Open Access Article
Open Access ArticleLignans and clerodane diterpenoids from Tinospora sinensis†
Chumao Wen‡
a,
Jun Li‡b,
Xiaoshi Xub,
Qingqing Lib,
Yitong Lib,
Yu Chen*c and
Guangzhong Yang *b
*b
aCollege of Biomedical Engineering, South-Central University for Nationalities, Wuhan 430074, P. R. China
bSchool of Pharmaceutical Sciences, South-Central University for Nationalities, Wuhan 430074, P. R. China. E-mail: yanggz888@126.com; Fax: +86 27 6784 1196; Tel: +86 27 6784 1196
cCollege of Chemistry and Material Sciences, South-Central University for Nationalities, Wuhan 430074, P. R. China. E-mail: chenyuwh888@126.com
First published on 28th July 2020
Abstract
Sixteen lignans with diverse carbon skeletons, including tetrahydrofuran lignans (1, 13–18), 7,9′-dinorlignan (2), dibenzylbutane lignans (9–10), aryl tetrahydronaphthalene lignans (11–12), dihydrobenzofuran neolignans (19–20), 8-O-4′ neolignans (21–22), and six clerodane diterpenoid glucosides (3–8) were isolated from the 70% ethanolic extract of the stems of Tinospora sinensis (Lour.) Merr. Their structures were established by spectroscopic analyses and chemical methods. Among all these isolates, compounds 1–2 and 3–4 were determined as previously undescribed lignans and clerodane diterpenoids respectively, and compounds 9–13 and 17–22 were discovered from T. sinensis for the first time. In addition, it was the first time to report three types of lignans including 7,9′-dinorlignan, dihydrobenzofuran neolignan and 8-O-4′ neolignan from T. sinensis. The biological activities of all isolated compounds were assessed in terms of their nitric oxide production inhibitory activity in LPS stimulated RAW264.7 cells. The results showed that compound 20 exhibited a mild NO production inhibitory effect with an IC50 value of 17.43 ± 2.06 μM.
1. Introduction
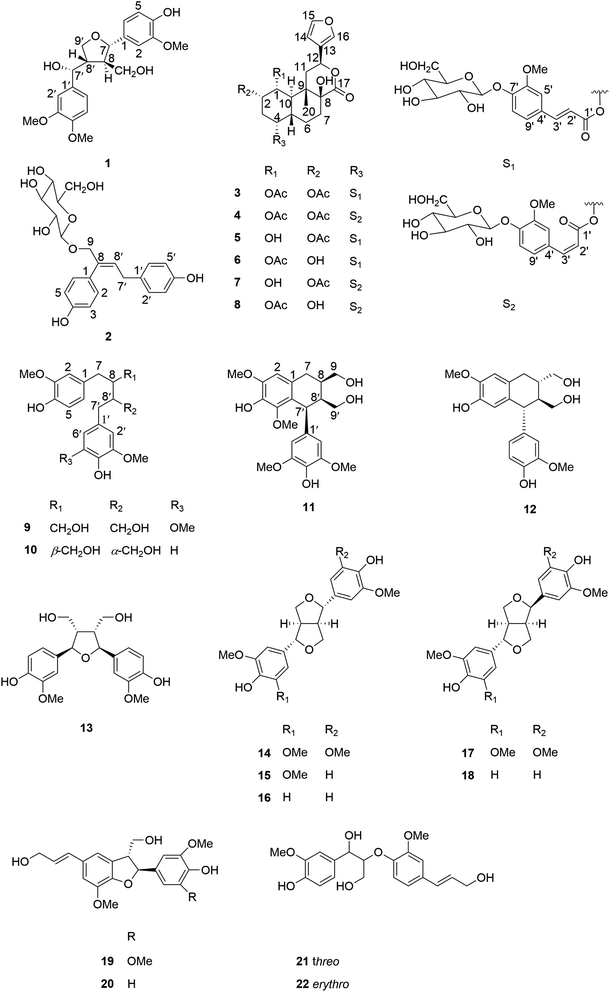
Tinospora sinensis (Lour.) Merr. is a well known medicinal plant belonging to the family Menispermaceae, which is mainly distributed in Tibet and southern China.1 The stems of T. sinensis have been used as Tibetan medicine, which were recorded in an ancient Chinese Tibetan medicinal encyclopedia “Jingzhu Bencao”.2 They have been widely used for the treatment of rheumatism, jaundice, and skin diseases for centuries.3 Previous studies demonstrated that the crude extracts of T. sinensis had potent immunomodulatory and anti-inflammatory activities.4,5 However, due to the complexities and diversities of its chemical constituents, the effective constituents and their molecular mechanisms for its anti-arthritis effect are still poorly understood.6,7 According to the previous report, the EtOAc fraction of ethanolic extract from the stems of T. sinensis exhibited an anti-arthritis effect, which might be attributed to inhibiting the production of pro-inflammatory cytokines and down-regulating the MAPK signaling pathway, and suggested that flavonoids including rutin, quercetin and isorhamnetin might be the mainly effective constituents in the crude extracts.3 But actually, the chemical compositions of T. sinensis were very complicated, which contained terpenoids such as sesquiterpenoids, diterpenoids and triterpenoids, lignans, alkaloids, flavonoids and so on.6 Among them, diterpenoids and lignans were abundant in T. sinensis. Therefore, in order to have a fully understanding of these two classes of compounds in T. sinensis, a phytochemical reinvestigation of T. sinensis was undertaken, resulting in the isolation of twenty-two compounds, including sixteen lignans (1–2, 9–22) and six clerodane diterpenoid glucosides (3–8). Among them, compound 1 was a previously undescribed tetrahydrofuran lignan, and compound 2 was a previously undescribed 7,9′-dinorlignan glucoside. Compounds 3–4 were previously undescribed clerodane diterpenoid glucosides. In this paper, we described structure elucidations of compounds 1–4 and their NO production inhibitory activities of all isolated compounds 1–22.2. Result and discussion
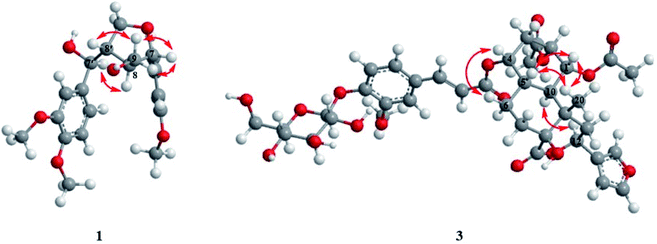
Compound 1 was obtained as amorphous powder. It exhibited the molecular formula C21H26O7 by HR-ESI-MS (m/z 413.1569 [M + Na]+, calcd for C21H26O7Na, 413.1571), indicating 9 degrees of unsaturation. The 1H NMR spectra (Table 1) of 1 showed two sets of AMX type trisubstituted aromatic proton signals at δH 6.89 (1H, d, J = 1.8 Hz), 6.88 (1H, d, J = 8.4 Hz), 6.84 (1H, dd, J = 8.4, 1.8 Hz); 6.93 (1H, d, J = 1.8 Hz), 6.76 (1H, d, J = 8.4 Hz) and 6.80 (1H, dd, J = 8.4,1.8 Hz), three methoxy groups at δH 3.85, 3.81 and 3.80, two oxygenated methines [δH 4.52 (1H, d, J = 8.4 Hz), 4.62 (1H, d, J = 7.8 Hz)], two methines [δH 1.88 (1H, m), 2.55 (1H, m)], two oxygenated methylenes [δH 4.25 (1H, dd, J = 9.0, 4.2 Hz), 3.93 (1H, dd, J = 9.0, 7.8 Hz); 3.29 (1H, m), 3.21 (1H, dd, J = 11.4, 6.0 Hz)], which were characteristic proton signals of a 7,9′ epoxy-tetrahydrofuran lignan. The 13C-NMR and DEPT spectra (Table 1) of 1 revealed 21 carbon signals, which were assigned as two 1,2,4-trisubstituted benzene rings, four methines, two oxygenated methylenes and three methoxy groups. Detailed analysis of the NMR data of 1 suggested that 1 was structurally related to tanegool.8 The significant difference between them was that 1 had one more methoxy group than tanegool, suggesting that 1 was 4′-O-methylated of tanegool which was further confirmed by the HMBC correlation (Fig. 2) of OCH3 (δH 3.80) with C-4′ (δC 150.1). The relative configuration of 1 was determined by the ROESY spectrum (Fig. 3). The ROESY correlations of H-7/H2-9/H-8′, H-8/H-7′ indicated that H-7, H2-9 and H-8′ were on the same side, H-8 and H-7′ were on the other side which were consistent with those of tanegool. Compund 1 and tanegool had the similar chromophores. By comparison of the ECD of 1 with those of tanegool, they displayed the same Cotton effects in the CD spectra.8 The absolute configuration of 1 was assigned as to be 7S, 7′R, 8R, 8′S which was identical to tanegool. Therefore, the structure of compound 1 was established as 4′-O-methylated tanegool and named as tinosporine A.| No. | 1 | 2 | ||
|---|---|---|---|---|
| δHa | δCb | δHa | δCb | |
| a Recorded at 600 MHz.b Recorded at 150 MHz. | ||||
| 1 | 134.8 | 131.1 | ||
| 2 | 6.93 (d, 1.8) | 111.1 | 7.15 (d, 8.4) | 131.2 |
| 3 | 149.1 | 6.78 (d, 8.4) | 116.1 | |
| 4 | 147.2 | 157.8 | ||
| 5 | 6.76 (d, 8.4) | 116.1 | 6.78 (d, 8.4) | 116.1 |
| 6 | 6.80 (dd, 8.4, 1.8) | 120.3 | 7.15 (d, 8.4) | 131.2 |
| 7 | 4.62 (d, 7.8) | 85.1 | ||
| 8 | 1.88 (m) | 53.7 | 138.1 | |
| 9 | 3.29 (m) | 62.3 | 4.54 (d, 12.0) | 74.7 |
| 3.21 (dd, 11.4, 6.0) | 4.34 (d, 12.0) | |||
| 1′ | 137.6 | 133.3 | ||
| 2′ | 6.89 (d, 1.8) | 111.7 | 6.95 (d, 8.4) | 130.4 |
| 3′ | 150.5 | 6.67 (d, 8.4) | 116.3 | |
| 4′ | 150.1 | 156.7 | ||
| 5′ | 6.88 (d, 8.4) | 112.7 | 6.67 (d, 8.4) | 116.3 |
| 6′ | 6.84 (dd, 8.4, 1.8) | 120.7 | 6.95 (d, 8.4) | 130.4 |
| 7′ | 4.52 (d, 8.4) | 76.6 | 3.26 (m) | 35.2 |
| 8′ | 2.55 (m) | 50.8 | 5.94 (t, 7.8) | 130.7 |
| 9′ | 4.25 (dd, 9.0, 4.2) | 71.5 | ||
| 3.93 (dd, 9.0, 7.8) | ||||
| 1′′ | 4.36 (d, 7.8) | 102.9 | ||
| 2′′ | 3.19 (m) | 75.2 | ||
| 3′′ | 3.20 (m) | 78.1 | ||
| 4′′ | 3.29 (m) | 71.7 | ||
| 5′′ | 3.33 (m) | 78.2 | ||
| 6′′ | 3.83 (dd, 12.0, 2.4) | 62.8 | ||
| 3.64 (dd, 12.0, 6.0) | ||||
| 3-OMe | 3.85 (s) | 56.6 | ||
| 3′-OMe | 3.81 (s) | 56.5 | ||
| 4′-OMe | 3.80 (s) | 56.5 | ||
Compound 2 had the molecular formula of C22H26O8 as assigned by HR-ESI-MS (m/z 441.1518 [M + Na]+, calcd 441.1520). The acid hydrolysis experiment revealed 2 contained a D-glucose moiety. Its 1H-NMR displayed two sets of AA'BB′ type aromatic proton signals at δH 7.15 (2H, d, J = 8.4 Hz), 6.78 (2H, d, J = 8.4 Hz), 6.95 (2H, d, J = 8.4 Hz), 6.67 (2H, d, J = 8.4 Hz), one olefinic proton signal at δH 5.94 (1H, t, J = 7.8 Hz), one oxygenated methylene at δH 4.34 (1H, d, J = 12.0 Hz), 4.54 (1H, d, J = 12.0 Hz), and a set of β-glucopyranosyl proton signals with the resonance for the anomeric proton at δH 4.36 (1H, d, J = 7.8 Hz). Apart from the glucopyranosyl signals, the remaining 13C-NMR and DEPT spectra of 2 (Table 1) exhibited 16 carbon signals, including two para-substituted benzene rings, one double bond and two methylenes respectively. The abovementioned NMR data were similar to those of squadinorlignoside,9 except for the chemical shift of C-9 shifting downfield from δC 67.9 in the latter to δC 74.7 in 2, which suggested that both of them had the same aglycon and 9-OH of the aglycon of squadinorlignoside was glucosylated in 2, instead of 4-OH of the aglycon being glycosylated in squadinorlignoside. This conclusion was further confirmed by the HMBC correlations of 2 (Fig. 2) from H2-9 to C-1′′ (δC 102.9). Accordingly, the structure of 2 was elucidated as showed in Fig. 1 and named as tinodinorlignoside A.
Compound 3 was isolated as white amorphous powder, and its molecular formula was exhibited to be C38H46O17 by HR-ESI-MS (m/z 797.2619 [M + Na]+, calcd 797.2627), suggesting 16 degrees of unsaturation. The acid hydrolysis experiment indicated that 3 had a D-glucose moiety. The 1H-NMR spectra (Table 2) of 3 displayed the presence of a 1,2,4-trisubstituted aromatic moiety [δH 7.28 (d, J = 1.2 Hz), 7.15 (d, J = 8.4, Hz), 7.19 (d, J = 8.4, 1.2 Hz)], one methoxy group [δH 3.82 (s)], three singlet methyls [δH 1.95 (s), 1.90 (s), 1.08 (s)], a trans double bond [δH 6.42 (d, J = 16.2 Hz), 7.64 (d, J = 16.2 Hz)], an anomeric sugar proton [4.95 (d, J = 7.2 Hz)], and the typical furan moiety signals [δH 6.52 (br s), 7.52 (t, J = 1.2 Hz), 7.64 (br s)]. The 13C NMR spectra and DEPT (Table 2) of 3 showed 38 carbon resonances, including one lactone carbonyl signal at δC 174.5, one 3-substituted furan moiety, two quaternary carbons, six methines, four methylenes and one methyl at δC 15.3, which implied that 3 possessed a clerodane diterpenoid core. Besides, the two carbonyl groups at δC 172.3, δC 171.3 and two methyls at δH 1.95, δH 1.90 in the NMR spectrum suggested the presence of two acetyl groups in 3. Apart from these signals, the remaining signals displayed a trans-feruloyl and glucopyranosyl group. The abovementioned data were closely similar to those of tinosinenoside D,10 except for the presence of one more acetyl group compared to tinosinenoside D. The HMBC correlation of 3 (Fig. 2) from H-1 (δH 5.11) to the carbonyl group of OAc (δC 171.3), from H-2 (δH 5.31) to the carbonyl group of OAc (δC 172.3) indicated that 3 could be a 1-acetylated of tinosinenoside D. The relative configuration of 3 was revealed by the coupling constants and the ROESY spectrum (Fig. 3). The coupling constants of H-1/H-2 (J1,2 = 3.0 Hz) and H1/H-10 (J1,10 = 11.4 Hz) suggested H-1, H-2 and H-10 were axial, equatorial and axial position respectively. The ROESY correlations of H-1/H-5 (δH 1.99)/H3-20 (δH 1.08) indicated that H-1, H-5 and H3-20 were β-oriented, and H-12 was assigned to be α-oriented by the correlation of H-12 (δH 5.70)/H-10. The correlation of H-4 (δH 4.85)/H-6β (δH 1.58) indicated H-4 was β-oriented. Consequently, the structure of 3 was established as depicted in Fig. 1 and named as 1-acetyltinosinenoside D.
| No. | 3 | 4 | ||
|---|---|---|---|---|
| δHa | δCb | δHa | δCb | |
| a Recorded at 600 MHz.b Recorded at 150 MHz. | ||||
| 1 | 5.11 (dd, 11.4, 3.0) | 74.6 | 5.13 (dd, 11.4, 3.0) | 74.4 |
| 2 | 5.31 (q, 3.0) | 70.3 | 5.30 (q, 3.0) | 70.3 |
| 3 | 2.32 (overlapped) | 32.1 | 2.36 (overlapped) | 32.1 |
| 1.98 (overlapped) | 1.99 (overlapped) | |||
| 4 | 4.85 (overlapped) | 72.8 | 4.91 (overlapped) | 72.5 |
| 5 | 1.99 (m) | 39.0 | 2.01 (m) | 38.8 |
| 6 | 1.58 (m) | 26.5 | 1.58 (m) | 26.7 |
| 1.47 (td, 13.8, 5.4) | ||||
| 7 | 2.31 (overlapped) | 30.2 | 2.34 (overlapped) | 30.3 |
| 1.72 (td, 13.2, 5.4) | 1.71 (td, 413.8, 4.2) | |||
| 8 | 76.7 | 76.6 | ||
| 9 | 40.8 | 40.9 | ||
| 10 | 2.67 (overlapped) | 36.6 | 2.54 (t, 11.4) | 36.7 |
| 11 | 2.67 (overlapped) | 37.2 | 2.66 (t, 13.2) | 37.1 |
| 1.99 (overlapped) | 1.99 (overlapped) | |||
| 12 | 5.70 (dd, 12.6, 3.6) | 73.6 | 5.65 (dd, 13.2, 3.6) | 73.4 |
| 13 | 127.1 | 127.0 | ||
| 14 | 6.52 (br s) | 109.7 | 6.55 (d, 1.2) | 109.7 |
| 15 | 7.52 (t, 1.2) | 145.5 | 7.53 (t, 1.2) | 145.4 |
| 16 | 7.64 (br s) | 141.4 | 7.65 (br s) | 141.4 |
| 17 | 174.5 | 174.6 | ||
| 20 | 1.08 (s) | 15.3 | 1.11 (s) | 15.4 |
| 1′ | 168.1 | 167.7 | ||
| 2′ | 6.42 (d, 16.2) | 117.4 | 5.93 (d, 13.2) | 118.6 |
| 3′ | 7.64 (d, 16.2) | 146.9 | 6.93 (d, 13.2) | 144.8 |
| 4′ | 130.4 | 130.9 | ||
| 5′ | 7.28 (d, 1.2) | 113.0 | 7.74 (d, 1.8) | 115.4 |
| 6′ | 151.1 | 150.1 | ||
| 7′ | 150.2 | 149.3 | ||
| 8′ | 7.15 (d, 8.4) | 117.4 | 7.14 (d, 8.4) | 116.8 |
| 9′ | 7.19 (d, 8.4, 1.2) | 123.5 | 7.18 (d, 8.4, 1.8) | 126.1 |
| 1-Ac | 1.90 (s) | 21.5 | 1.92 (s) | 21.5 |
| 171.3 | 171.4 | |||
| 2-Ac | 1.95 (s) | 21.3 | 2.02 (s) | 21.2 |
| 172.3 | 172.4 | |||
| 6′-OCH3 | 3.82 (s) | 56.9 | 3.86 (s) | 56.7 |
| Glc-1′′ | 4.95 (d, 7.2) | 102.2 | 4.98 (d, 7.8) | 102.3 |
| 2′′ | 3.48 (m) | 74.9 | 3.50 (m) | 74.9 |
| 3′′ | 3.45 (m) | 77.9 | 3.48 (m) | 78.0 |
| 4′′ | 3.37 (m) | 71.4 | 3.40 (m) | 71.4 |
| 5′′ | 3.42 (m) | 78.4 | 3.44 (m) | 78.4 |
| 6′′ | 3.66 (dd, 12.0, 5.4) | 62.5 | 3.69 (dd, 12.0, 5.4) | 62.6 |
| 3.85 (dd, 12.0, 1.8) | 3.88 (overlapped) | |||
Compound 4 shared the same molecular formula of C38H46O17 as 3, which was determined by HR-ESI-MS at m/z 797.2624 [M + Na]+ (calcd for C38H46O17Na, 797.2627) and 13C NMR data. Its NMR data (Table 2) exhibited close similarities to those of compound 3, except for the absence of a trans double bond signal and the presence of a cis double bond signal [δH 6.93 (d, J = 13.2 Hz), 5.93 (d, J = 13.2 Hz); δC 144.8, 118.6]. So it suggested that the trans-feruloyl group at C-4 in 3 was substituted by a cis-feruloyl group in 4, which was further confirmed by the HMBC spectrum (Fig. 2). The relative stereochemistry of 4 was determined by the ROESY correlations, which was consistent with those of 3. Thus, the structure of 4 was defined and named as 1-acetyltinosinenoside E.
The remaining sixteen known compounds were identified as tinosinenoside D (5),10 tinosinenoside F (6),10 tinosinenoside E (7),10 tinosinenoside K (8),11 2-(4-hydroxy-3,5-dimethoxybenzyl)-3-(4-hydroxy-3-methoxybenzyl)butane-1,4-diol (9),12 secoisolariciresinol (10),13 1,2,3,4-tetrahydro-7-hydroxy-1-(4-hydroxy-3,5-dimethoxyphenyl)-6,8-dimethoxy-2,3-naphthalenedimethanol (11),14 isolariciresinol (12),15 rel-(2R,3S,4R,5S)-tetrahydro-2,5-bis-(4-hydroxy-3-methoxyphenyl)-3,4-furandimethanol (13),16 syringaresinol (14),13 medioresinol (15),17 pinoresinol (16),18 lirioresinol-A (17),18 epipinoresinol (18),19 dehydrodiconiferyl alcohol (19),20 simulanol (20),21 threo-guaiacylglycerol-β-O-4′-coniferyl ether (21),22 erythro-guaiacylglycerol-β-O-4′-coniferyl ether (22)22 based on the comparison of their NMR data with those reported in the literatures.
In addition, all the isolated compounds were assessed for their inhibitory activities against NO production in LPS stimulated RAW264.7 cells with dexamethasone as a positive control (IC50 = 9.58 ± 3.34 μM). As a result, compound 20 exhibited mild NO production inhibitory effects with IC50 values of 17.43 ± 2.06 μM, however, the remaining compounds did not display obvious NO production inhibitory activity (IC50 > 20 μM). These results showed that six clerodane diterpenoid glucosides had no obvious inhibitory activities against NO production which was consistent with the previous report,10 suggesting that clerodane diterpenoid glucosides might not be the effective constituents of T. sinensis in the treatment of inflammation.
3. Experimental
3.1 General experimental procedures
Optical rotations were recorded on an Autopol IV polarimeter (Rudolph Research Analytical, Hackettstown, NJ, USA). UV spectra were obtained on a UH5300 UV-VIS Double Beam spectrophotometer (Hitachi Co., Tokyo, Japan). NMR spectra were measured with a Bruker AVANCE IIITM 600 MHz spectrometer (Bruker, Ettlingen, Germany) in C5D5N using tetramethylsilane (TMS) as an internal reference standard. Chemical shifts (δ) have been shown in ppm and the coupling constants (J) have been expressed in Hz. High-resolution electrospray mass spectroscopy was performed on an Thermo Scientific Q Exactive Orbitrap LC-MS/MS System (HR-ESI-MS) (Thermo Scientific, Waltham, MA, USA). High-performance liquid chromatography (HPLC) was conducted on an Ultimate 3000 HPLC system (Dionex Co., Sunnyvale, CA, USA) equipped with an Ultimate 3000 pump and Ultimate 3000 Variable Wavelength detector, as well as a semi-preparative YMC-Pack ODS-A column (250 × 10 mm, 5 μm), column chromatography (CC) was conducted with silica gel (200–300 mesh and 300–400 mesh, Qingdao Haiyang Chemical Industry Co., Ltd, Qingdao, China). Chromatographic grade acetonitrile was purchased from Chang Tech Enterprise Co., Ltd (Taiwan, China). RAW264.7 murine macrophages were purchased from the cell bank of Chinese Academy of Sciences (Shanghai, China). Dexamethasone and lipopolysaccharides (LPS) were purchased from Sigma Chemical Co. Ltd (St. Louis, MO, USA). Cell Counting Kit (CCK-8) was purchased from Beyotime Biotechnology (Shanghai, China). Dulbecco modified Eagle medium (DMEM) and penicillin-streptomycin solution were purchased from GE Healthcare Life Sciences (Logan, UT, USA). Fetal bovine serum (FBS) was purchased from Gibco, Life Technologies (Grand Island, NY, USA). Reagent grade dimethyl sulfoxide (DMSO) was purchased from Vetec, Sigma Chemical Co. (St. Louis, MO, USA). The absorbance was read on a Multiskan GO microplate reader (Thermo Fisher Scientific Inc. Waltham, MA, USA).3.2 Plant material
The stems of T. sinensis were purchased from Xining, Qinghai province, P. R. China and identified by Associate professor Xinqiao Liu, School of Pharmaceutical Sciences, South Central University for Nationalities.3.3 Extraction and isolation
The air-dried stems of Tinospora sinensis (Lour.) Merr. (9.4 kg) were extracted by 70% EtOH for three times (each 24 h) at room temperature, yielding a crude extract (1.0 kg) after evaporation, then the extract was chromatographed over a polyamide column (80–100 mesh) eluting with a gradient of EtOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 7
0, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, 0
7, 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain five fractions (Fr. A–Fr. E). Fr. B (41.3 g) was subjected to ODS column chromatography (MeOH–H2O, 2
1) to obtain five fractions (Fr. A–Fr. E). Fr. B (41.3 g) was subjected to ODS column chromatography (MeOH–H2O, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, 4
8, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, 6
6, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 8
4, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) to afford fifteen subfractions (Fr. B1–Fr. B15). Fr. B11 was chromatographed by silica gel column eluting with a gradient of CH2Cl2–MeOH (19
0) to afford fifteen subfractions (Fr. B1–Fr. B15). Fr. B11 was chromatographed by silica gel column eluting with a gradient of CH2Cl2–MeOH (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 9
1, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and then further purified by semi-preparative HPLC to afford compounds 1 (4.0 mg), 11 (3.0 mg), 12 (8.7 mg), 13 (4.2 mg), 21 (29.0 mg), 22 (11.5 mg). Fr. B13 was chromatographed by silica gel column eluting with a gradient of CH2Cl2–MeOH (19
1), and then further purified by semi-preparative HPLC to afford compounds 1 (4.0 mg), 11 (3.0 mg), 12 (8.7 mg), 13 (4.2 mg), 21 (29.0 mg), 22 (11.5 mg). Fr. B13 was chromatographed by silica gel column eluting with a gradient of CH2Cl2–MeOH (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 9
1, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8
1, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 7
2, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and then further purified by semi-preparative HPLC to afford compounds 2 (5.0 mg), 3 (9.9 mg), 4 (2.2 mg), 5 (24.8 mg), 6 (1.8 mg), 7 (9.0 mg), 8 (3.5 mg), 9 (4.0 mg), 10 (123.1 mg), 14 (10.3 mg), 15 (1.5 mg), 16 (30.8 mg), 17 (1.4 mg), 18 (5.7 mg), 19 (8.8 mg), 20 (11.0 mg).
1), and then further purified by semi-preparative HPLC to afford compounds 2 (5.0 mg), 3 (9.9 mg), 4 (2.2 mg), 5 (24.8 mg), 6 (1.8 mg), 7 (9.0 mg), 8 (3.5 mg), 9 (4.0 mg), 10 (123.1 mg), 14 (10.3 mg), 15 (1.5 mg), 16 (30.8 mg), 17 (1.4 mg), 18 (5.7 mg), 19 (8.8 mg), 20 (11.0 mg).
3.4 Spectroscopic data
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 235 (3.60), 275 (3.58) nm; ECD (c 8 × 10−4 M, MeOH) λ (θ): 281 (+0.47), 236 (+1.11), 205 (+8.31). 1H and 13C NMR data see Table 1; HR-ESI-MS m/z 413.1569 [M + Na]+ (calcd for C21H26O7Na, 413.1571).
ε): 235 (3.60), 275 (3.58) nm; ECD (c 8 × 10−4 M, MeOH) λ (θ): 281 (+0.47), 236 (+1.11), 205 (+8.31). 1H and 13C NMR data see Table 1; HR-ESI-MS m/z 413.1569 [M + Na]+ (calcd for C21H26O7Na, 413.1571).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 220 (3.95), 280 (3.89) nm; 1H and 13C NMR data see Table 1; HR-ESI-MS m/z 441.1518 [M + Na]+ (calcd for C22H26O8Na, 441.1520).
ε): 220 (3.95), 280 (3.89) nm; 1H and 13C NMR data see Table 1; HR-ESI-MS m/z 441.1518 [M + Na]+ (calcd for C22H26O8Na, 441.1520).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 240 (2.67), 295 (2.63), 320 (2.67) nm; 1H and 13C NMR data see Table 2; HR-ESI-MS m/z 797.2619 [M + Na]+ (calcd for C38H46O17Na, 797.2627).
ε): 240 (2.67), 295 (2.63), 320 (2.67) nm; 1H and 13C NMR data see Table 2; HR-ESI-MS m/z 797.2619 [M + Na]+ (calcd for C38H46O17Na, 797.2627).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 235 (3.55), 300 (3.52) nm; 1H and 13C NMR data see Table 2; HR-ESI-MS m/z 797.2624 [M + Na]+ (calcd for C38H46O17Na, 797.2627).
ε): 235 (3.55), 300 (3.52) nm; 1H and 13C NMR data see Table 2; HR-ESI-MS m/z 797.2624 [M + Na]+ (calcd for C38H46O17Na, 797.2627).3.5 Acid hydrolysis of compounds 2–423
Compounds 2 (1.1 mg), 3 (1.2 mg) and 4 (0.9 mg) were respectively dissolved in 2.0 mL of 4.0 M CF3COOH, three reaction mixtures were refluxed at 95 °C for 3 h and then extracted with EtOAc for three times (each 2 mL). The aqueous layers were evaporated to obtain the sugar fractions. The absolute configuration of sugar was determined by the following method. Pyridine (0.5 mL) and L-cysteine methyl ester hydrochloride (1.0 mg) were added to the sugar fractions and standard of D-glucose respectively, and four reaction mixtures were refluxed at 60 °C for 1 h. After cooling, 2-methylphenyl isothiocyanate was added to four reaction mixtures, and continued to react for 1 h at 60 °C. Then four reaction mixtures were subjected to HPLC analysis (column: YMC-Pack ODS-A 250 × 4.6 mm I. D; mobile phase: CH3CN–H2O, 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75; flow rate: 0.8 mL min−1; wavelength: 250 nm). The retention times of three sugar fractions were observed at 20.78, 20.87 and 20.81 min respectively, which were closely consistent with that of D-glucose (20.81 min). Therefore, the sugars moiety in 2–4 were identified to be D-glucose.
75; flow rate: 0.8 mL min−1; wavelength: 250 nm). The retention times of three sugar fractions were observed at 20.78, 20.87 and 20.81 min respectively, which were closely consistent with that of D-glucose (20.81 min). Therefore, the sugars moiety in 2–4 were identified to be D-glucose.
3.6 NO production measurement and cell viability assay24
The NO production was measured by the Griess reaction, and the cell viability was evaluated by the CCK-8 method.4. Conclusions
In summary, our study on the chemical constituents of Tinospora sinensis has led to the isolation of a previously undescribed tetrahydrofuran lignan, tinosporine A (1), a previously undescribed 7,9′-dinorlignan glucoside, tinodinorlignoside A (2), two previously undescribed clerodane diterpenoid glucosides, 1-acetyltinosinenoside D (3) and 1-acetyltinosinenoside E (4), together with four known clerodane diterpenoid glucosides (5–8) and fourteen known lignans (9–22). According to these structure types, these compounds might be divided into lignans including tetrahydrofuran lignans (1, 13–18), 7,9′-dinorlignan (2), dibenzylbutane lignans (9–10), aryl tetrahydronaphthalene lignans (11–12), dihydrobenzofuran neolignans (19–20), 8-O-4′ neolignans (21–22), and six clerodane diterpenoid glucosides (3–8). Among them, compounds 9–13, 17–22 were discovered from the genus Tinospora for the first time. Moreover, it was the first time to report three types of lignans including 7,9′-dinorlignan, dihydrobenzofuran neolignan and 8-O-4′ neolignan from T. sinensis, which could be regard as characteristic maker constituents of T. sinensis. Three types lignans such as tetrahydrofuran, dibenzylbutane and aryl tetrahydronaphthalene lignans and clerodane diterpenoids are the most prominent secondary metabolism in the genus of Tinospora, such as T. capillipes and T. cordifolia.2 In contrast, it was noteworthy that the clerodane diterpenoid glucosides with a feruloyl group, such as compounds 3–8, had been only found in T. sinensis, which suggested that this type of compounds might be used to distinguish T. sinensis from other species of Tinospora. These findings together with those previous reports imply that lignans and clerodane diterpenoids could serve as chemotaxonomic markers of species in series Tinospora, but also need to be further research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Major New Drugs Innovation and Development (2017Z09301060) and the Special Fund for Basic Scientific Research of Central Colleges, South-Central University for Nationalities (CZP18004).References
- W. Bai, Y. Xie, Y. Wei, Y. Ma, C. Ge Sang and L. Huang, J. Chin. Med. Mater., 2017, 40, 600–603 Search PubMed.
- S. S. Chi, G. M. She, D. Han, W. H. Wang, Z. Liu and B. Liu, J. Evidence-Based Complementary Altern. Med., 2016, 2, 1–32 Search PubMed.
- H. Xiong, X. Ding, H. Wang, H. Q. Jiang, X. Y. Wu, C. Y. Tu, C. Q. Wu, Y. Pi, G. Z. Yang, Z. Q. Zhao and Z. N. Mei, Phytomedicine, 2019, 57, 271–281 CrossRef PubMed.
- P. N. Manjrakar, C. I. Jolly and S. Narayanan, Fitoterapia, 2000, 71, 254–257 CrossRef.
- R. W. Li, G. David Lin, S. P. Myers and D. N. Leach, J. Ethnopharmacol., 2003, 85, 61–67 CrossRef PubMed.
- Q. S. Jiao, L. L. Xu, J. Y. Zhang, Z. J. Wang, Y. Y. Jiang and B. Liu, Molecules, 2018, 23, 274–293 CrossRef PubMed.
- S. H. Lam, P. H. Chen, H. Y. Hung, T. L. Hwang, C. C. Chiang, T. D. Thang, P. C. Kuo and T. S. Wu, Molecules, 2018, 23, 2541–2553 CrossRef PubMed.
- (a) F. A. Macías, A. López, R. M. Varela, A. Torres and J. M. G. Molinillo, J. Agric. Food Chem., 2004, 52, 6443–6447 CrossRef PubMed; (b) W. Li, K. Koike, L. Liu, L. Lin, X. Fu, Y. Chen and T. Nikaido, Chem. Pharm. Bull., 2004, 52, 638–640 CrossRef CAS PubMed.
- Y. L. Yang, F. R. Chang and Y. C. Wu, Helv. Chim. Acta, 2005, 88, 2731–2737 CrossRef CAS.
- H. Jiang, G. J. Zhang, Y. F. Liu, H. S. Wang and D. Liang, J. Nat. Prod., 2017, 80, 975–982 CrossRef CAS PubMed.
- H. Jiang, G. J. Zhang, H. B. Liao and D. Liang, Fitoterapia, 2018, 131, 127–133 CrossRef CAS PubMed.
- R. Kraus and G. Spiteller, Phytochemistry, 1990, 29, 1653–1659 CrossRef CAS.
- H. B. Park, K. H. Lee, K. H. Kim, I. K. Lee, H. J. Noh, S. U. Choi and K. R. Lee, Nat. Prod. Sci., 2009, 15, 17–21 CAS.
- S. Yoshihide, K. Seiichi, N. Haruo, O. Kenji, N. Yoshinori and A. Yukihiro, PCT Int. Appl. WO 2006068254 A1 20060629, 2006.
- J. X. Zhu, J. Ren, J. J. Qin, X. R. Cheng, Q. Zeng, F. Zhang, S. K. Yan, H. Z. Jin and W. D. Zhang, Arch. Pharm. Sci. Res., 2012, 35, 1739–1747 CrossRef CAS PubMed.
- R. Kraus and G. Spiteller, Liebigs Ann. Chem., 1990, 10, 1205–1213 CrossRef.
- X. H. Yu, P. P. Li, Y. Y. Geng, Y. J. Huang and Y. Hua, Chem. Ind. For. Prod., 2017, 37, 149–153 CAS.
- Q. Liu, J. Li, X. Y. Chai, Y. Jiang and P. F. Tu, J. Chin. Pharm. Sci., 2013, 22, 427–430 CAS.
- Y. C. Yang, X. X. Wu, Y. L. Meng, Y. Qin, C. F. Xia, Q. F. Hu and Y. K. Li, Chin. Tradit. Pat. Med., 2015, 37, 2455–2457 CAS.
- S. J. In, K. H. Seo, N. Y. Song, D. S. Lee, Y. C. Kim and N. I. Baek, Arch. Pharm. Sci. Res., 2015, 38, 26–34 CrossRef CAS PubMed.
- Y. P. Yang, M. J. Cheng, C. M. Teng, Y. L. Chang, I. L. Tsai and I. S. Chen, Phytochemistry, 2002, 61, 567–572 CrossRef CAS PubMed.
- H. Q. Yu, H. J. Bai, W. L. Mei, W. J. Zuo, H. Wang, J. L. Yang and H. F. Dai, J. Trop. Subtrop. Bot., 2015, 23, 323–328 CAS.
- T. Tanaka, T. Nakashima, T. Ueda, K. Tomii and I. Kouno, Chem. Pharm. Bull., 2007, 55, 899–901 CrossRef CAS PubMed.
- H. D. Teng, Y. S. Ren, Z. Y. Ma, X. Tan, J. Xu, Y. Chen and G. Z. Yang, Fitoterapia, 2019, 137, 104245–104251 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04917d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |