Open Access Article
Open Access ArticleA sensitive fluorometric sensor for Ag+ based on the hybridization chain reaction coupled with a glucose oxidase dual-signal amplification strategy†
Yubin Li *,
Ling Xie,
Jiaming Yuan and
Huazhong Liu
*,
Ling Xie,
Jiaming Yuan and
Huazhong Liu
College of Chemistry and Environment, Guangdong Ocean University, Zhanjiang, 524088, China. E-mail: 2007liyubin@163.com
First published on 13th July 2020
Abstract
In this work, an efficient and sensitive fluorometric sensor was developed to detect silver ions (Ag+). It is based on the cytosine–Ag+–cytosine (C–Ag+–C) structure via a dual-signal amplification strategy using glucose oxidase (GOx) and the hybridization chain reaction (HCR). A silver-coated glass slide (SCGS) acts as an ideal material for separation. Cytosine rich (C-rich) capture DNA (C-DNA) assembled themselves on the SCGS via Ag–S bonds and hybridized with signal DNA (S-DNA) to trigger the HCR. With specific base-pairing, the S-DNA and HCR products bind on the SCGS. Then, the GOx–biotin–streptavidin (SA) complexes bind to the HCR products through SA–biotin interactions. Owing to the formation of a particular C–Ag+–C structure between two neighboring C-rich C-DNA on the SCGS, the C-DNA/S-DNA/HP1-GOx/HP2-GOx complex gradually moved away from the SCGS as the concentration of Ag+ increased and the combined GOx fell into the buffer. H2O2 could be generated during the oxidation of glucose, catalyzed by GOx in the buffer. Afterward, H2O2 could oxidize the substrate (3-(p-hydroxyphenyl)-propanoic acid) when Horseradish peroxidase was present, giving rise to blue fluorescence. The proposed strategy reached a limit of detection (LOD) of 1.8 pmol L−1 with a linear detection range of 5 to 1000 pmol L−1 for Ag+. Moreover, this assay has been commendably used for the detection of Ag+ in actual samples with fairly good results.
1 Introduction
Environmental pollution and human health problems caused by heavy metal ions have aroused extensive attention.1,2 The silver ion (Ag+) is a particularly notorious heavy metal ion because it is highly poisonous and widespread in water, soil, atmosphere, crops, and fishery products.3–6 Ag+ can inactivate various metabolites and sulfhydryl enzymes by interacting with thiol, amino, and carboxyl groups to produce cytopathogenic effects.7,8 Taking these adverse factors into account, it is urgent and necessary to develop a highly sensitive and selective assay for Ag+ monitoring. Currently, many strategies have been established for the detection of Ag+, including atomic absorption spectroscopy,9,10 inductively coupled plasma mass spectroscopy,11,12 and electrochemical analysis.13,14 However, traditional atomic absorption spectroscopy cannot be used for the determination of trace content samples and it has a narrow linear range, which is inconvenient for practical applications. Besides, inductively coupled plasma mass spectrometry requires costly instruments and a well-trained analyst, and is sensitive to the sample medium and temperature. Although the accuracy and sensitivity of electrochemical analysis methods are high, they are susceptible to a variety of interference currents and have poor selectivity in complex samples. Methods based on real-time fluorescence PCR have both pros and cons. Despite their high sensitivity, they have shortcomings including being prone to false-positive signals and requiring expensive equipment and complicated operating procedures. Since Ono et al. reported that Ag+ specifically interacts with cytosine (C) to form a stable C–Ag+–C structure,15,16 several C–Ag+–C based sensors for Ag+ analysis have been reported, such as fluorescent, colorimetric, and electrochemical sensors.17–19 Moreover, the peculiar C–Ag+–C interactions guarantee prominent selectivity because C–C mismatches bind to Ag+ beyond everything else. Due to the limitations of the above methods and the specific C–Ag+–C structure, this study proposes a fluorescence assay for Ag+ based on the C–Ag+–C structure, which has many advantages including simplicity, high sensitivity, high selectivity, and easy operation.Up to now, to advance the determination sensitivity of sensors, methods based on signal amplification strategies have been extensively investigated, comprising polymerase chain reaction (PCR),20,21 hybridization chain reaction (HCR),22,23 rolling circle amplification (RCA)24,25 and strand displacement amplification (SDA).26,27 Among these strategies, HCR is regarded as a promising tool for signal amplification by virtue of its enzyme-free nature, simplicity, economy, isothermality, and excellent sensitivity. Moreover, HCR has been extensively used for the determination of oligonucleotides,28 proteins,29 and tumor cells.30
Oligonucleotides and some biomolecules can be immobilized on solid substrates to isolate bound biomolecules from free molecules. This strategy has been successful in the study of surface functionalization on glass, nano-magnetic microspheres, nanoparticles, electrodes, and so forth.31–35 Nevertheless, these immobilized means using in the above researches still exist downsides like costly, inconvenient, inefficient and so on. This paper presents a preferable separation material, namely, silver-coated glass slide (SCGS),36–39 which was readily obtained by the silver mirror reaction.40–42 In addition, thiolated DNA can assemble themselves on the surface of silver through covalent forces using Ag–S bonds.43–45
GOx, an inexpensive and stable aerobic dehydrogenase, serves as a signal amplifier for the fabrication of sensors to detect glucose, proteins, nucleic acid, and biological molecules.46–49 Inspired by the aforementioned work, herein, based on C–Ag+–C interactions, we present an efficient and sensitive fluorometric sensor for the detection of Ag+ using HCR/GOx dual signal amplification and SCGS. This paper may offer a novel way for the determination of Ag+ in the future.
2 Experimental section
2.1 Reagents and chemicals
Horseradish peroxidase (HRP), bovine serum albumin powder (BSA), GOx, streptavidin (SA), and all of the synthetic oligonucleotides (Table S1†) used in this work were obtained commercially from Sangon Bioengineering Technology and Services Co. Ltd (Shanghai, China). 3-(p-Hydroxyphenyl)-propanoic acid (HPPA), trichloromethyl phosphate (TCEP), argentum nitricum, glucose, 1% ammonia spirit, and the inorganic reagents were obtained from Sinopharm Chemical Reagent Co. Ltd (Beijing, China). The human sera were obtained from Hongquan biological technology Co., Ltd (Guangzhou, China). Ultra-pure water (18.1 MΩ cm, at 25 °C) was utilized in all of the experiments. All of the reagents used in the experiments were analytically pure. All of the oligonucleotides were dissolved in phosphate-buffered saline (PBS, pH 7.4, 20 mmol L−1, 50 mmol L−1 NaNO3). The 5% BSA solution was prepared by dissolving 5.0 g bovine serum albumin powder in 100 mL PBS buffer solution. All of the working solutions listed above were kept in a fridge at 4 °C until use.The method of preparing SCGS36,37 and GOx–biotin38,39 is outlined by Li et al. Hairpin probes, HP1 and HP2, were annealed in a water bath at 95 °C for 5 min, which was followed by slowly cooling to the indoor temperature to form the stable stem-loop structure before use.
2.2 Apparatus
A pHS-3E pH meter (Lei-ci Instrument Factory, Shanghai, China) was used for pH measurements. The AAS measurements were performed using a Z-2000 series polarized Zeeman atomic absorption spectrometer (Hitachi, Japan). The fluorescence spectra were acquired using an F-4600 Hitachi spectrofluorimeter (Japan) equipped with excitation at 320 nm and the emission spectra were obtained from 360 to 460 nm. The excitation and emission slits were 5 nm. The fluorescence intensity of the oxidation products of HPPA at 400 nm was utilized to appraise the performance of the presented method.2.3 Fabrication of the sensor
Firstly, according to a reported method, the captured DNA (C-DNA) was immobilized onto a silver-coated glass slide by Ag–S bonds.40,41 Then, the sample was soaked in 5% BSA buffer solution for 15 minutes to plug any potential residual active sites and prevent non-specific adsorption. Then, the C-DNA immobilized SCGS was immersed in a 100 nmol L−1 signal DNA (S-DNA) solution for 1 hour at 25 °C. After that, the C-DNA/S-DNA modified SCGS and 400 nmol L−1 HP1 and HP2 were mixed in a buffer solution for 80 min at 37 °C to initiate the HCR reaction and form long-nicked double helices. After the addition of 2 μg mL−1 SA and 50 μg mL−1 GOx–biotin, the C-DNA/S-DNA/HP1-GOx/HP2-GOx complex immobilized SCGSs were submerged in buffer solutions of different Ag+ concentrations for 1 h at 37 °C. Afterward, the aforementioned solution was mixed with 50 mmol L−1 glucose and reacted at 37 °C for 1 h to generate H2O2. Ultimately, 0.5 μg mL−1 HRP and 50 μmol L−1 HPPA were added to the aforementioned solution and incubated at 37 °C for 30 min in the dark, and the eventual solution was investigated via fluorescence spectrophotometry.2.4 Real sample application
The concentration of Ag+ in Ruiyun Lake water from Zhanjiang city and tap water were monitored by fluorescence spectrophotometry. The water samples were filtered first using a 10 μm filter and then using a 0.45 μm filter to facilitate the removal of debris and impurities. In addition, the determination of Ag+ in biological samples is crucial for monitoring health. Hence, human sera samples were also analyzed to examine the feasibility of the presented sensor. The human sera were transferred into two sterile centrifuge tubes and centrifuged at 4000 rpm for 12 min. The supernatant was collected and filtered (0.45 μm filter).The standard addition method was adopted for quantifying Ag+. Ag+ stock solutions with concentrations ranging from 5 to 1000 pmol L−1 were added to the aliquots of the above-mentioned samples. Eventually, each sample was diluted 10 times with phosphate-buffered saline and then the proposed method was performed to detect Ag+.
3 Results and discussion
3.1 Working mechanism
Scheme 1 depicts the basic principle of the Ag+ assay. The hairpin sequences (HP1 and HP2) modified with biotin at the 5′ end are stabilized in the absence of S-DNA. Since it can specifically bind to biotin, SA is utilized to couple HP1 and HP2 with GOx–biotin, which serves as a signal amplification component. In our strategy, C-rich C-DNA can assemble themselves on the surface of SCGSs via Ag–S bonds and hybridize with S-DNA. HCR efficiently primes the sample to form long-nicked duplex DNA (S-DNA/HP1-biotin/HP2-biotin structures) in the presence of S-DNA. GOx–biotin adheres to the HCR product via SA–biotin interactions. With specific base-pairing between C-DNA and S-DNA, the S-DNA/HP1-GOx/HP2-GOx complexes are attracted to the surface of the SCGSs. The SCGSs modified with the above complexes were then immersed in a buffer solution containing Ag+. Then, Ag+ enters the binding site of the mismatched C–C bases to form a stable “C–Ag+–C” structure between two adjacent C-rich C-DNA on the SCGS and the complexes move away from the SCGS, resulting in the combined GOx falling into the buffer solution. Thus, with higher Ag+ concentrations in the buffer solution, more GOx can be found in the buffer solution. In the presence of HRP, HPPA is oxidized by H2O2, which is generated during the GOx-catalyzed oxidation of glucose and gives rise to the fluorescence signals. Therefore, the concentration of Ag+ could be appraised from the fluorescence intensity.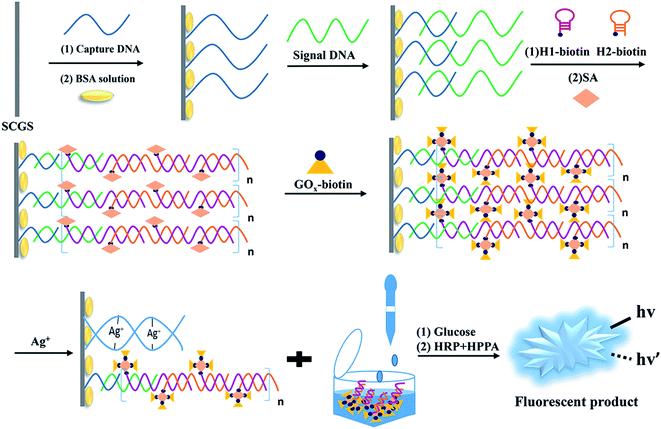 | ||
| Scheme 1 Schematic of the Ag+ detection method based on the C–Ag+–C structure, which utilizes a HCR/GOx dual-signal amplification strategy and SCGS as an ideal separation material. | ||
3.2 Feasibility study on the proposed sensor
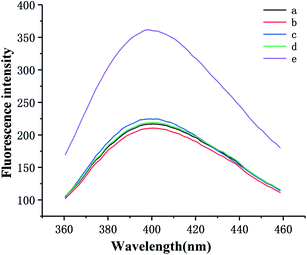
To explore the feasibility of this approach, the fluorescence emission spectra of the different mixtures were recorded. As displayed in Fig. 1, the fluorescence intensity is feeble without C-DNA because the C-DNA/S-DNA/HP1/HP2 structure has not formed (curve a). The same situation occurs in the absence of S-DNA and HP1. Likewise, the fluorescence spectrum displays a weak signal without SA–biotin–GOx (curve b), indicating that the fluorescent product cannot be yielded in the absence of GOx. Although the nicked double-helix polymer cannot be produced without HP2, the C-DNA/S-DNA/HP1-GOx structure can still be formed, causing a slight increase in fluorescence (curve c). The fluorescence intensity was weak in the absence of Ag+ (curve d). Nevertheless, as Ag+ can enter the binding site of the mismatched C–C bases and form a stable “C–Ag+–C” structure between two adjacent C-rich C-DNA, the C-DNA/S-DNA/HP1-GOx/HP2-GOx complexes gradually move away from the SCGSs with increasing Ag+ concentration. Thus, the fluorescence intensity increased significantly (curve e). The above results powerfully demonstrate the feasibility of this sensor for Ag+ determination.3.3 Sensitivity of the proposed sensor
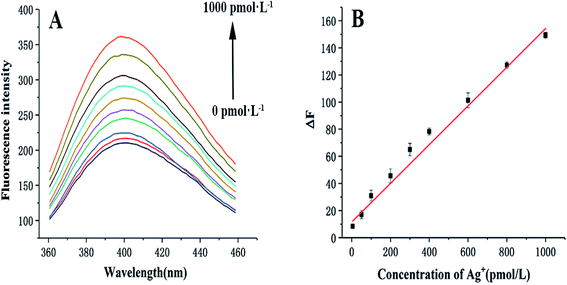
Different concentrations of Ag+ were added to investigate the sensitivity of the sensor under optimal conditions. As illustrated in Fig. 2A and B, the fluorescence intensity enhanced with increasing Ag+ concentration. It shows a positive linear concentration range between 5 pmol L−1 and 1000 pmol L−1 of Ag+, with a linear regression equation of ΔF = 0.14C + 15.51 (R2 = 0.993, C refers to the concentration of Ag+ (pmol L−1), ΔF represents the change in fluorescence intensity, assessed by the equation: ΔF = Ftarget − Fblank, where Fblank represents the fluorescence signal of the mixture without Ag+, Ftarget denotes the fluorescence signal of the mixture in the presence of Ag+ at different concentrations.). The LOD of this sensor is 1.8 pmol L−1 (N = 3, RSD = 3.2%), which was calculated using the equation: DL = 3σ/slope (σ represents the relative standard deviation of the blank sample and slope represents the slope of the linear regression equation).Compared with several other assays (Table 1), the dynamic range and LOD of the proposed sensor are better than those of other relevant studies. Moreover, there are other prominent characteristics of this sensor, such as the use of SCGSs as an ideal separation material, the dual-signal amplification strategy based on HCR/GOx, and the C–Ag+–C structure as a recognition element.
| Analytical method | Linear range | LOD | Reference |
|---|---|---|---|
| Electrochemical biosensor based on DNA–Au bio-bar codes and silver | 5 pmol L−1 to 50 μmol L−1 | 2 pmol L−1 | 2 |
| Colorimetric assay based on gold nanoparticle oligomers | 0.5 pmol L−1 to 5 nmol L−1 | 0.246 pmol L−1 | 4 |
| Gold nanoparticle-based fluorescence polarization | 50–750 nmol L−1 | 9.5 nmol L−1 | 5 |
| Fluorescent chemosensor based on thionine | 5–800 fmol L−1 | 5.0 fmol L−1 | 50 |
| Room-temperature phosphorescence assay | 0.1–10 μmol L−1 | 36 nmol L−1 | 51 |
| Electrochemical biosensor based on magnetic nanoparticles | 1 fmol L−1 to 10 pmol L−1 | 0.5 fmol L−1 | 52 |
| Biosensor based on nanoporous gold and anionic intercalator | 0.1 nmol L−1 to 1 μmol L−1 | 0.48 pmol L−1 | 53 |
| Sensor based on HCR and GOx | 5–1000 pmol L−1 | 1.8 pmol L−1 | This work |
3.4 Selectivity of the proposed sensor
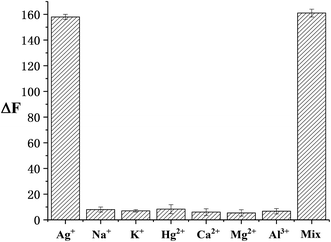
The selectivity of this sensor was investigated using other environmentally related cations. A mixture of all of the cations (including Ag+) was tested under the same conditions. As demonstrated in Fig. 3, a prominent enhancement in fluorescence intensity is observed in the presence of Ag+. This outcome uncovers that this method exhibits prominent selectivity towards Ag+ monitoring despite high concentrations of other cations.3.5 Real sample detection
The extensive range of applicability of the presented assay was assessed by recovery studies using lake water, tap water, and human sera. The results were then compared with those obtained through atomic absorption spectrometry (AAS), which served as a reference standard. The results of the experiments are presented in Table 2. The high recovery and satisfactory RSD values proved that this sensor is both practical and reliable for the determination of Ag+ in authentic samples.| Sample | Added (pmol L−1) | Proposed method meana ± SDb (pmol L−1) | AAS meana ± SDb (pmol L−1) |
|---|---|---|---|
| a Mean of three determinations.b SD, standard deviation. | |||
| Ruiyun Lake water | 5.0 | 5.3 ± 0.3 | 5.1 ± 0.4 |
| Ruiyun Lake water | 50.0 | 52.1 ± 2.6 | 53.7 ± 3.1 |
| Ruiyun Lake water | 500.0 | 513.7 ± 29.0 | 487.3 ± 34.7 |
| Ruiyun Lake water | 1000.0 | 969.7 ± 39.0 | 1063.1 ± 38.4 |
| Tap water | 5.0 | 5.4 ± 0.3 | 5.7 ± 0.2 |
| Tap water | 50.0 | 55.2 ± 2.7 | 49.7 ± 3.7 |
| Tap water | 500.0 | 488.4 ± 22.1 | 491 ± 28.3 |
| Tap water | 1000.0 | 979.1 ± 36.4 | 1012.7 ± 38.9 |
| Human sera | 5.0 | 5.2 ± 0.3 | 5.3 ± 0.3 |
| Human sera | 50.0 | 47.3 ± 3.4 | 50.7 ± 4.1 |
| Human sera | 500.0 | 488.9 ± 26.1 | 488.3 ± 27.6 |
| Human sera | 1000.0 | 971.2 ± 33.7 | 983.7 ± 46.8 |
4 Conclusion
In this study, an efficient and sensitive fluorescence method to determine Ag+ was established based on a C–Ag+–C structure and GOx/HCR dual-signal amplification using SCGSs as the separation material. This approach has some prominent characteristics. First, the proposed approach has been successfully used to detect Ag+ in complicated systems with many components including biological media. Further, the LOD of this assay is low because of the dual-signal amplification strategy based on the HCR and GOx. Moreover, this assay has been commendably used for the detection of Ag+ in actual samples with fairly good results. We firmly believe that this assay may provide a novel way for the determination of Ag+ in the future.Live subject statement
All of the experiments in this study were performed in compliance with the guidelines of the ethics committee of Guangdong Ocean University, China. It was approved by the ethical committee of Guangdong Ocean University. Informed consent was obtained from the human subjects prior to obtaining the plasma sera.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work is supported by the project of Enhancing School with Innovation of Guangdong Ocean University, China (no. 2020KZDZX1108), the foundation and applied foundation research Joint-Youth Fund Project of Guangdong Province, China (no. 2019A1515110648), the Youth Innovation Talents Project of Guangdong Province Universities (no. 2017KQNCX096), and the Science and Technology Project on Special Fund for Public Welfare Research and Ability Construction of Guangdong Province (no. 2017A010105010).References
- J. Wang, J. Guo, J. Zhang, W. Zhang and Y. Zhang, Signal-on electrochemical sensor for the detection of two analytes based on the conformational changes of DNA probes, Anal. Methods, 2016, 8, 8059–8064 RSC.
- Y. Zhang, H. Li, J. Xie, M. Chen, D. Zhang, P. Pang, H. Wang, Z. Wu and W. Yang, Electrochemical biosensor for silver ions based on amplification of DNA–Au bio–bar codes and silver enhancement, J. Electroanal. Chem., 2017, 785, 117–124 CrossRef CAS.
- Y. Li, J. Yuan and Z. Xu, A Sensitive Fluorescence Biosensor for Silver Ions (Ag+) Detection Based on C-Ag+-C Structure and Exonuclease III-Assisted Dual-Recycling Amplification, J. Anal. Methods Chem., 2019, 2019, 1–8 CAS.
- X. Jiang, W. Xu, X. Chen and Y. Liang, Colorimetric assay for ultrasensitive detection of Ag(I) ions based on the formation of gold nanoparticle oligomers, Anal. Bioanal. Chem., 2019, 411, 2439–2445 CrossRef CAS PubMed.
- G. Wang, S. Wang, C. Yan, G. Bai and Y. Liu, DNA-functionalized gold nanoparticle-based fluorescence polarization for the sensitive detection of silver ions, Colloids Surf., B, 2018, 167, 150–155 CrossRef CAS PubMed.
- X. Zuo, H. Zhang, Q. Zhu, W. Wang, J. Feng and X. Chen, A dual-color fluorescent biosensing platform based on WS 2 nanosheet for detection of Hg2+ and Ag+, Biosens. Bioelectron., 2016, 85, 464–470 CrossRef CAS PubMed.
- S. Choi, G. Lee, I. S. Park, M. Son, W. Kim and H. Lee, Detection of silver ions using dielectrophoretic tweezers-based force spectroscopy, Anal. Chem., 2016, 88, 10867–10875 CrossRef CAS PubMed.
- W. Zhou, J. Ding and J. Liu, 2-Aminopurine-modified DNA homopolymers for robust and sensitive detection of mercury and silver, Biosens. Bioelectron., 2017, 87, 171–177 CrossRef CAS PubMed.
- T. Daşbaşı, Ş. Saçmacı, A. Ülgen and Ş. Kartal, A simple dispersive liquid–liquid microextraction method for determination of Ag(I) by flame atomic absorption spectrometry, J. Ind. Eng. Chem., 2015, 28, 316–321 CrossRef.
- J. A. López-López, B. Herce-Sesa and C. Moreno, Solventbar micro-extraction with graphite atomic absorption spectrometry for the determination of silver in ocean water, Talanta, 2016, 159, 117–121 CrossRef PubMed.
- A. V. Chugaev and I. V. Chernyshev, High-noble measurement of 107Ag/109Ag in native silver and gold by multicollector inductively coupled plasma mass spectrometry (MC-ICP-MS), Geochem. Int., 2012, 50, 899–910 CrossRef CAS.
- M. Roman, C. Rigo, H. Castillo-Michel, I. Munivrana, V. Vindigni, I. Mičetić and W. R. L. Cairns, Hydrodynamic chromatography coupled to single-particle ICP-MS for the simultaneous characterization of AgNPs and determination of dissolved Ag in plasma and blood of burn patients, Anal. Bioanal. Chem., 2015, 408, 5109–5124 CrossRef PubMed.
- S. Yazdanparast, A. Benvidi, M. Banaei, H. Nikukar, M. D. Tezerjani and M. Azimzadeh, Dual-aptamer based electrochemical sandwich biosensor for MCF-7 human breast cancer cells using silver nanoparticle labels and a poly(glutamic acid)/MWNT nanocomposite, Mikrochim. Acta, 2018, 185, 405–414 CrossRef PubMed.
- M. Ebrahimi, J. B. Raoof and R. Ojani, Sensitive electrochemical DNA-based biosensors for the determination of Ag+ and Hg2+ ions and their application in analysis of amalgam filling, J. Iran. Chem. Soc., 2018, 15, 1871–1880 CrossRef CAS.
- A. Ono, H. Torigoe and Y. Tanaka, Binding of metal ions by pyrimidine base pairs in DNA duplexes, Cheminform, 2011, 40, 5855–5866 CAS.
- A. Ono, S. Cao and H. Togashi, Specific interactions between silver(i) ions and cytosine–cytosine pairs in DNA duplexes,, Chem. Commun., 2008, 39, 4825–4827 RSC.
- G. Liu, Y. Yuan, S. Wei and D. Zhang, Impedimetric DNA-Based Biosensor for Silver Ions Detection with Hemin/G-Quadruplex Nanowire as Enhancer, Electroanal, 2014, 26, 2732–2738 CrossRef CAS.
- H. Xi, M. Cui and W. Li, Colorimetric detection of Ag+, based on C-Ag+-C binding as a bridge between gold nanoparticles, Sens. Actuators, B, 2017, 250, 641–646 CrossRef CAS.
- Z. Zhang and J. Yan, A signal-on electrochemical biosensor for sensitive detection of silver ion based on alkanethiol–carbon nanotube-oligonucleotide modified electrodes, Sens. Actuators, B, 2014, 202, 1058–1064 CrossRef CAS.
- Y. Duan, L. Wang, Z. Gao, H. Wang, H. Zhang and H. Li, An aptamer-based effective method for highly sensitive detection of chloramphenicol residues in animal-sourced food using real-time fluorescent quantitative PCR, Talanta, 2017, 165, 671–676 CrossRef CAS PubMed.
- J. Chen, Z. Zhao and Y. Chen, Development and application of a SYBR green real-time PCR for detection of the emerging avian leukosis virus subgroup K, Poult. Sci., 2018, 97, 2568–2574 CrossRef CAS PubMed.
- T. Hou, W. Li and X. Liu, Label-Free and Enzyme-Free Homogeneous Electrochemical Biosensing Strategy Based on Hybridization Chain Reaction: A Facile, Sensitive and Highly Specific MicroRNA Assay, Anal. Chem., 2015, 87, 11368–11374 CrossRef CAS PubMed.
- R. M. Dirks, N. A. Pierce and S. L. Mayo, Triggered amplification by hybridization chain reaction, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- L. L. Yang, Y. Z. Tao, G. Y. Yue, R. B. Li, B. Qiu, L. H. Guo, Z. Y. Lin and H. H. Yang, Highly selective and sensitive electrochemiluminescence biosensor for p53 DNA sequence based on nicking endonuclease assisted target recycling and hyperbranched rolling circle amplification, Anal. Chem., 2016, 88, 5097–5103 CrossRef CAS PubMed.
- J. Teng, Y. Ye, L. Yao, C. Yan, K. Cheng, F. Xue, D. Pan, B. Li and W. Chen, Rolling circle amplification based amperometric aptamer/immuno hybrid biosensor for ultrasensitive detection of Vibrio parahaemolyticus, Microchim. Acta, 2017, 184, 3477–3485 CrossRef CAS.
- D. Wu, H. Xu, H. M. Shi, W. H. Li, M. Z. Sun and Z. S. Wu, A label-free colorimetric isothermal cascade amplification for the detection of disease-related nucleic acids based on double-hairpin molecular beacon, Anal. Chim. Acta, 2017, 957, 55–62 CrossRef CAS PubMed.
- D. Zhu, L. Zhang, W. Ma, S. Lu and X. Xing, Detection of microRNA in clinical tumor samples by isothermal enzyme-free amplification and label-free graphene oxide-based SYBR Green I fluorescence platform, Biosens. Bioelectron., 2015, 65, 152–158 CrossRef CAS PubMed.
- Z. Chen, Y. Liu, C. Xin, J. Zhao and S. Liu, A cascade autocatalytic strand displacement amplification and hybridization chain reaction event for label-free and ultrasensitive electrochemical nucleic acid biosensing, Biosens. Bioelectron., 2018, 113, 1–8 CrossRef CAS PubMed.
- T. Bao, M. Wen, W. Wen, X. Zhang and S. Wang, Ultrasensitive electrochemical biosensor of interferon-gamma based on gold nanoclusters-graphene@zeolitic imidazolate framework-8 and layered-branched hybridization chain reaction, Sens. Actuators, B, 2019, 296, 126606–126613 CrossRef CAS.
- Y. Zhang, Z. W. Chen, Y. Tao, Z. Z. Wang, J. S. Ren and X. G. Qu, Hybridization chain reaction engineered dsDNA for Cu metallization: an enzyme-free platform for amplified detection of cancer cells and microRNAs, Chem. Commun., 2015, 51, 11496–11499 RSC.
- W. T. Lu, R. Arumugam and D. Senapati, Multifunctional oval-shaped gold-nanoparticle-based selective detection of breast cancer cells using simple colorimetric and highly sensitive two-photon scattering assay, ACS Nano, 2010, 4, 1739–1749 CrossRef CAS PubMed.
- N. Duan, S. Wu and X. Chen, Selection and characterization of aptamers against Salmonella typhimurium using whole-bacterium Systemic Evolution of Ligands by Exponential Enrichment (SELEX), J. Agric. Food Chem., 2013, 61, 3229–3234 CrossRef CAS PubMed.
- L. Wang and W. Tan, Multicolor FRET silica nanoparticles by single wavelength excitation, Nano Lett., 2006, 6, 84–88 CrossRef CAS PubMed.
- X. Ma, Y. Jiang, F. Jia, Y. Yu, J. Chen and Z. Wang, An aptamer-based electrochemical biosensor for the detection of Salmonella, J. Microbiol. Methods, 2014, 98, 94–98 CrossRef CAS PubMed.
- S. Lian, P. Zhang and P. Gong, A universal quantum dots-aptamer probe for efficient cancer detection and targeted imaging, J. Nanosci. Nanotechnol., 2012, 12, 7703–7708 CrossRef CAS PubMed.
- Y. Li, S. Liu and L. Ling, Sensitive Fluorescent Sensor for Recognition of HIV-1 dsDNA by Using Glucose Oxidase and Triplex DNA, J. Anal. Methods Chem., 2018, 2018, 1–8 Search PubMed.
- Y. Li, S. Liu, Q. Deng and L. Ling, A sensitive colorimetric DNA biosensor for specific detection of the HBV gene based on silver-coated glass slide and G-quadruplex-hemin DNAzyme, J. Med. Virol., 2018, 90, 699–705 CrossRef CAS PubMed.
- Y. Li and L. Ling, Aptamer-based fluorescent solid-phase thrombin assay using a silver-coated glass substrate and signal amplification by glucose oxidase, Microchim. Acta, 2015, 182, 1849–1854 CrossRef CAS.
- Y. Li, H. Zhang, H. Zhu and L. Ling, A sensitive fluorescence method for sequence-specific recognition of single-stranded DNA by using glucose oxidase, Anal. Methods, 2015, 7, 5436–5440 RSC.
- L. Qu and L. Dai, Novel silver nanostructures from silver mirror reaction on reactive substrates, J. Phys. Chem. B, 2005, 109, 13985–13990 CrossRef CAS PubMed.
- Y. Zhou, M. Li, B. Su and Q. Lu, Superhydrophobic surface created by the silver mirror reaction and its drag-reduction effect on water, J. Mater. Chem., 2009, 19, 3301–3306 RSC.
- O. W. Lau and B. Shao, Determination of glucose using a piezoelectric quartz crystal and the silver mirror reaction, Anal. Chim. Acta, 2000, 407, 17–21 CrossRef CAS.
- G. Braun, S. Lee, M. Dante, T. Nguyen, M. Moskovits and N. Reich, Surface-enhanced Raman spectroscopy for DNA detection by nanoparticle assembly onto smooth metal films, J. Am. Chem. Soc., 2007, 129, 6378–6379 CrossRef CAS PubMed.
- J. S. Lee, A. K. R. Lytton-Jean and S. J. Hurst, Silver Nanoparticle Oligonucleotide Conjugates Based on DNA with Triple Cyclic Disulfide Moieties, Nano Lett., 2007, 7, 2112–2115 CrossRef CAS PubMed.
- D. G. Thompson, A. Enright, K. Faulds, W. E. Smith and D. Graham, Ultrasensitive DNA detection using oligonucleotide-silver nanoparticle conjugates, Anal. Chem., 2008, 80, 2805–2810 CrossRef CAS PubMed.
- S. Lu, T. Hu, S. Wang, J. Sun and X. Yang, Ultra-Sensitive Colorimetric Assay System Based on the Hybridization Chain Reaction-Triggered Enzyme Cascade Amplification, ACS Appl. Mater. Interfaces, 2017, 9, 167–175 CrossRef CAS PubMed.
- X. L. Huo, N. Zhang, J. J. Xu and H. Y. Chen, Ultrasensitive electrochemiluminescence immunosensor with wide linear range based on a multiple amplification approach, Electrochem. Commun., 2019, 98, 33–37 CrossRef CAS.
- L. Kong, D. Wang, Y. Chai, Y. Yuan and R. Yuan, Electrocatalytic Efficiency Regulation between Target-Induced HRP-Mimicking DNAzyme and GOx with Low Background for Ultrasensitive Detection of Thrombin, Anal. Chem., 2019, 91, 10289–10294 CrossRef CAS PubMed.
- X. Guo, S. Liu, M. Yang, H. Du and F. Qu, Dual signal amplification photoelectrochemical biosensor for highly sensitive human epidermal growth factor receptor-2 detection, Biosens. Bioelectron., 2019, 139, 111312–111317 CrossRef CAS PubMed.
- A. D. Arulraj, R. Devasenathipathy, S. Chen, V. S. Vasantha and S. F. Wang, Highly selective and sensitive fluorescent chemosensor for femtomolar detection of silver ion in aqueous medium, Sens. Biosensing Res., 2015, 6, 19–24 CrossRef.
- L. Q. Lu, P. Deng, Y. H. Ma, L. L. Wang, F. Peng and X. K. Tian, Room-Temperature Phosphorescence Detection of Silver Ions Based on Glutathione Capped Mn-Doped ZnS Quantum Dots, J. Nanosci. Nanotechnol., 2017, 17, 6759–6764 CrossRef CAS.
- Y. Zhang, H. Li, M. Chen, X. Fang, P. Pang, H. Wang, Z. Wu and W. Yang, Ultrasensitive electrochemical biosensor for silver ion based on magnetic nanoparticles labeling with hybridization chain reaction amplification strategy, Sens. Actuators, B, 2017, 249, 431–438 CrossRef CAS.
- Y. Zhou, L. Tang, G. Zeng, J. Zhu, H. Dong, Y. Zhang, X. Xie, J. Wang and Y. Deng, A novel biosensor for silver(I) ion detection based on nanoporous gold and duplex-like DNA scaffolds with anionic intercalator, RSC Adv., 2015, 5, 69738–69744 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04202a |
| This journal is © The Royal Society of Chemistry 2020 |