Open Access Article
Open Access ArticlePortable environment-signal detection biosensors with cell-free synthetic biosystems†
Xiaomei Lin,
Yuting Li,
Zhixia Li,
Rui Hua,
Yuyang Xing and
Yuan Lu *
*
Key Laboratory of Industrial Biocatalysis, Ministry of Education, Department of Chemical Engineering, Tsinghua University, Beijing 100084, China. E-mail: yuanlu@tsinghua.edu.cn
First published on 26th October 2020
Abstract
By embedding regulated genetic circuits and cell-free systems onto a paper, the portable in vitro biosensing platform showed the possibility of detecting environmental pollutants, namely arsenic ions and bacterial quorum-sensing signal AHLs (N-acyl homoserine lactones). This platform has a great potential for practical environmental management and diagnosis.
The problems of environment and food threaten the human survival and health seriously. Many environmental or food pollutions have been attributed to chemicals or biological pathogens.1–4 The significant level of pollutants can result in many human health diseases.5 Strict regulations for the limited concentration of poisonous heavy metals in the environment have been established all over the world. In these cases, rapid medical diagnosis and pollution detection methods are urgently required. Recently, much progress has been made in the global healthcare detection for rapid, low-cost, sensitive, easy-to-use, and hand-hold platforms that combined new technologies such as the Internet-of-things (IoT) facilities and smartphones.6–8 Among the new hand-hold or wearable detection platforms, biosensors with the advantages of better specificity and sensitivity are becoming much widely used in the detection.
A biosensor is an analytical device that integrates the recognition of biologically derived molecules, with a suitable physicochemical transducer.9 Over the past half century, many signs of progress have been witnessed in biosensors from commercial glucose analyzers, immunosensors, DNA biosensors, nano-biosensors to sweat microfluidic biosensors. However, the state of this considerable new progress is still a proof-of-concept. It still faces the challenges of biosafety and biological stability for practical applications. Notably, with the development of synthetic biology, the whole-cell biosensors have been developed.9 In the whole-cell biosensors, living cells can detect particular biochemical molecules or physical stimulation after intergrading one or a couple of logic genetic circuits. Increasing number of logic genetic circuits based on cellular regulations were developed for environmental management or clinical diagnosis.10–14 However, the cell-based approaches were limited to the laboratory for the concerns of biosafety.15 A sterile technology is emerging in synthetic biology for these wide applications. A cell-free protein synthesis is the production of proteins using biological machinery in vitro without using living cells. Compared to the in vivo cellular protein synthesis, the cell-free systems are safe and easy to regulate. Combined with the synthetic logic genetic circuits, the cell-free systems can provide a versatile venue for the biological detection.16–25
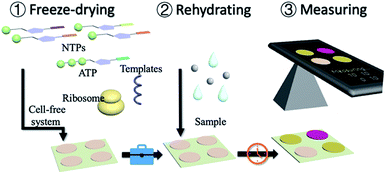
Herein, we present a portable in vitro sensing platform with synthetic genetic networks. With the basic materials of transcription and translation, the cell-free systems were embedded onto papers by freeze-drying for sterile applications (Fig. 1 and S1, ESI†). This system can be stably stored at room temperature and can be easily activated by adding water. The synthetic genetic networks, which can be regulated by the environmental inorganic pollutants, namely arsenic ions and pathogenic bacterial quorum sensing molecular AHLs, were employed for a demonstration.
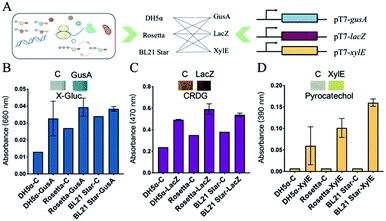
To successfully develop cell-free biosensing systems for testing and reporting, in vitro transcription and translation systems with suitable reporters were first investigated in small solution-phase reactions (Fig. S2, ESI†). For the visible output, three enzymes capable of mediating the color change were employed for the response to a conditional input. The three engaged enzymes, namely GusA (β-glucuronidase), LacZ (β-galactosidase), and XylE (catechol-2,3-dioxygenase) can cleave the colorless substrate X-Gluc into a blue product, the yellow substrate chlorophenol red-β-D-galactopyranoside (CRDG) into a purple chlorophenol red product, and the colorless low-cost substrate pyrocatechol into a yellow product, respectively. The color output can be measured by an ultraviolet spectrophotometer at 660 nm, 470 nm, and 390 nm for GusA, LacZ, and XylE cleaving products, respectively.26–28 Many attempts were also made to improve the visible performance by improving the enzyme yield and weakening the background of the cell-based endogenous enzymatic response. Three E. coli lysate-based cell-free systems were employed for in vitro input–output modification (Fig. 2A). The three cell-free extracts were basically derived from a genomic-modified strain DH5α that alone had the incomplete coding sequence of LacZ, as well as two commonly high-yield strain Rosetta (DE3) and BL21 Star (DE3).29,30 After the cell-free expression of the three enzymes, a color change developed after supplying the substrates. However, the results of GusA were not ideal because of the high background (Fig. 2B).
Although the background of Rosetta-based and BL21 Star-based systems was obviously stronger than that of the DH5α-based system, there was a distinct color contrast with 1.69-fold for the strong expression in the Rosetta-based system of the LacZ-based reporting system (Fig. 2C). Similarly, XylE-based reactions presented a maximum of 22.9-fold change with zero background (Fig. 2D and S3, ESI†). From these results, the two enzymes LacZ and XylE revealed the potential for expressing reporters with Rosetta-based cell-free expression systems.
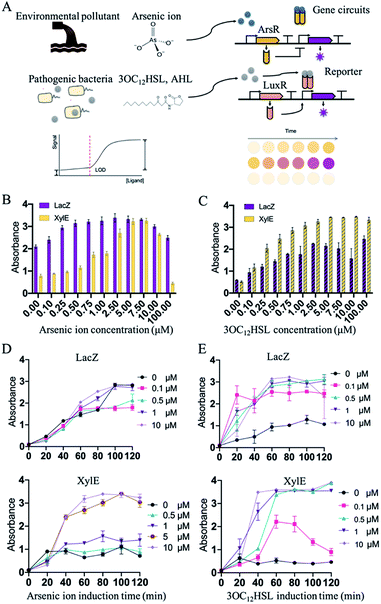
After the selection of an appropriate cell-free biosensing reporting system, the platform was then applied for the molecular detection of environmental or clinical samples. Two conventional molecules, namely arsenic ions and AHLs, known as environmental pollutants and Gram-negative bacterial population signals, respectively, were selected for the confirmation. Regulation of these expression systems were mediated by the ArsR repressor (arsenic ion) or LuxR (3OC12HSL, AHL) that bind the promoters, thus preventing transcription or activation (Fig. 3A). The Ars and Lux systems were commonly designed for the detection of 3OC12HSL and arsenic ions.25 Such in vitro regulations required constitutive arsR or luxR to be encoded into the genetic circuits. Expression of reporters (XylE or LacZ) was then induced by the inducer arsenic ion or 3OC12HSL. To optimize the detecting features, RBS (ribosome binding site) in different strengths related to LacZ or XylE was selected and characterized in the induction. RBS of LacZ or XylE was predicted and selected from the Salis RBS tools software in a range of predicted expression rates relative to LacZ or XylE.31,32 The constructs with the obvious signal development were selected for further construction of the detection system (Fig. S4–S8, ESI†).
Initially, the constructed plasmids were reacted in the cell-free solution-phase expression systems for the characterization of the detection limit and detection time. By varying only the concentration of arsenic ion, the cell-free reactions were incubated at 37 °C. Under the arsenic ion induction, the cell-free reaction system could quickly display the color changes after adding the substrate (Fig. S9, ESI†). From 0 μM to 100 μM incubation, the sensing systems with reporters of LacZ and XylE exhibited sensitivity towards arsenic ions by at least 1 μM with a visible color change (Fig. 3B and D). The arsenic detection limit of the XylE-based system was slightly higher than that of the LacZ-based system, and can contribute to the weak expression of XylE to reach the detection threshold. Moreover, high arsenic concentration (above 10 μM) showed toxicity towards the cell-free systems and the signal output decreased (Fig. 3B).
In addition to sensing inorganic ions presented above, this in vitro synthetic biology-based approach also held tremendous potential for sensing organic molecules. Similarly, the sensing system with the reporter of LacZ and XylE could detect 3OC12HSL of at least 0.1 μM (Fig. 3C and E). From the results, the cell-free systems in solution-phase could effectively detect the arsenic ions and 3OC12HSL with the LacZ reporter or XylE reporter.
As the biosensing systems could effectively detect the target molecules, the detection time was further investigated. The substrates were added into the cell-free reactions that were induced with arsenic ion or 3OC12HSL. After being induced with arsenic ion, the XylE-based system displayed visible output after 40 min incubation, and the LacZ-based system displayed the output within 60 min (Fig. 3D). Similarly, for the 3OC12HSL detection, the XylE-based system displayed visible output after 40 min incubation, and the LacZ-based system displayed the output within 20 min (Fig. 3E). The total time to perform the biosensor assay was within 1 h (from inducing to the visible output reading), which is an order of magnitude less than that with the whole-cell biosensor.
Although the cell-free systems were capable of detecting environmental pollutants (arsenic ions) and bacterial secretions (AHLs), it was more important for the adoption and commercial applications of these sensing systems. To realize it, a paper disc embedded with freeze-dried cell-free systems, which could perform biosensing after rehydration, was then carried out. Based on the assumption mentioned above, we embedded the cell-free systems without using a synthetic genetic network onto the 4 mm paper discs by the freeze-drying method (Fig. S10, ESI†). The paper discs containing cell-free systems could be stored at room temperature.
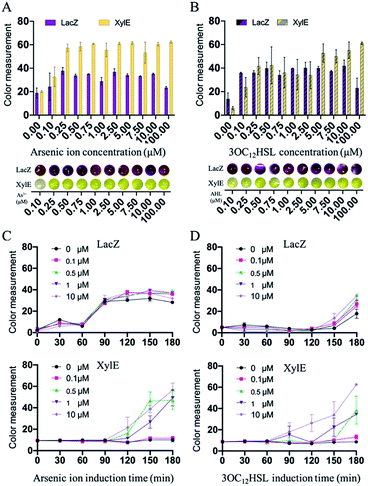
After the preparation of paper discs, the synthetic genetic networks with the inducer were added onto the paper discs to activate the sensing systems (Fig. 1 and S1, ESI†). The rehydrated paper discs were incubated in a 96-well plate at 37 °C. Due to the endogenous background of cell lysates in LacZ-based sensing systems, the yellow substrate (CRDG) was supplied to the reactions after incubation to avoid the chromogenic reactions performed by the endogenous enzymes before the expression (Fig. S11, ESI†). Pyrocatechol could be supplied to the reactions prior to the freeze-drying and cleaved along with incubation in the XylE-based systems, which were more convenient. After hours of incubation and colorimetric reaction, the papers gradually exhibited visible color outputs, and were captured by the camera of a smartphone, which could be easily quantitatively analyzed by the ImageJ software. The paper-based rehydrated cell-free biosensing system produced a visible inducer-dependent response with arsenic ions or 3OC12HSL (Fig. 4A and B), with maximum expression rates occurring within 3 h (Fig. 4C and D). Among the signal development, the XylE-based system detected 0.5 μM arsenic ions in 2 h (Fig. 4C). The inducer of 3OC12HSL could activate the signal output in two sensing systems within 0.5 μM with the 3 h induction (Fig. 4D). Although they showed a longer incubation time than the LacZ-based system in the solution-phase, the XylE-based reporting systems still showed advantages in paper testing for the pre-added substrates. In the future, the incubation can be monitored real time using a smartphone or IoT technology for the environmental sample detection.
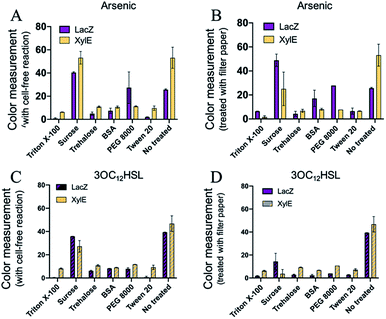
The freeze-drying technology has revealed significant potential for the cell-free biosensing systems in terms of storage and transportation for the diagnostics and environmental management. However, there might be a protein denaturation or loss of enzymatic activity during the free-drying and rehydration processes. Moreover, the cellulose matrix of the paper can also impede the activity of the reactions. To address these concerns, several protectants were employed for either freeze-drying protective agents or blocking.33,34 Six blocking reagents were selected for protection, including Triton X-100, sucrose, trehalose, BSA (Bovine Serum Albumin), PEG 8000 (polyethylene glycol 8000), and Tween 20, which were commonly used for biological molecule protection or blocking. The reagents were supplied with a cell-free component mixture before freeze-drying or treating the filter papers (Fig. S1, ESI†). However, there was reduction in the colorimetric outputs of the three protectants (Triton X-100, trehalose, and Tween 20) after either pre-treating cell-free components or treating the paper. For BSA and PEG 8000, it seemed that only the arsenic ion sensing was functional after treating the paper. For sucrose, the sensing systems could maintain the sensing activity. Except for the supply of sucrose, the cell-free systems pre-added with reagents exhibited ineffective sensing (Fig. 5 and S12, ESI†). Moreover, the paper-based cell-free reactions were rehydrated with more water to improve the activity. However, a weak signal still developed for the three protectants (Triton X-100, trehalose, and Tween-20), as shown in Fig. S13.† These can be attributed to the high concentration of reagents that could affect the processes of in vitro protein syntheses, resulting in reduced protein yields. Among the six protectants, sucrose was the alternative with the protection function for a higher colorimetric output (Fig. 5A and B).
In conclusion, we successfully constructed portable in vitro synthetic biology-based biosensors by combining the cell-free systems with inducible synthetic genetic circuits. For widely available applications outside the laboratory, the colorimetric outputs were employed for the sensing systems, which could be visible to the naked eyes. Three enzymes (GusA, LacZ, and XylE) were characterized by the cell-free reporting systems that could mediate the colorimetric outputs by cleaving specific substrates. Within the genetic circuits that respond to arsenic ions and AHLs, 0.5 μM arsenic ions or AHLs could be detected with these in vitro sensing systems within 3 h of incubation. Quantification of these reactions can also be done easily by measuring the intensity of the color channels from the images produced by most cameras, including those widely available in smartphones. In future, the development of data-shared platforms through an IoT-based cloud server for real-time data updation or sharing is also expected. Integrating with synthetic biology, the cell-free systems provide highly prospective portable and user-friendly biosensors for environmental monitoring or clinical diagnosis.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFA0901700), National Natural Science Foundation of China (21706144, 21878173), and Laboratory Innovation Fund of Tsinghua University.Notes and references
- G. Kebede, D. Mushi, R. B. Linke, O. Dereje, A. Lakew, D. S. Hayes, A. H. Farnleitner and W. Graf, Ecol. Indic., 2020, 108, 105733 CrossRef CAS.
- Y. Li, Q. Zhou, B. Ren, J. Luo, J. Yuan, X. Ding, H. Bian and X. Yao, Rev. Environ. Contam. Toxicol., 2020, 251, 1–24 CAS.
- H. Ming, Y. Ma, Y. Gu, J. Su, J. Guo, J. Li, X. Li, Y. Jin and J. Fan, Ecol. Indic., 2020, 110, 105938 CrossRef.
- I. Tessnow-von Wysocki and P. Le Billon, Environ. Sci. Policy, 2019, 100, 94–104 CrossRef.
- Z. Zhang, K. Zhu and G. J. D. Hewings, Energy Econ., 2017, 64, 13–23 CrossRef.
- F. Aktas, C. Ceken and Y. E. Erdemli, J. Med. Biol. Eng., 2018, 38, 966–979 CrossRef.
- M.-Y. Chuang, C.-C. Chen, H.-W. Zan, H.-F. Meng and C.-J. Lu, ACS Sens., 2017, 2, 1788–1795 CrossRef CAS.
- S. J. Davies, P. Španěl and D. Smith, Bioanalysis, 2014, 6, 843–857 CrossRef CAS.
- J. Kim, A. S. Campbell, B. E.-F. de Ávila and J. Wang, Nat. Biotechnol., 2019, 37, 389–406 CrossRef CAS.
- J. W. Kotula, S. J. Kerns, L. A. Shaket, L. Siraj, J. J. Collins, J. C. Way and P. A. Silver, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 4838–4843 CrossRef CAS.
- V. Siciliano, B. DiAndreth, B. Monel, J. Beal, J. Huh, K. L. Clayton, L. Wroblewska, A. McKeon, B. D. Walker and R. Weiss, Nat. Commun., 2018, 9, 1881 CrossRef.
- J. R. van der Meer and S. Belkin, Nat. Rev. Microbiol., 2010, 8, 511–522 CrossRef CAS.
- B. Wang, M. Barahona and M. Buck, Biosens. Bioelectron., 2013, 40, 368–376 CrossRef CAS.
- Y. Hu, Y. Yang, E. Katz and H. Song, Chem. Commun., 2015, 51, 4184–4187 RSC.
- M. H. Do, H. H. Ngo, W. Guo, S. W. Chang, D. D. Nguyen, Y. Liu, S. Varjani and M. Kumar, Sci. Total Environ., 2020, 712, 135612 CrossRef CAS.
- T. Kawaguchi, Y. P. Chen, R. S. Norman and A. W. Decho, Appl. Environ. Microbiol., 2008, 74, 3667–3671 CrossRef CAS.
- K. Y. Wen, L. Cameron, J. Chappell, K. Jensen, D. J. Bell, R. Kelwick, M. Kopniczky, J. C. Davies, A. Filloux and P. S. Freemont, ACS Synth. Biol., 2017, 6, 2293–2301 CrossRef CAS.
- K. Pardee, A. A. Green, T. Ferrante, D. E. Cameron, A. Daleykeyser, P. Yin and J. J. Collins, Cell, 2014, 159, 940–954 CrossRef CAS.
- P. Sadat Mousavi, S. J. Smith, J. B. Chen, M. Karlikow, A. Tinafar, C. Robinson, W. Liu, D. Ma, A. A. Green, S. O. Kelley and K. Pardee, Nat. Chem., 2020, 12, 48–55 CrossRef CAS.
- Y. Tang, H. Li and B. Li, Chem. Commun., 2020, 56, 2483–2486 RSC.
- A. S. M. Salehi, S. O. Yang, C. C. Earl, M. J. Shakalli Tang, J. Porter Hunt, M. T. Smith, D. W. Wood and B. C. Bundy, Toxicol. Appl. Pharmacol., 2018, 345, 19–25 CrossRef CAS.
- X. Wang, K. Zhu, D. Chen, J. Wang, X. Wang, A. Xu, L. Wu, L. Li and S. Chen, Ecotoxicol. Environ. Saf., 2021, 207, 111273 CrossRef CAS.
- Y. H. Yang, T. W. Kim, S. H. Park, K. Lee, H. Y. Park, E. Song, H. S. Joo, Y. G. Kim, J. S. Hahn and B. G. Kim, Appl. Environ. Microbiol., 2009, 75, 6367–6372 CrossRef CAS.
- J. K. Jung, K. K. Alam, M. S. Verosloff, D. A. Capdevila, M. Desmau, P. R. Clauer, J. W. Lee, P. Q. Nguyen, P. A. Pastén, S. J. Matiasek, J. F. Gaillard, D. P. Giedroc, J. J. Collins and J. B. Lucks, Nat. Biotechnol., 2020, 1–9 Search PubMed.
- P. Zhang, H. Feng, J. Yang, H. Jiang, H. Zhou and Y. Lu, J. Biotechnol., 2019, 300, 78–86 CrossRef CAS.
- X. He, M. Bhateja and C. Fuqua, J. Microbiol. Methods, 2005, 60, 281–283 CrossRef CAS.
- H. P. Schweizer, Gene, 1993, 134, 89–91 CrossRef CAS.
- D. C. Stein, Gene, 1992, 117, 157–158 CrossRef CAS.
- B. J. L. Dopp, D. D. Tamiev and N. F. Reuel, Biotechnol. Adv., 2019, 37, 246–258 CrossRef CAS.
- J.-H. Ahn, H.-S. Chu, T.-W. Kim, I.-S. Oh, C.-Y. Choi, G.-H. Hahn, C.-G. Park and D.-M. Kim, Biochem. Biophys. Res. Commun., 2005, 338, 1346–1352 CrossRef CAS.
- H. M. Salis, in Methods in Enzymology, Academic Press Inc., 2011, vol. 498, pp. 19–42 Search PubMed.
- H. M. Salis, E. A. Mirsky and C. A. Voigt, Nat. Biotechnol., 2009, 27, 946–950 CrossRef CAS.
- M. T. Smith and S. D. Berkheimer, Biotechniques, 2014, 56, 186–193 CrossRef CAS.
- D. K. Karig, S. Bessling, P. Thielen, S. Zhang and J. Wolfe, J. R. Soc., Interface, 2017, 14, 20161039 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, methods and supplementary data. See DOI: 10.1039/d0ra05293k |
| This journal is © The Royal Society of Chemistry 2020 |