14b–e showed higher activity compared to curcumin | 38 |
| 15a–e | 2–3 h heating of precursors in EtOH at 60 °C in presence of acetic acid | 91–94 | Antibacterial/antifungal and anti-inflammatory | Only 15e showed higher activity compared to curcumin | 38 |
| 16a–f and 17a–f | Reflux of precursors in EtOH in presence of acetic acid | 71.5–92.5 | Antibacterial/antifungal and anti-inflammatory | 16a was most potent compound | 39 |
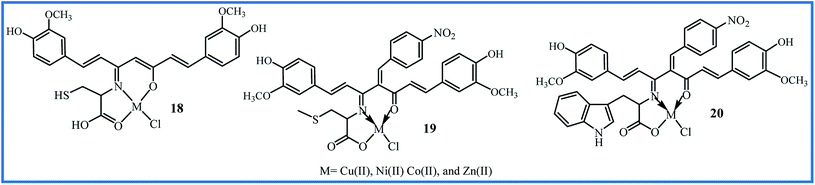
The synthetic methodology, yields, and distinct biological activity of 18–20 are summarized in Table 3.
| Compound | Condition of synthesis | Yield (%) | Activity | Outcome | Reference |
|---|---|---|---|---|---|
| 18 | 6 h reflux of precursors in EtOH | 60 | Antibacterial/antifungal and DNA cleavage | Cu(II) complex showed the highest activities but was not compared with curcumin | 41 |
| 19 | 6 h reflux of precursors in EtOH | 60 | Antibacterial/antifungal and DNA cleavage | Cu(II) complex was the most potent compound but was not compared with curcumin | 42 |
| 20 | 6 h reflux of precursors in EtOH | 63 | Antibacterial/antifungal and DNA cleavage | Cu(II) complex showed the highest activities but was not compared with curcumin | 43 |
2.3. Schiff base derivatives of curcumin's analogues
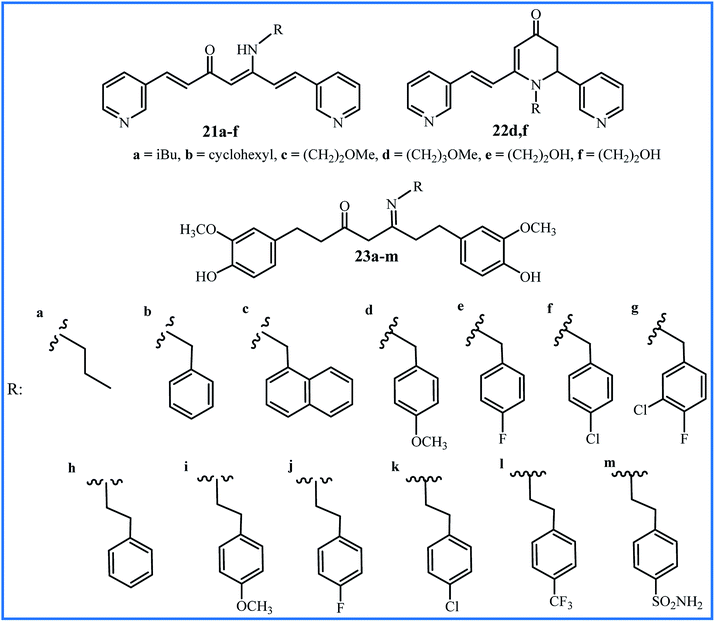
Improving the bioavailability and stability of curcumin can be one of the goals to form the new analogues of curcumin. Recently, Camp, D'hooghe and co-workers focused on a class of aza-aromatic curcuminoids, including pyridine-, indole-, and pyrrole-based curcumin analogues with slightly improved water solubility compared to the curcumin. Subsequently, they selected the bispyridine analogue to be used in the Schiff base reaction and preparation of the corresponding β-enaminone derivatives (21a–f and 22d,f in Fig. 6). It was founded that β-enaminone 21a–f and 22d,f (bearing a polar aliphatic moiety) indicated remarkably increase in water solubility in the range of 4 to 1600-fold compared to curcumin. The evaluation of antioxidant activities using DPPH and FRAP assays indicated a decrease compared to curcumin, which can be attributed to the absence of hydroxyl groups in the aromatic rings of these compounds. The evaluation of cytotoxic effects of compounds by MTT assay and cancer cell lines HepG2, EA.hy926, Caco-2, HT-29, and CHO-K1 demonstrated that 21a–d exhibited moderate growth inhibition, while 21e and 21f indicated no activity at concentration 75 μM. The β-enaminone 21a exhibited the highest anti-proliferative activity that was similar to the activity of curcumin. Intracellular ROS for compounds was dependent on the cell types. Compounds 21a–e indicated moderate to high ROS generation at 10 μM only in Caco-2 cells. In general, N-alkyl β-enaminone curcuminoids presented an acceptable balance between good water solubility and notable bioactivity. The authors proposed that β-enaminone curcuminoids with nonpolar aliphatic side chains can be used for further progress in medicinal chemistry.44Recently, Mahal and co-workers used the tetrahydrocurcumin (THC), a metabolite of curcumin, to prepare thirteen Schiff base derivatives 23a–m.45 All compounds showed moderate to good anti-cancer activity against cancer cell lines A549, HeLa and MCF-7, except compounds 23a and 23m (IC50 > 50 μM). Compound 23l with a CF3 group at the para-position of the phenyl ring showed the highest activity (IC50 values 4.8 μM against MCF-7, 11.9 μM against A549 and 12.7 μM against HeLa cell lines).
The synthetic methodology, yields, and distinct biological activity of 21–23 are summarized in Table 4.
| Compound | Condition of synthesis | Yield (%) | Activity | Outcome | Reference |
|---|---|---|---|---|---|
| 21a–f and 22d,f | Heating of precursors in 2-methyl-THF or EtOH under microwave irradiation for 75–105 min at 80–85 °C in presence of MK10 clay and acetic acid | 6–24 | Anticancer and antioxidant | Enhanced water-solubility, comparable anticancer and lower antioxidant activities to curcumin | 44 |
| 23a–m | 10 h reflux of precursors in EtOH in presence of acetic acid | 45–94 | Anticancer | 23l was the most potent compound but was not compared with curcumin | 45 |
2.4. Schiff base macro-structures for release of curcumin
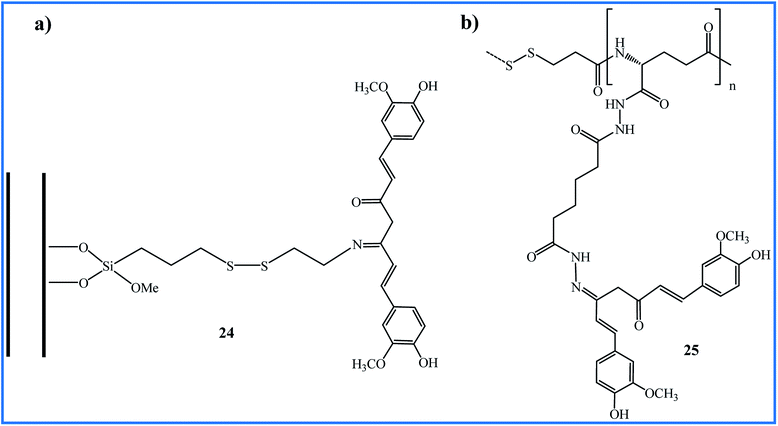
The use of triggered carrier can improve the circulation-stability and bioavailability of curcumin. One of the interesting syntheses of Schiff base compounds from curcumin has been reported by Riela et al. They functionalized the halloysite nanotubes (HNTs) with curcumin through Schiff base formation as a stimuli-responsive linkage. The resulting HNT–curcumin 24 prodrug has dual stimuli-responsive ability upon contact to the glutathione-rich or acidic environment due to the presence of disulfide and imine linkages (Fig. 7a). The anti-proliferative in vitro studies against HA22T/VGH and Hep3B cell lines indicated that HNT–Cur prodrug induced high cytotoxicity (% viability < 20 with a curcumin concentration up to 50 μM) compared to the free curcumin or pristine HNT (% viability > 90). The prodrug preserved its antioxidant characteristics and showed excellent stability in normal physiological conditions.46In 2017, Jayakumar and co-workers, during the four-step reactions from γ-polyglutamic acid (γ-PGA), 3-mercaptopropionic acid (3-MPA), and adipic acid dihydrazide (ADH) prepared the (γ-PGA-SS-ADH) polymer. This polymer was conjugated with curcumin via hydrazone to obtain γ-PGA-SS-ADH-Cur (25 in Fig. 7b). In the presence of the stabilized aqueous medium, polymeric conjugate 25 was formulated into self-assembled core–shell nanoparticles (NPs) that showed pH and redox-responsive curcumin delivery. The microscopic study of the uptake of NPs by HT-29 cells revealed the presence of NPs in the lysosomes via the endocytosis mechanism. NPs exhibited higher cytotoxicity against HT-29 cells at lower pH and the absence of H2O2, which confirms that conjugated 25 can be used as a developing drug delivery system in cancer therapy.47
Sathianarayanan et al. synthesized the curcumin conjugated chitosan microspheres (CCCMs) by Schiff base reaction between amino groups of chitosan biopolymer and carbonyl groups of curcumin. CCCMs showed the sustained release of curcumin and maximum release observed after 48 h. Antimicrobial activity of CCCMs against S. aureus and E.coli was relatively higher compared to free curcumin. The results of DPPH and H2O2 assays showed the IC50 values of 216 μg mL−1 and 228 μg mL−1 for CCCMs, respectively, which indicated the antioxidant activity of CCCMs is more or less equal to the free curcumin. The anti-inflammatory potential of CCCMs, obtained by the bovine serum albumin method, was similar to that of curcumin with IC50 about 45 μg mL−1. The prepared microspheres did not exhibit any toxicity in normal NIH 3T3 (mouse embryonic fibroblast) cells, suggesting that CCCMs can be useful for biomedical fields.48
In 2018, a delivery nanosystem was designed with the folate (FA)-receptor targeting mesoporous silica nanoparticles to load the curcumin through Schiff base reaction. This designed curcumin-loaded nanoparticles showed higher cellular uptake in FA-receptor-rich MCF-7 cells compared to FA-receptor-poor HEK-293T cell lines. They did not show remarkable toxicity even while the incubated concentration increased to 640 μg mL−1.49
The synthetic methodology, curcumin loading (wt%), and distinct biological activity of 24 and 25 are summarized in Table 5.
| Compound | Condition of synthesis | Curcumin loading (wt%) | Activity | Outcome | Reference |
|---|---|---|---|---|---|
| 24 | Stirring of precursors in EtOH under microwave irradiation for 1 h at 80 °C | 2.9 | Anticancer | The prodrug showed the higher cytotoxicity compared to curcumin | 46 |
| 25 | Stirring of precursors in MeOH/H2O for 72 h at 37 °C in presence of acetic acid | 5 ± 2 | Anticancer | Compound showed a pH and redox-responsive and higher cytotoxicity compared to curcumin | 47 |
3. Hydrazone derivatives of curcumin
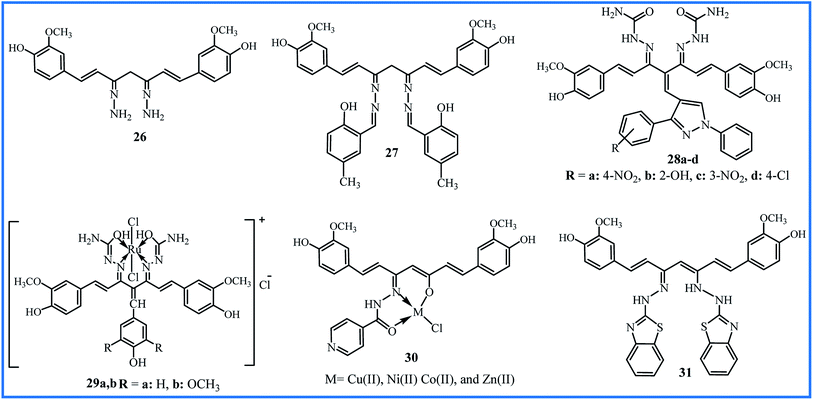
Hydrazines (compounds with NH2–NH2 functional group) can easily contribute to Schiff base reaction and form the hydrazone compounds. Venkatesh et al. synthesized the curcumin derivative 26 (Fig. 8) by the condensation reaction of curcumin and hydrazine hydrate in a methanol solution. Schiff base ligand 27 was then obtained by further condensation of derivative 26 with methyl salicylaldehyde. The metal chloride salts were used to obtain the Cu(II), Ni(II), and Zn(II) complexes of ligand 27. This study was useful to considerate the interaction mechanism of the complexes with DNA, and it can be valuable in the new chemotherapy drug progress. The in vitro DNA binding studies were performed by the absorption titration of metal complexes with CT-DNA concentrations. The spectroscopic changes indicated the intrinsic binding constants 4.72 × 104 M−1, 3.89 × 104 M−1, and 3.13 × 104 M−1 for Cu(II), Ni(II), and Zn(II) complexes, respectively. Fluorescence measurements showed a strong quenching in the emission spectrum of DNA-EB by the addition of metal complexes, which proved the binding of complexes to DNA (EB is a molecular fluorophore that emits fluorescence in the presence of CT-DNA). These results revealed that all complexes interact with the DNA through intercalating binding mode, and Cu(II) complex showed better binding affinity than Ni(II), and Zn(II) complexes.50In 2013, an enlarged study was performed on the synthesis and pharmacological properties of a series of Knoevenagel condensates and disemicarbazones derivatives of curcumin.51 The ortho, meta and para-substituted pyrazolealdehydes were synthesized and applied to prepare of Knoevenagel's condensates with curcumin. The final disemicarbazones products 28a–d (Fig. 8) were obtained by reaction of Knoevenagel's condensates and semicarbazide hydrochloride. The cytotoxicity of the compounds was evaluated by in vitro hemolytic assay by Mammalian RBCs, and the interaction of compounds with DNA was studied by UV-vis spectroscopy. The compounds 28a–d showed higher Kb (2.6 × 104 to 8.1 × 105 M−1) compared to initial pyrazolealdehydes (1.4 × 103 to 7.6 × 104 M−1) and Knoevenagel's condensates derivatives (1.4 × 104 to 7.8 × 105 M−1). The less hemolytic than doxorubicin (7% to 25%), the low cell line viability even at low concentration 1.0 μg mL−1, and high DNA binding constant indicated that compounds 28a–d could be good anticancer agents. The in vitro anticancer profiles of compounds have determined by MTT assay against human breast cancer cell line MCF-7, and results showed the viabilities 59% to 79% at 1.0 L g mL−1. The initial pyrazolealdehydes derivatives showed higher anticancer activities, which can be attributed to the smaller size and ability to pass the membrane.
The incorporation of metals into the structure of ligands and the formation of complexes enhance the biological activity. Imran's research team synthesized two Knoevenagel derivatives, two ligands and finally two ruthenium(III) complexes of curcumin derivatives (29a,b in Fig. 8) with octahedral geometries. Compared to other metal complexes, ruthenium complexes show improved antitumor activity and higher binding capacities to DNA due to octahedral structures.52 The binding constants, calculated by UV-vis spectroscopy, were 1.46 × 104 and 3.54 × 104 M−1 for 29a and 29b. The fluorometry spectroscopy also showed the strong binding of compounds with DNA. Determination of the toxicity of complexes was performed by hemolytic assays based on the effects of the compound on human RBCs. The complexes showed lower toxicity than the standard anticancer drug letrazole. Also, they indicated good cytotoxicity against cervical cancer (HeLa) cell line (IC50 values of 0.004 to 20.35 nM) and moderate cytotoxicity against other cell lines, including liver hepatocellular carcinoma, breast cancer, and human colon adenocarcinoma.
In 2019, the biological activities of curcumin-derived hydrazone 30 and its metal complexes were studied as in silico and in vitro. The UV-vis spectroscopy verified a groove mode of binding between the CT DNA and metal complexes. The Cu(II) complex with Kb value of 2.9 × 10−4 M showed the highest binding efficiency. The agarose gel electrophoresis technique in the presence of oxidant H2O2 revealed that complexes could cleave pUC 19 (Plasmid University of California) DNA. So, they can be purposed as the potent anticancer agents. DFT analysis using the Gaussian 09 W Program demonstrated that the copper complex has a higher dipole moment, leading to a better interaction with the biological systems. This higher biological availability is experimentally verified by antimicrobial assays. The metal complexes showed MIC between 3.1 and 5.6 (× 10−3 μM) against S. aureus, B. subtilis, K. pneumonia, and E. coli bacteria, while MIC values for ligand 30 were between 10.3 and 13.0 (× 10−3 μM). Also, MIC values were between 11.1 and 15.4 (× 10−3 μM) against A. niger, A. flavus, C. lunata, R. bataticola, and C. albicans fungi for metal complexes. MIC values for the antifungal activity of ligand 30 were between 20.4 and 22.7 (× 10−3 μM). The most significant antibacterial and antifungal activities were shown by Cu(II) complex.53
Recently, the hydrazone compound 31, as a tautomeric form, was prepared by reaction of 2-hydrazinobenzothiazole and curcumin in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. The copper(II) coordination complexes in 1
1 molar ratio. The copper(II) coordination complexes in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratios were synthesized using the copper chloride, bromide, acetate and nitrate by template method. The antioxidant capacity was evaluated by comparing the activity of compounds with TROLOX as the standard by photo chemiluminescence method. The total antioxidant capacity of ligand 31 (16.88%) revealed to be slightly lower than the capacity of curcumin (17.07%). All copper complexes, with the total antioxidant capacity between 22.34% and 28.70%, were better antioxidant agents than curcumin attributed to redox reactions within the Cu2+/Cu+ couple. The synthesized complexes by copper nitrate that have 2
2 molar ratios were synthesized using the copper chloride, bromide, acetate and nitrate by template method. The antioxidant capacity was evaluated by comparing the activity of compounds with TROLOX as the standard by photo chemiluminescence method. The total antioxidant capacity of ligand 31 (16.88%) revealed to be slightly lower than the capacity of curcumin (17.07%). All copper complexes, with the total antioxidant capacity between 22.34% and 28.70%, were better antioxidant agents than curcumin attributed to redox reactions within the Cu2+/Cu+ couple. The synthesized complexes by copper nitrate that have 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 metal
1 metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand molar ratio, exhibited the highest antioxidant capacity.54
ligand molar ratio, exhibited the highest antioxidant capacity.54
The synthetic methodology, yields, and distinct biological activity of 26–31 are summarized in Table 6.
| Compound | Condition of synthesis | Yield (%) | Activity | Outcome | Reference |
|---|---|---|---|---|---|
| 26 and 27 | Stirring of precursors in MeOH for 6 h, then 8 h reflux | No reported | DNA cleavage | Cu(II) complex showed the highest Kb | 50 |
| 28a–d | 24 h reflux of precursors in CHCl3/MeOH in presence of piperidine | 60–67 | DNA cleavage and anticancer | All compounds showed the good DNA interacting tendency | 51 |
| 29 | Stirring of precursors in MeOH for 24 h in presence of piperidine | No reported | DNA cleavage and anticancer | Two ruthenium(III) complexes were potent for treatment of cervical cancer | 52 |
| 30 | Reflux of precursors in EtOH for 4 h in presence of acetic acid | 80 | Antibacterial/antifungal and DNA cleavage | Cu(II) complex showed the highest activities | 53 |
| 31 | Reflux of precursors in EtOH | No reported | Antioxidant | All Cu(II) complexes showed the higher activity than curcumin | 54 |
4. Oxime and oxime ether derivatives of curcumin
Oximes, an abbreviation of oxy-imine, and oxime ethers are the advantaged groups in chemistry that show a broad range of biological and pharmaceutical activities.55,56 In this section, we reviewed the performed efforts to incorporate these groups into the structure of the curcumin or its analogues.The first oxime analogue of curcumin was synthesized by Dutta et al. in 2001 through condensation of curcumin and hydroxylamine hydrochloride. They also prepared the copper complex of the obtained oxime analogue and showed that copper conjugation strangely improved the antioxidant activity of the parent oxime ligand.57 In 2008, Simoni et al. synthesized two dioxime ether analogues of curcumin by O-methyl or O-benzyl hydroxylamine hydrochloride. The antitumor effects against MCF-7 and multidrug-resistant (MDR) variant MCF-7R cancer cell lines showed that benzyl oxime derivative has a high antitumor activity even against MDR tumors.58
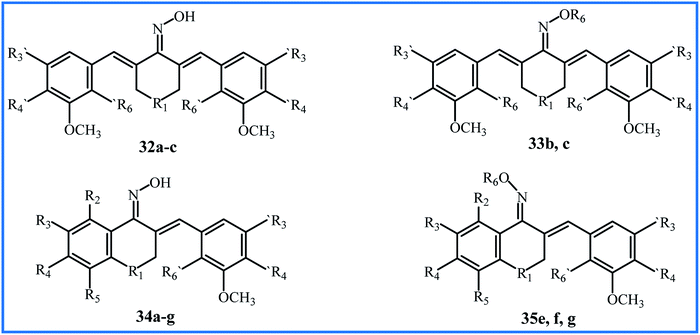
Qin et al. conducted extensive research on sixty-nine α,β-unsaturated carbonyl-based compounds, oxime and oxime ether analogues in 2016. Fifteen new oxime and oxime ether compounds 32–35 are shown in Fig. 9, Tables 7 and 8. MTT assay by MCF-10A cells showed that all compounds were nontoxic with cell viability of more than 90%. The structure–activity relationship (SAR) of compounds was determined for their anti-proliferative activity against PC-3, HT-29, MCF-7, H-460, A-549, PaCa-2 and Panc-1 cancer cell lines. All oxime analogues 32a–c and 34a–g exhibited more inhibition of cancer cells than parent α,β-unsaturated carbonyl-based compounds, and all oxime ether analogues 33b,c and 35e,f,g exhibited less inhibition of cancer cells compared to parent oximes. Oxime compound 34g showed the highest anti-proliferative activity against most human cancer cell lines, with an IC50 value of 0.02 μM. From all compounds, the ten most effective 32b,c, 33c and 34a–g were chosen for further anticancer mechanistic studies. Oxime compound 34f with nitro substitution at position R3 exhibited the most potent BRAFV600E inhibitory activity (IC50 = 0.9 μM), and Oxime 32c with N–CH3 substitution at the R1 position exhibited the highest EGFR inhibitory activity with an IC50 value of 0.07 μM, which was equivalent to the positive control Erlotinib.59 Later, Masand et al. applied the multiple quantitative structure–activity relationships (QSARs) models for the anticancer activity of the compounds 32–35. The models showed that the internal electronic environment of compounds and a specific combination of nitrogen and oxygen atoms connected by five bonds have an association with the anticancer activity.60
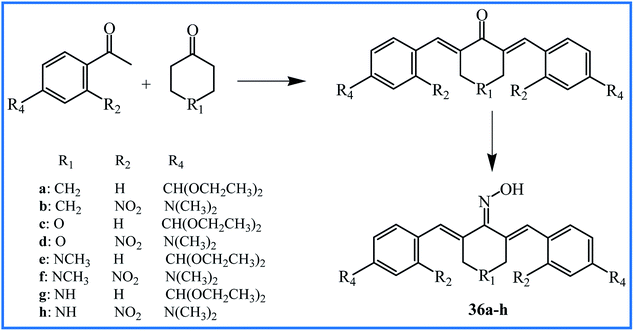
Two years later, Qin's research team reported the synthesis of new curcumin-like oxime analogues 36a–h (Fig. 10) using α,β-unsaturated carbonyl based anticancer compounds. MTT assay by MCF-10A cells showed that all oxime analogues were nontoxic with cell viability of more than 88%. The results of the anti-proliferative assay against above mention cell lines indicated that all analogues exhibited strong anti-proliferative activity with IC50 values between 0.02 and 2.7 μM. Analogues 36g and 36h were most potent agents. The SAR analysis showed that compounds with a combination of 4-N(CH3)2 and 2-NO2 groups exhibited considerably potent anticancer activity compared to those having –CH(OCH2CH3)2 group at position 4 of rings. The mechanistic study revealed that compounds 36c and 36d were the most potent inhibitors of tubulin assembly; compounds 36e and 36f were the most potent inhibitors of EGFR, and all compounds were potent inhibitors of BRAFV600E with IC50 values between 1.3–3.1 μM.61
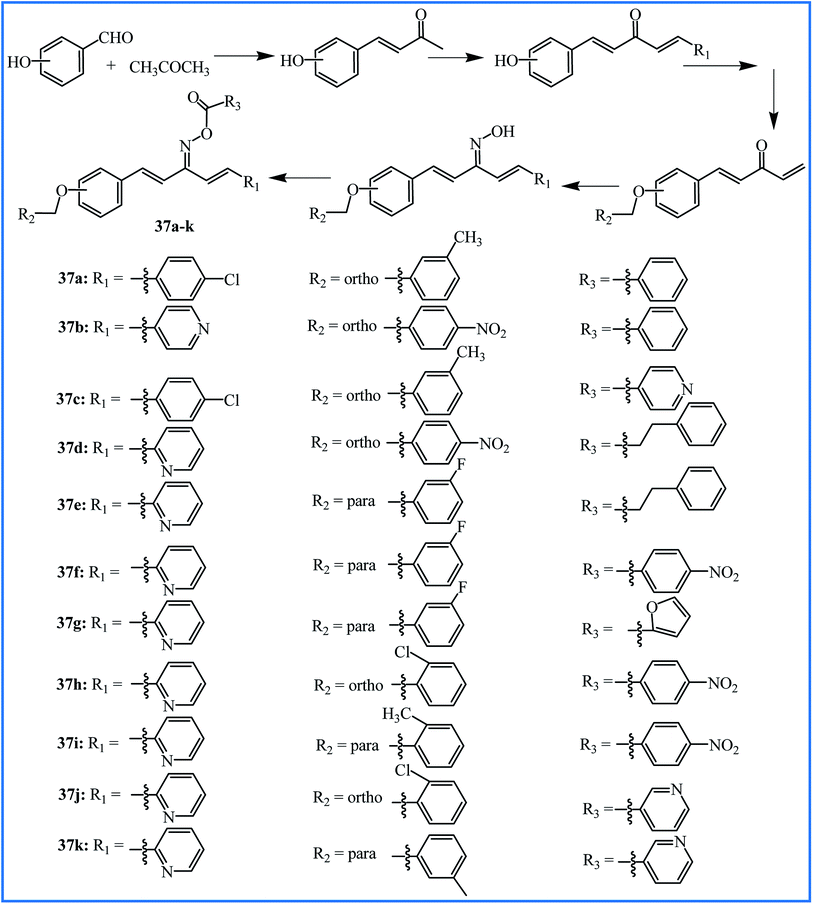
To progress the new anti-inflammatory agents, Li, Zhang and co-workers designed and synthesized twelve pentadienone oxime ester derivatives, which can be considered as the curcumin analogues (37a–k in Fig. 11). Among all the derivatives, compound 37j showed good anti-inflammatory activity with the highest inhibition of LPS-induced nitric oxide (NO) and IL-6 release in murine monocyte–macrophage RAW 264.7 cells with IC50 values of 6.66 and 5.07 μM, respectively. The pro-inflammatory mediators NO and IL-6 have significant roles in the progression of inflammation-related diseases. The SAR analysis revealed that pyridine ring substituent at R3 is better than that of benzene, and substituent R2 of benzene possessed a significant influence on inhibition activity.62
The synthetic methodology, yields, and distinct biological activity of 32–37 are summarized in Table 9.
| Compound | Condition of synthesis | Yield (%) | Activity | Outcome | Reference |
|---|---|---|---|---|---|
| 32–35 | 32 and 34: 6–8 h reflux of α,β-unsaturated carbonyl compounds with NH2OH·HCl in EtOH | 42–62 | Anticancer | 34f and 34g were the most potent anticancer agents | 60 |
| 33 and 35: 8 h reflux of 32 and 34 with haloalkylamine in dry acetone in presence of K2CO3 | |||||
| 36a–h | Reflux of α,β-unsaturated carbonyl compounds with NH2OH·HCl in EtOH | 39–51 | Anticancer | 36g and 36h were the most potent anticancer agents | 61 |
| 37a–k | Stirring of precursors for 5 h at 0–5 °C in dry acetone in presence of K2CO3 | 48.2–76 | Anti-inflammatory | 37j was the most potent anti-inflammatory agent | 62 |
5. Summary and future perspectives
Schiff base reaction is an interesting route for many chemists due to simple, efficient, and high yield protocol. In the last decade, several attempts have been made by many researchers to improve the pharmacological activities of curcumin by Schiff base reaction. The hydroxy and methoxy groups play a vital role in the antioxidant and anticancer properties of curcumin, and modification of curcumin through carbonyl groups does not interfere with these groups. So, Schiff base reaction is a valuable method to the formation of new curcumin derivatives.The discussed examples in this review were investigated for antibacterial activity (compounds 1, 2, 8–11, 13–20, and 30), antifungal activity (compounds 2, 9–11, 14–20, and 30), antioxidant activity (compounds 4–7, 12, 21, 22, and 31), anti-inflammatory activity (compounds 14–17 and 37), DNA cleavage efficacy (compounds 3, 9, 10, 18–20, and 26–30), and anticancer activity (compounds 5–7, 11, 12, 21–25, 28, 29, 32–36).
For several compounds (4–8, 13–17, 21, 22, 24, 25, and 31), the planned activities have been compared with those of curcumin, and in most of them, an improvement in activity can be seen. This instruction can't apply to antioxidant properties, and compounds 4, 5g–i, 6e–h, 7, 21, 22, and 31 exhibited lower antioxidant activity compared to curcumin. This decrease in antioxidant activity is very notable in compounds 21 and 22 that have been synthesized from a curcumin analogue without hydroxy and methoxy groups on the aromatic rings. Among the compound, only 6a–d exhibited a slightly higher antioxidant activity compared to curcumin.
In several studies, including compounds 1–3, 8–11, 18–20, 29, 30 and 31, the complexes of ligands with various metals such as Cu(II), Ni(II), Co(II), and Zn(II) have also been synthesized. All metal complexes showed improved activities (including antibacterial, antifungal and DNA cleavage) compared to parent ligands. About the antibacterial and antifungal activities, the zinc complex of 1 and copper complexes of 2, 3, 8–11, and 18–20 showed the highest activity. The authors explained this advancement of activity by Overtone's concept63 and the Tweedy's chelation theory.64 Based on Overtone's concept, the lipophilicity of the cell membrane is a significant factor that controls cell permeability. According to chelation, the overlap of the orbital of ligand and partial distribution of the positive charge of the metal ion with donor groups leads to a principal decrease of the polarity of the metal ion and the increase in the delocalization of π-electrons over the chelate ring. The decreased polarity of the metal ion enhances the lipophilicity and penetration of the complexes into the membrane of the cell. The complexes can block the metal-binding sites in the enzymes of microorganisms and disturb the vital cellular processes such as the respiration process and synthesis of proteins. In most reports, the copper complex shows higher activity than other complexes, which is due to its greater size that decreases the polarization. It is also stated that the enhanced activity of Cu(II) complex can be attributed to effective binding with DNA.32
Designing the drug delivery systems that bind to curcumin through Schiff base reaction is a useful strategy because it can result in stimuli-responsive release of curcumin in the target tissue. Cancer microenvironment is reported to have lower pH than the normal tissues, and the Schiff base bond as a dynamic chemical bond can be hydrolyzed in the acidic environment to release curcumin. Therefore, the use of hydrophilic polymers including the amino groups is suggested to develop the stimuli-sensitive delivery systems. These systems not only lead to targeted delivery of curcumin, but can also significantly increase the water-solubility of this compound. It is also apparent from the studies that presence of polar groups including hydroxy and methoxy groups in the introducing amine compound (6a–h) also increases the water-solubility of curcumin.
Comparing the results obtained for 14–17 derivatives indicated that formation Schiff base on both carbonyl groups of curcumin decrease the biological activities. An overview of all compounds also proved that most compounds with outstanding activities (such as 5a–f, 5i, 6a–d, 14b–e, 16a and 23l) have an unblocked carbonyl group.
The results of studies also revealed that the combination of curcumin with other drug families such as sulfonamides leads to a synergistic effect and improves biological properties. It seems that the use of amine compounds containing different pharmacological groups to attach the curcumin by Schiff base reaction can be an effective way to obtain new analogues with improved properties. We complete this review by hoping that it will stimulate researchers to develop the new and different derivatives of curcumin to obtain qualified compounds in anticancer, antimicrobial, antioxidant, and other medicinal and biological fields.
Conflicts of interest
There are no conflicts of interest.Acknowledgements
The authors are appreciative for budgetary help from Iran National Science Foundation (INSF) as well as Lorestan University for facilitations.References
- Q. Yao, Z. q. Ke, S. Guo, X. s. Yang, F. x. Zhang, X. f. Liu, X. Chen, H. g. Chen, H. y. Ke and C. Liu, J. Mol. Cell. Cardiol., 2018, 124, 26–34 CrossRef CAS PubMed.
- D. Bagchi, S. Chaudhuri, S. Sardar, S. Choudhury, N. Polley, P. Lemmens and S. K. Pal, RSC Adv., 2015, 5, 102516–102524 RSC.
- A. I. Joseph, R. L. Edwards, P. B. Luis, S. H. Presley, N. A. Porter and C. Schneider, Org. Biomol. Chem., 2018, 16, 3273–3281 RSC.
- X. X. Yang, C. M. Li, Y. F. Li, J. Wang and C. Z. Huang, Nanoscale, 2017, 9, 16086–16092 RSC.
- S. Izui, S. Sekine, K. Maeda, M. Kuboniwa, A. Takada, A. Amano and H. Nagata, J. Periodontol., 2016, 87, 83–90 CrossRef CAS PubMed.
- G. Djiokeng Paka, S. Doggui, A. Zaghmi, R. Safar, L. Dao, A. Reisch, A. Klymchenko, V. G. Roullin, O. Joubert and C. Ramassamy, Mol. Pharm., 2016, 13, 391–403 CrossRef CAS PubMed.
- J. Chen, Z. M. He, F. L. Wang, Z. S. Zhang, X. z. Liu, D. D. Zhai and W. D. Chen, Eur. J. Pharmacol., 2016, 772, 33–42 CrossRef CAS PubMed.
- H. Soleimani, A. Amini, S. Taheri, E. Sajadi, S. Shafikhani, L. A. Schuger, V. B. Reddy, S. K. Ghoreishi, R. Pouriran and S. Chien, J. Photochem. Photobiol. B Biol., 2018, 181, 23–30 CrossRef CAS PubMed.
- M. Qureshi, E. A. Al-Suhaimi, F. Wahid, O. Shehzad and A. Shehzad, Neurol. Sci., 2018, 39, 207–214 CrossRef PubMed.
- F. N. Kaufmann, M. Gazal, C. R. Bastos, M. P. Kaster and G. Ghisleni, Eur. J. Pharmacol., 2016, 784, 192–198 CrossRef CAS PubMed.
- P. Kumaraswamy, S. Sethuraman and U. M. Krishnan, J. Agric. Food Chem., 2013, 61, 3278–3285 CrossRef CAS PubMed.
- R. K. Singh, D. Rai, D. Yadav, A. Bhargava, J. Balzarini and E. De Clercq, Eur. J. Med. Chem., 2010, 45, 1078–1086 CrossRef CAS PubMed.
- M. G. El-Gazzar, N. H. Zaher, E. M. El-Hossary and A. F. Ismail, J. Photochem. Photobiol. B Biol., 2016, 162, 694–702 CrossRef CAS PubMed.
- Y. P. Bharitkar, M. Das, N. Kumari, M. P. Kumari, A. Hazra, S. S. Bhayye, R. Natarajan, S. Shah, S. Chatterjee and N. B. Mondal, Org. Lett., 2015, 17, 4440–4443 CrossRef CAS PubMed.
- M. Singh, A. Hazra, Y. P. Bharitkar, R. Kalia, A. Sahoo, S. Saha, V. Ravichandiran, S. Ghosh and N. B. Mondal, RSC Adv., 2018, 8, 18938–18951 RSC.
- L. Rišiaňová, E. Fischer-Fodor, J. Valentová, C. Tatomir, N. C. Decea, P. Virag, I. Pechová, F. Devínsky and N. Miklášová, Bioorg. Med. Chem. Lett., 2017, 27, 2345–2349 CrossRef PubMed.
- A. Arezki, G. G. Chabot, L. Quentin, D. Scherman, G. Jaouen and E. Brulé, Med. Chem. Commun., 2011, 2, 190–195 RSC.
- F. C. Rodrigues, N. A. Kumar and G. Thakur, Eur. J. Med. Chem., 2019, 177, 76–104 CrossRef CAS PubMed.
- S. A. Noureddin, R. M. El-Shishtawy and K. O. Al-Footy, Eur. J. Med. Chem., 2019, 182, 111631 CrossRef CAS PubMed.
- S. Zhao, C. Pi, Y. Ye, L. Zhao and Y. Wei, Eur. J. Med. Chem., 2019, 180, 524–535 CrossRef CAS PubMed.
- P. Y. Khor, M. F. F. M. Aluwi, K. Rullah and K. W. Lam, Eur. J. Med. Chem., 2019, 183, 111704 CrossRef CAS PubMed.
- M. A. Tomeh, R. Hadianamrei and X. Zhao, Int. J. Mol. Sci., 2019, 20, 1033 CrossRef CAS PubMed.
- M. Thomas, A. Kulandaisamy and A. Manohar, Int. J. Chem. Tech. Res., 2012, 4, 247–257 CAS.
- S. Sumathi, P. Tharmaraj, C. Sheela and R. Ebenezer, J. Coord. Chem., 2012, 65, 506–515 CrossRef CAS.
- M. Jayandran, M. Haneefa and V. Balasubramanian, Int. J. Chem. Nat. Sci., 2014, 2, 157–163 Search PubMed.
- M. Jayandran, M. Haneefa and V. Balasubramanian, Indian J Sci Technol, 2016, 9, 1–9 CAS.
- J. Rajesh, A. Gubendran, G. Rajagopal and P. Athappan, J. Mol. Struct., 2012, 1010, 169–178 CrossRef CAS.
- M. H. Afsarian, M. Farjam, E. Zarenezhad, S. Behrouz and M. N. S. Rad, Acta Chim. Slov., 2019, 66, 874–887 CrossRef CAS.
- R. Revathi, D. A. Ananth, A. Rameshkumar and T. Sivasudha, Asian J. Chem., 2015, 27, 1489–1494 CrossRef CAS.
- R. De Vreese, C. Grootaert, S. D’hoore, A. Theppawong, S. Van Damme, M. Van Bogaert, J. Van Camp and M. D’hooghe, Eur. J. Med. Chem., 2016, 123, 727–736 CrossRef CAS PubMed.
- A. Theppawong, R. De Vreese, L. Vannecke, C. Grootaert, J. Van Camp and M. D’hooghe, Bioorg. Med. Chem. Lett., 2016, 26, 5650–5656 CrossRef CAS PubMed.
- A. Kareem, M. Arshad, S. A. Nami and N. Nishat, J. Photochem. Photobiol. B Biol., 2016, 160, 163–171 CrossRef CAS PubMed.
- N. Raman, T. Chandrasekar, G. Kumaravel and L. Mitu, Appl. Organomet. Chem., 2018, 32, e3922 CrossRef.
- J. Porkodi and N. Raman, Appl. Organomet. Chem., 2018, 32, e4030 CrossRef.
- A. Palanimurugan and A. Kulandaisamy, Asian J. Chem., 2018, 30, 1262–1268 CrossRef CAS.
- A. Kareem, M. S. Khan, S. A. Nami, S. A. Bhat, A. U. Mirza and N. Nishat, J. Mol. Struct., 2018, 1167, 261–273 CrossRef CAS.
- N. G. Gogoi and J. G. Handique, J. Chem. Sci., 2019, 131, 74 CrossRef.
- J. Lal, S. K. Gupta, D. Thavaselvam and D. D. Agarwal, Eur. J. Med. Chem., 2013, 64, 579–588 CrossRef CAS PubMed.
- M. Ahmed, M. A. Qadir, M. I. Shafiq, M. Muddassar, Z. Q. Samra and A. Hameed, Arab. J. Chem., 2019, 12, 41–53 CrossRef CAS.
- M. Ahmed, M. A. Qadir, A. Hameed, M. N. Arshad, A. M. Asiri and M. Muddassar, Biochem. Biophys. Res. Commun., 2017, 490, 434–440 CrossRef CAS PubMed.
- T. Chandrasekar, N. Pravin and N. Raman, Inorg. Chem. Commun., 2014, 43, 45–50 CrossRef CAS.
- T. Chandrasekar, N. Pravin and N. Raman, J. Mol. Struct., 2015, 1081, 477–485 CrossRef CAS.
- T. Chandrasekar and N. Raman, J. Mol. Struct., 2016, 1116, 146–154 CrossRef CAS.
- A. Theppawong, T. Van de Walle, C. Grootaert, M. Bultinck, T. Desmet, J. Van Camp and M. D'hooghe, ChemistryOpen, 2018, 7, 381–392 CrossRef CAS PubMed.
- A. Mahal, P. Wu, Z. H. Jiang and X. Wei, ChemistrySelect, 2019, 4, 366–369 CrossRef CAS.
- M. Massaro, R. Amorati, G. Cavallaro, S. Guernelli, G. Lazzara, S. Milioto, R. Noto, P. Poma and S. Riela, Colloids Surf. B, 2016, 140, 505–513 CrossRef CAS PubMed.
- S. Pillarisetti, S. Maya, S. Sathianarayanan and R. Jayakumar, Colloids Surf. B, 2017, 159, 809–819 CrossRef CAS PubMed.
- T. Saranya, V. Rajan, R. Biswas, R. Jayakumar and S. Sathianarayanan, Int. J. Biol. Macromol., 2018, 110, 227–233 CrossRef CAS PubMed.
- C. Chen, W. Sun, X. Wang, Y. Wang and P. Wang, Mater. Sci. Eng. C, 2018, 85, 88–96 CrossRef CAS PubMed.
- N. Priyadharshini, I. S. Pillai, C. Joel, R. B. Bennie, S. Subramanian and P. Venkatesh, Indo Am. j. pharm. sci., 2015, 2, 1491–1505 CAS.
- I. Ali, A. Haque, K. Saleem and M. F. Hsieh, Bioorg. Med. Chem., 2013, 21, 3808–3820 CrossRef CAS PubMed.
- I. Ali, K. Saleem, D. Wesselinova and A. Haque, Med. Chem. Res., 2013, 22, 1386–1398 CrossRef CAS.
- P. Jeyaraman, A. Alagarraj and R. Natarajan, J. Biomol. Struct. Dyn., 2020, 38, 488–499 CrossRef CAS PubMed.
- M. Călinescu, M. Fiastru, D. Bala, C. Mihailciuc, T. Negreanu-Pîrjol and B. Jurcă, J. Saudi Chem. Soc., 2019, 23, 817–827 CrossRef.
- C. Canario, S. Silvestre, A. Falcao and G. Alves, Curr. Med. Chem., 2018, 25, 660–686 CAS.
- D. S. Reddy, M. Kongot, S. P. Netalkar, M. M. Kurjogi, R. Kumar, F. Avecilla and A. Kumar, Eur. J. Med. Chem., 2018, 150, 864–875 CrossRef CAS PubMed.
- S. Dutta, A. Murugkar, N. Gandhe and S. Padhye, Metal based drugs, 2001, 8, 183–188 CrossRef CAS PubMed.
- D. Simoni, M. Rizzi, R. Rondanin, R. Baruchello, P. Marchetti, F. P. Invidiata, M. Labbozzetta, P. Poma, V. Carina and M. Notarbartolo, Bioorg. Med. Chem. Lett., 2008, 18, 845–849 CrossRef CAS PubMed.
- H.-L. Qin, J. Leng, C.-P. Zhang, I. Jantan, M. W. Amjad, M. Sher, M. Naeem-ul-Hassan, M. A. Hussain and S. N. A. Bukhari, J. Med. Chem., 2016, 59, 3549–3561 CrossRef CAS PubMed.
- V. H. Masand, N. N. El-Sayed, M. U. Bambole and S. A. Quazi, J. Mol. Struct., 2018, 1157, 89–96 CrossRef CAS.
- H. L. Qin, J. Leng, B. G. Youssif, M. W. Amjad, M. A. G. Raja, M. A. Hussain, Z. Hussain, S. N. Kazmi and S. N. A. Bukhari, Chem. Biol. Drug Des., 2017, 90, 443–449 CrossRef CAS PubMed.
- Q. Li, J. Zhang, L. Z. Chen, J. Q. Wang, H. P. Zhou, W. J. Tang, W. Xue and X. H. Liu, J. Enzyme Inhib. Med. Chem., 2018, 33, 130–138 CrossRef CAS PubMed.
- R. Ramesh and S. Maheswaran, J. Inorg. Biochem., 2003, 96, 457–462 CrossRef CAS PubMed.
- Y. Anjaneyulu and R. P. Rao, Synth React Inorg Met Org Chem, 1986, 16, 257–272 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |