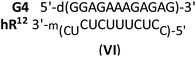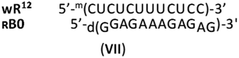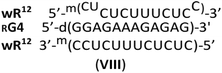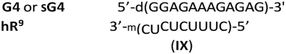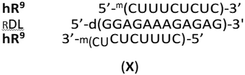Open Access Article
Open Access ArticleLNA units present in [RP-PS]-(DNA#LNA) chimeras enhance the thermal stability of parallel duplexes and triplexes formed with (2′-OMe)-RNA strands†‡
Katarzyna Jastrzębska *,
Barbara Mikołajczyk and
Piotr Guga
*,
Barbara Mikołajczyk and
Piotr Guga
Department of Bioorganic Chemistry, Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, Sienkiewicza 112, 90-363 Łódź, Poland. E-mail: kjastrz@cbmm.lodz.pl
First published on 10th June 2020
Abstract
The results of CD measurements indicate that 2-4 LNA units distributed along 12 nt P-stereodefined phosphorothioate [RP-PS]-(DNA#LNA) chimeras impose a C3′-endo conformation on the 2′-deoxyribonucleosides. Under neutral and slightly acidic conditions homopurine [RP-PS]-(DNA#LNA) hybridizes with 9–12 nt Hoogsteen-paired (2′-OMe)-RNA strands to form parallel duplexes, which are thermally more stable than the reported earlier analogous complexes containing LNA-free [RP-PS]-DNA oligomers (ΔTm = 7 °C per LNA unit at pH 5.4). Upon addition of the corresponding Watson–Crick-paired (2′-OMe)-RNA strands, parallel triplexes are formed with further increased thermal stability.
Introduction
Nucleic acid triple-helical forms have been known for sixty years.1 Their formation seems to play a role in sequence-specific recognition of a double helix,2–4 as well as in chromatin organization, DNA repair, transcriptional regulation and RNA processing.5,6 The third strand binds to an antiparallel duplex either in a parallel or an antiparallel orientation with respect to the purine strand, utilizing either a Hoogsteen or reverse Hoogsteen hydrogen bonding scheme, respectively.7 Hoogsteen G-C+ base pairing requires protonation of the N3 nitrogen atoms in cytosines8 (for CMP and dCMP, pKa of 4.3 and 4.6 was found at 25 °C, respectively9). Triplexes may comprise RNA and/or DNA strands, with consequences for thermal stability.10,11,12 Usually, the Hoogsteen paired (Hp) parallel duplexes are thermally much less stable than the Watson–Crick paired (WCp) duplexes, but their stability increases when a Hp chain is linked to a pyrimidine chain (of inverted polarity) by a 3′–3′ or 5′–5′ linker.13–16 There are a few reports on parallel stretches in native DNA17–19 and in Escherichia coli mRNA.20 Hp-DNA duplexes seem to be involved in regulation of cell processes and evolution of neurodegenerative diseases.21–23Since natural DNA molecules are easily degraded by phosphodiesterases, several modifications of the sugar-phosphate backbone have been proposed. Among them, phosphorothioate analogs of DNA (PS-DNA) were found to be very useful because their electronic and steric properties are remarkably close to those of DNA.24–26 However, as short as 10–12 nt PS-DNA oligomers prepared by standard chemical methods (a phosphoramidite or an H-phosphonate approach) exist as mixtures of hundreds or even thousands of P-diastereomers.27 Developed in this laboratory an oxathiaphospholane approach28,29 (utilizing the OTP monomers 1, Chart 1) allows for preparation of P-stereodefined PS-DNA.
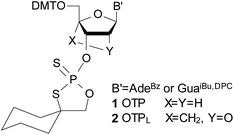 | ||
| Chart 1 Structure of the 2-thio-1,3,2-oxathiaphospholane derivatives of nucleosides of the DNA (1) and LNA (2) type. | ||
It was found that the antiparallel duplexes formed by [RP-PS]-DNA (RDNA) or the SP-counterparts (SDNA) with DNA or RNA oligomers were thermally less stable than analogous complexes formed by unmodified oligomers.30 However, we discovered that homopurine RDNA form thermodynamically highly stable parallel duplexes RNA&RDNA (e.g. I, “&” indicates the parallel orientation of strands) and even more stable parallel triplexes RNA&RDNA:RNA (e.g. II).31–33
Since only the homopurine [RP-PS]-DNA oligomers form the complexes I and II, we hypothesized (based on IR measurements and molecular modeling) that the Hp-strand is anchored not only by the hydrogen bonding but also by water bridge(s) (via charge assisted hydrogen bonds34) between the sulfur atoms of the RP-phosphorothioate moieties and the O2 atoms in pyrimidine nucleobases.§ The overall A-like conformation of the RNA&RDNA and RNA&RDNA:RNA complexes, which is required for the interactions of that type, was imposed by the RNA strand(s), and was confirmed by CD measurements. The conformational factor is important because the complexes I and II are more stable when formed with the participation of (2′-OMe)-RNAs (mRNA), which are known for adopting more profound C3′-endo conformation than RNA molecules. To analyze further this phenomenon we employed LNA units35 (often abbreviated as BNA, Bridged Nucleic Acids36), in which the sugar rings adopt a profoundly rigid 3′-endo conformation with a very limited pseudorotational flexibility. Our experiments showed that 2-4 pyrimidine LNA units present in 9-12 nt Hp-mRNA strands gave rise to significantly enhanced thermal stability of the (mRNA#LNA)&RDNA complexes.37 We wanted to check if a few LNA units (here denoted AL or GL) present in homopurine [RP-PS]-(DNA#LNA) chimeric oligomers (RDL, 3, Scheme 1) would impose a C3′-endo conformation on the 2′-deoxyribonucleosides in the phosphorothioate strand. If so, an energetic toll for the C2′-endo → C3′-endo transition of the phosphorothioate central strand (Scheme 1) should be smaller giving rise to increased thermal stability of the parallel complexes. For that purpose we used recently developed P-diastereomerically pure OTPL monomers 2 (Chart 1).38
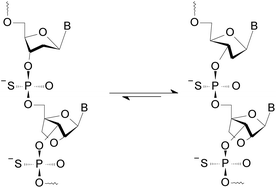 | ||
| Scheme 1 An equilibrium leading to a C3′-endo conformation of the 2′-deoxyribonucleosides in an RDL strand (3). | ||
Results and discussion
Synthesized oligomers
In the codes shown in Table 1 and used throughout the text, the prefixes R/S indicate RP-/SP-PS analogs, respectively, and the ending digit indicates the number of LNA units in a 12 nt oligomer. Accordingly, a code RA2 indicates an [RP-PS]-(DNA#LNA) chimera bearing 10 2′-deoxyribonucleosides (dA, dG) and 2 AL units, whereas SG4 stands for an [SP-PS]-(DNA#LNA) analog bearing 4 GL units. Four reference, non-palindromic, homopurine 12 nt oligonucleotides were synthesized (Table 1): (1) B0, a basal phosphate, LNA-free oligomer d(GGAGAAAGAGAG), (2) G4, an analog of B0 containing four GL units, (3) RB0, an RP-PS analog of B0; and (4) SG4, an SP-PS analog of G4.| Code | Sequence (5' → 3′ direction) | M. mass calc. | m/z founda | |
|---|---|---|---|---|
| a The relevant mass spectra for RDL and SG4 are given in Fig. S1–S5 (ESI). | ||||
| Reference | B0 | [PO]-d(GGA/GAAAGAGAG) | 3793 | 3793.0 |
| G4 | [PO]-d(GGLA/GLAAAGLAGLAG) | 3905 | 3903.8 | |
| RB0 | [RP-PS]-d(GGA/GAAAGAGAG) | 3969 | 3962.9 | |
| SG4 | [SP-PS]-d(GGLA/GLAAAGLAGLAG) | 4081 | 4078.2 | |
| wD12 | [PO]-d(CTCTCTTTCTCC) | 3498 | 3491.8 | |
| RDL | RA2 | [RP-PS]-d(GGA/GALAAGALGAG) | 4025 | 4019.5 |
| RA4 | [RP-PS]-d(GGAL/GAALAGALGALG) | 4081 | 4075.1 | |
| RG2 | [RP-PS]-d(GGA/GLAAAGLAGAG) | 4025 | 4019.3 | |
| RG4 | [RP-PS]-d(GGLA/GLAAAGLAGLAG) | 4081 | 4075.5 | |
| mRNA | wR12 | (2′-OMe)-CUCUCUUUCUCC | 3774 | 3770.4 |
| hR12 | (2′-OMe)-CCUCUUUCUCUC | 3774 | 3770.6 | |
| wR9 | (2′-OMe)-CUCUCUUUC | 2814 | 2809.7 | |
| hR9 | (2′-OMe)-CUUUCUCUC | 2814 | 2810.1 | |
The [PS]-(DNA#LNA) chimeras RA2, RA4, RG2, RG4, and SG4 (all are isosequential to B0; Table 1) were manually synthesized on solid support using pure P-diastereomers of the oxathiaphospholane derivatives of DNA (1, OTP (ref. 29)) or LNA nucleosides (2, OTPL (ref. 38)).
Homopyrimidine 12 nt WCp- and Hp-mRNAs (wR12 and hR12, respectively)¶ and their 9-nt congeners (wR9 and hR9), as well as the oligomers B0, G4 and 12-nt WCp-DNA (wD12, d(CTCTCTTTCTCC), the fifth reference oligomer) were synthesized on an automated synthesizer using commercially available phosphoramidite monomers. The sequences of wR9 and hR9 are subsets of wR12 and hR12, truncated by three nucleotides either from the 3′-end (wR9) or from the 5′-end (hR9). The 9 nt segments located after the slashes in the sequences of homopurine oligomers (Table 1) are expected to interact with wR9 and hR9 in the mismatch-free manner, as shown in the complexes III, IV, and V. Other alignment of these strands is also possible but would lead to less stable, imperfect complexes, indicated as (im). As shown in Table 1, in the complexes formed by 9 nt mRNAs with RA2 or RG2, both LNA nucleotides hybridize within the 2- or 3-strand structures, whereas in RA4 and RG4 only three LNA units do this, as AL-10 and GL-11 (numbered from the 3′-end) stay outside. This partial involvement is denoted as RA4{3} and RG4{3}.
CD spectra for single-stranded chimeric RDL oligonucleotides
The impact of the LNA units on conformation of the RDL oligomers (single strands) was assessed by CD spectroscopy (Fig. 1). As a reference we used a spectrum recorded for wD12, in which the 2′-deoxyribonucleosides exist in a 2′-endo form. It was found that in the spectra recorded for RA4, RG2, and RG4 the isoelliptic points shifted from 263 nm to 255–257 nm, so the conformations changed towards that observed for the mRNA oligomer hR12 (the isoelliptic point 250 nm, λmax = 273 nm, Θ = 5.66 mdeg), in which the nucleosides adopt a 3′-endo form. That change is more developed for RA4 and RG4 as the intensities of the bands around 270 nm are ca. 50% higher than for RG2.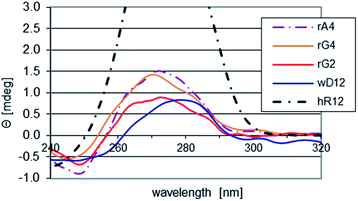 | ||
| Fig. 1 CD spectra for the chimeric RDL and the reference hR12 and wD12 oligomers, recorded at pH 5.4 at room temperature. | ||
Melting and CD experiments in pH 7.2 buffer
For the annealing/melting and CD experiments, equimolar amounts of the homopurine oligomers (RDL, SG4, or the reference oligomers) and the 12 nt mRNAs (wR12 and/or hR12) or the reference wD12 were mixed. The temperatures of association (Ta, determined during annealing) and melting (Tm) were determined using the first order derivative method. The hysteresis values (Tm–Ta) for the two component mixtures did not exceed 3 °C, unless otherwise stated (Table 2).| Homopurine oligomer | wR12 | hR12 | hR12 + wR12 |
|---|---|---|---|
| a Roman numerals in parentheses refer to the proposed structures of complexes.b Numbers in brackets indicate the temperature of association found during the annealing in the cases where the hysteresis value exceeded 3 °C. | |||
| B0 | 51 | 47 | 53 |
| G4 | 64 | [57]b/61 (VI)a | [65]/69 |
| RB0 | 50 (VII) | 62 (I) | [67]/>80 (II) |
| RA2 | [65]/73 | [64]/79 | [65]/>80 (II) |
| RG4 | 77 (VIII) | 75 (I) | [80]/>80 (II) |
| sG4 | 57 | 52 | 60 |
The complex formed by the phosphate oligomer G4 with hR12 was less stable than G4:wR12 (Tm = 61 °C and 64 °C, respectively; Fig. S6, ESI†) and this indicates the formation of an imperfect duplex (VI).
The WCp duplex sG4:wR12 was more stable than B0:wR12 (ΔTm = 11 °C) thus, the earlier identified stabilizing effect of the LNA units39 overrode the commonly observed destabilizing effect of the phosphorothioate modification (vide supra30). The Tm values found for the intended hR12&SG4 duplex (52 °C) and the hR12&SG4:wR12 triplex (60 °C) were close to that for sG4:wR12 (57 °C), indicating that the parallel complexes were not formed.
As mentioned earlier, RDNA or SDNA and WCp DNA or RNA matrices form the corresponding antiparallel duplexes of significantly lower thermal stability than natural DNA. Thus, taking into account Tm = 50 °C noted for the putative RB0:wR12 complex, which was higher than for B0:wR12 (Tm = 46 °C), and Tm = 62 °C noted for the mismatch-free hR12&RB0, one can conclude that the imperfect (im)wR12&RB0 complex (VII, 3 mismatches) was formed, rather than the RB0:wR12. This phenomenon is supported by the relevant CD spectrum, in which an intense negative signal around 210 nm (similar to that for hR12&RB0) was noted (Fig. S7, ESI†) and indicates an enormous stabilizing effect of the [RP-PS]-modification.
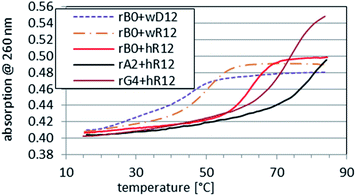
As expected, the parallel duplexes hR12&RA2 and hR12&RG4 were thermally substantially more stable (Tm > 75 °C, Fig. 2) than hR12&RB0 (ΔTm = 17 °C and ΔTm = 13 °C, respectively). The latter effect, formally generated by 4 GL units, was assumed to be more marked, but neutral pH apparently renders the C&GL interactions less effective.
Interestingly, whereas (im)wR12&RB0 (VII) was less stable than hR12&RB0 (ΔTm = −12 °C, vide supra), a Tm value for RG4:wR12 was higher than for hR12&RG4 (77 °C vs. 75 °C). This phenomenon might be explained by strong stabilizing effect of four GL units at the WC interface, analogously to sG4:wR12. However, the CD spectrum for RG4:wR12 (Fig. 3, a green line) contained an intense negative signal around 210 nm and the isoelliptic point at 249 nm, and was similar to that for hR12&RG4:wR12 (an orange line). Therefore, we suggest an additional favorable factor, where the GL units effectively promote the C2′-endo → C3′-endo conformational change of the central strand and allow formation of imperfect VIII.
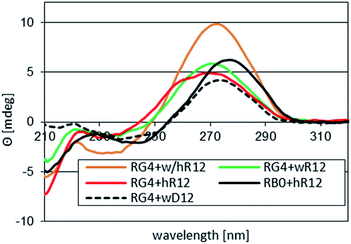 | ||
| Fig. 3 CD spectra recorded for complexes formed by RG4 with 12 nt mRNAs in pH 7.2 buffer. Profiles for hR12&RB0 ([RP-PS]-DNA, no LNA units) and for RG4:wD12 are given as the references. | ||
This process seems to occur for RA2 mixed with wR12 or hR12 because the observed Tm = 73 °C or 79 °C, respectively, were close to those for RG4:wR12 and hR12&RG4 (Tm = 77 °C and 75 °C, respectively). This suggestion is supported by an 8 °C hysteresis, which for a bimolecular association should be very low (typically <3 °C), whereas it exceeds 13–15 °C for the trimolecular systems hR12&RB0:wR12 and hR12&RA2:wR12. Unlike commonly known triplexes containing a PO-DNA central strand, which dissociate in two discrete steps, those formed by RDNA undergo a single-step melting transition, followed by return of RDNA to the C2′-endo conformation. Thus, the reverse process is “more trimolecular” and in terms of entropy it is less favored than a bimolecular one, giving rise to the hysteresis. For wR12&RG4:wR12 and hR12&RG4:hR12 the observed hysteresis values were low because 4 GL units substantially shift the conformation of the central PS-strand in the C3′-endo direction and make it “ready” for rapid association.
At pH 7.4, the triple-stranded complex hR12&RB1:wR12 and its congeners containing RA2 or RG4 were thermally so stable that the inflection points could not be determined (the melting profiles not shown).
The above presented data indicate, that at neutral pH, the LNA units present in the RDL oligomers make the Hoogsteen interactions with mRNAs significantly stronger compared to the analogous RDNA. However, concomitantly, the rigid LNA nucleotides enhance hybridization at the WC interface strongly enough to promote the formation of the imperfect triplex VIII. This hybridization may result in the binding of non-target RNAs. This loss of specificity may be avoided using RDL oligomers with Watson–Crick base pairings being hampered due to the presence of N6–Me deoxyadenosine (m6dA) units. We reported recently that the m6dA units present in RDNA stabilize (by up to 4.5 °C per modified unit) parallel duplexes formed by with Hp mRNAs compared to the analogous reference duplex containing only unmodified nucleobases, and prevent the formation of the corresponding parallel triplexes.40
Melting and CD experiments performed using pH 5.4 buffer
Because the Tm values for complexes of the RDL oligomers with wR12 or hR12 formed in pH 5.4 buffer could not be determined, next melting experiments were done for the complexes with wR9 and/or hR9.Compared to the reference RB0:wR9, the Tm values for complexes formed by RDL and SG4 with wR9 increased by 2–10 °C (Table 3 and Fig. 4) and this increase may be attributed to the stabilizing effect of the LNA nucleosides.
| Homopurine oligomer | wR9 | hR9 | hR9 + wR9 | |
|---|---|---|---|---|
| 1 | G4 | 65 | 63 (IX) | 68 |
| 2 | RB0 | 47 | 54 (III) | 77 (V) |
| 3 | RG2 | 54 | 67 (III or X) | >80 (V) |
| 4 | RA4{3} | 50 | 75 (III or X) | >80 (V) |
| 5 | RG4{3} | 49 | 75 (III or X) | >80 (V) |
| 6 | sG4 | 57 | 50 (IX) | 57 |
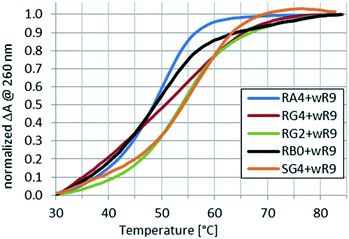 | ||
Fig. 4 Normalized melting profiles recorded for RA4, RG2, RG4, or SG4 mixed with wR9 at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio in pH 5.4 buffer. A profile for RB0 ([RP-PS]-DNA, no LNA units) is given as a reference. 1 molar ratio in pH 5.4 buffer. A profile for RB0 ([RP-PS]-DNA, no LNA units) is given as a reference. | ||
More interesting results were obtained for the complexes formed with hR9. The complex formed with SG4 (hR9&SG4{3} or (im)SG4:hR9) was 25 °C less stable than hR9&RG4{3}. Also, a Tm value found for the putative hR9&SG4{3}:wR9 was equal to that for SG4:wR9, so the SP internucleotide bonds present in SG4 did not promote the parallel hybridization, neither hR9 interacted with remarkably stable SG4{3}:wR9 to a measurable extent. The putative complexes hR9&G4{3} (no PS units) and hR9&SG4{3} (Fig. 5) were less stable (Tm = 63 °C and 50 °C, respectively) than the corresponding G4:wR9 (Tm = 65 °C) and SG4:wR9 (Tm = 57 °C) indicating the formation of the imperfect WCp duplexes (IX).
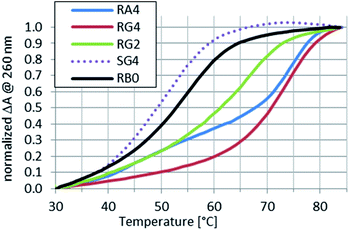 | ||
Fig. 5 Normalized melting profiles recorded for RA4, RG2, RG4, or SG4 mixed with hR9 at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio in pH 5.4 buffer. A profile for RB0 ([RP-PS]-DNA, no LNA units) is given as a reference. 1 molar ratio in pH 5.4 buffer. A profile for RB0 ([RP-PS]-DNA, no LNA units) is given as a reference. | ||
The profiles shown in Fig. 5 indicate that hR9&RG2, hR9&RA4{3}, and hR9&RG4{3} were remarkably more stable than hR9&RB0 (ΔTm = 13 °C, 21 °C, and 21 °C respectively) with an average increase ΔTm = 7 °C per the “active” LNA unit. But one cannot exclude partial formation of imperfect triplexes X, where a 7 nt part of hR9 acts as a WCp strand.
Of course, complexes analogous to X may be formed with participation of any fully or partially WCp RNA. This non-specific binding may be avoided using already mentioned RDL oligomers with the hampered Watson–Crick base pairings.
Despite very short 9 nt mRNAs used, the Tm values for the investigated triplexes hR9&RDL:wR9 exceeded 80 °C and could not be more precisely determined (data not shown). This phenomenon indicates that the C2′-endo → C3′-endo conformational shift caused by the LNA units is of great importance for the thermal stability of investigated parallel duplexes and triplexes, which may be beneficial in in vitro experiments with precisely selected RNA oligomers, but in cellular experiments may lead to unspecific hybridization with non-target oligomers present in the RNA pool.
Conclusions
The CD spectra recorded for single-stranded P-stereodefined phosphorothioate [RP-PS]-(DNA#LNA) chimeras (RDL) indicate that 2-4 LNA units distributed along 12 nt oligomer changed the C2′-endo conformation (typical for PS-DNA) into the C3′-endo conformation. The results of melting experiments suggest that this change reduced an energetic toll for the C2′-endo → C3′-endo transition of the RDL strand giving rise to an increase in thermal stability of the parallel complexes with (2′-OMe)-RNAs compared to those formed by LNA-free [RP-PS]-DNA oligomers (ΔTm = 7 °C per the LNA unit at pH 5.4). Thermally more stable complexes are formed upon addition of the corresponding Watson–Crick-paired (2′-OMe)-RNA strands due to the formation of highly stable parallel triplexes. The LNA nucleotides enhance hybridization at the WC interface and may promote the formation of imperfect triplexes VIII. This hybridization may result in the binding of non-target RNAs. This loss of specificity may be avoided using RDL oligomers with the Watson–Crick base pairing hampered due to the presence of N6–Me-2′-deoxyadenosine (m6dA) units.40 Work on such modified oligonucleotides is in progress.Experimental section
MALDI-TOF MS analyses of oligonucleotides were performed with the detection of negative ions, using a Voyager-Elite instrument (PerSeptive Biosystems Inc., Framingham, MA) operating in the reflector mode, or a Shimadzu Biotech Axima Performance instrument operating in the linear mode.Routine UV spectra were recorded on a CINTRA 10e spectrophotometer (GBC, Dandenong, Australia), using a quartz cuvette of 1 cm path length. UV monitored melting experiments were carried out in 1 cm path length cells, using a spectrophotometer CINTRA 4040 (GBC), equipped with a 6 × 1 Peltier thermocell.
For the UV-monitored (at 260 nm) thermal dissociation experiments the oligonucleotide samples were dissolved in pH 7.2 or pH 5.4 buffer containing 10 mM Tris–HCl, 100 mM NaCl, and 10 mM MgCl2. The annealing was done from 85 °C to 15 °C with a temperature gradient of 1 °C min−1. The melting profiles were recorded over a 15 → 85 °C range (0.5 °C min−1). The temperatures of association (Ta) and melting (Tm) were calculated using the first order derivative method.
CD measurements were done on a Jasco J-815 dichrograph at room temperature, using a 0.5 cm path-length quartz cell (Hellma). The spectra were recorded over a 210–320 nm range with a 1.0 nm bandwidth, a scanning speed 50 nm min−1, and a data pitch of 1 nm. After 3 spectra were accumulated, the baseline was subtracted and the resultant spectrum was smoothed with a Savitzky–Golay algorithm (the convolution width 7).
P-stereodefined oligonucleotides of the PS-DNA and PS-(DNA#LNA) series were synthesized manually at a 1 μmol scale, according to the previously published protocols.29,38 The first nucleoside unit was anchored to the solid support by a sarcosinyl linker.41 Routine coupling steps were performed using 20 mg of the OTP monomers. In the cycles where LNA-OTPL monomers 2 were incorporated, double coupling was executed (20 mg + 20 mg) and in both steps the coupling time was extended to 20 minutes. All synthesized oligomers were routinely purified by two-step reverse phase HPLC (DMT-on and DMT-off). Relevant chromatograms for RDL and SG4 are shown in Fig. S8–S12, ESI†), and their identity was assessed by MALDI-TOF MS and purity by polyacrylamide gel electrophoresis (PAGE).
Unmodified DNA and RNA oligonucleotides were synthesized on a Gene-World synthesizer (K&A Laborgeraete GbR, Schaafheim, Germany).
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Prof. Maria Bryszewska and Dr Katarzyna Miłowska (Lodz University) are gratefully acknowledged for providing access to a dichrograph. This work was financially supported by National Centre of Science, Poland, a grant UMO-2015/19/B/ST5/03116 to PG.Notes and references
- G. Fensenfeld, V. Davies and A. Rich, J. Am. Chem. Soc., 1957, 79, 2023 CrossRef.
- J. R. Goñi, X. de la Cruz and M. Orozco, Nucleic Acids Res., 2004, 32, 354 CrossRef PubMed.
- P. Jenjaroenpun, C. S. Chew, T. P. Yong, K. Choowongkomon, W. Thammasorn and V. A. Kuznetsov, Nucleic Acids Res., 2015, 43, D110–D116 CrossRef CAS PubMed.
- J. A. Brown, M. L. Valenstein, T. A. Yario, K. T. Tycowski and J. A. Steitz, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 19202 CrossRef CAS PubMed.
- F. A. Buske, J. S. Mattick and T. L. Bailey, RNA Biol., 2011, 8, 427 CrossRef CAS PubMed.
- A. Bacolla, G. Wang and K. M. Vasquez, PLoS Genet., 2015, 11(12), e1005696 CrossRef PubMed.
- C. Hélène, in Antisense Research and Application, ed. S. T. Crook and B. Lebleu, and references cited therein, CRC Press Inc. Boca Raton, Ann Arbor, London, Tokyo, 1993, pp. 375–385 Search PubMed.
- G. Raghunathan, H. T. Miles and V. Sasisekharan, Biopolymers, 1994, 12, 1573 CrossRef PubMed.
- L. Lavelle and J. R. Fresco, Nucleic Acids Res., 1995, 23, 2692 CrossRef CAS PubMed.
- R. W. Roberts and D. M. Crothers, Science, 1992, 258, 1463 CrossRef CAS PubMed.
- H. Han and P. B. Dervan, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3806 CrossRef CAS PubMed.
- C. Escude, J.-C. Francois, J.-S. Sun, G. Ott, M. Sprinzl, T. Garestier and C. Hélène, Nucleic Acids Res., 1993, 21, 5547 CrossRef CAS PubMed.
- A. Aviñó, M. Frieden, J. C. Morales, B. G. de la Torre, R. Güimil-García, F. Azorín, J. L. Gelpí, M. Orozco, C. González and R. Eritja, Nucleic Acids Res., 2002, 30, 2609 CrossRef PubMed.
- A. K. Shchyolkina, O. F. Borisova, M. A. Livshits, G. E. Pozmogova, B. K. Chernov, R. Klement and T. M. Jovin, Biochemistry, 2000, 39, 10034 CrossRef CAS PubMed.
- E. R. Kandimalla and S. Agrawal, Biochemistry, 1996, 35, 15332 CrossRef CAS PubMed.
- J. H. van de Sande, N. B. Ramsing, M. W. Germann, W. Elhorst, B. W. Kalisch, E. von Kitzing, R. T. Pon, R. C. Clegg and T. M. Jovin, Science, 1988, 241, 551 CrossRef CAS PubMed.
- N. A. Tchurikov, B. K. Chernov, Y. B. Golova and Y. D. Nechipurenko, FEBS Lett., 1989, 257, 415 CrossRef CAS PubMed.
- N. A. Tchurikov, A. K. Shchyolkina, O. F. Borissova and B. K. Chernov, FEBS Lett., 1992, 297, 233 CrossRef CAS PubMed.
- O. F. Borisova, A. K. Shchyolkina, B. K. Chernov and N. A. Tchurikov, FEBS Lett., 1993, 322, 304 CrossRef CAS PubMed.
- N. A. Tchurikov, L. G. Chistyakova, G. B. Zavilgelsky, I. V. Manukhov, B. K. Chernov and Y. B. Golova, J. Biol. Chem., 2000, 275, 26523 CrossRef CAS PubMed.
- A. K. Jain and S. Bhattacharya, Bioconjugate Chem., 2010, 21, 1389 CrossRef CAS PubMed.
- A. K. Shchyolkina, O. F. Borisova, M. A. Livshits and T. M. Jovin, Mol. Biol., 2003, 37, 223 CrossRef CAS.
- M. W. Germann, C. N. Johnson and A. M. Spring, Chimia, 2009, 63, 731 CrossRef CAS.
- S. Verma and F. Eckstein, Annu. Rev. Biochem., 1998, 67, 99 CrossRef CAS PubMed.
- P. Guga, Curr. Topics in Med. Chem., 2007, 7, 695 CrossRef CAS PubMed.
- P. Guga and M. Koziołkiewicz, Chem. Biodiversity, 2011, 8, 1642 CrossRef CAS PubMed.
- W. J. Stec and A. Wilk, Angew. Chem., Int. Ed. Engl., 1994, 33, 709 CrossRef.
- W. J. Stec, B. Karwowski, M. Boczkowska, P. Guga, M. Koziołkiewicz, M. Sochacki, M. Wieczorek and J. Błaszczyk, J. Am. Chem. Soc., 1998, 120, 7156 CrossRef CAS.
- P. Guga and W. J. Stec, in Current Protocols in Nucleic Acid Chemistry, ed. S. L. Beaucage, D. E. Bergstrom, G. D. Glick, and R. A. Jones, John Wiley & Sons, Hoboken, N.J., 2003, pp. 4.17.1 Search PubMed.
- M. Boczkowska, P. Guga and W. J. Stec, Biochemistry, 2002, 41, 12483 CrossRef CAS PubMed.
- W. J. Stec, A. Grajkowski, B. Karwowski, A. Kobylańska, M. Koziołkiewicz, K. Misiura, A. Okruszek, A. Wilk, P. Guga and M. Boczkowska, J. Am. Chem. Soc., 1995, 117, 12019 CrossRef CAS.
- P. Guga, M. Boczkowska, M. Janicka, A. Maciaszek, S. Kuberski and W. J. Stec, Biophys. J., 2007, 92, 2507 CrossRef CAS PubMed.
- P. Guga, M. Janicka, A. Maciaszek, B. Rębowska and G. Nowak, Biophys. J., 2007, 93, 3567 CrossRef CAS PubMed.
- J. J. Novoa, I. Nobeli, F. Grepioni and D. Braga, New J. Chem., 2000, 24, 5 RSC.
- A. A. Koshkin, S. K. Singh, P. Nielsen, V. K. Rajwanshi, R. Kumar, M. Meldgaard, C. E. Olsen and J. Wengel, Tetrahedron, 1998, 54, 3607 CrossRef CAS.
- (a) S. Obika, D. Nanbu, Y. Hari, K. Morio, Y. In, T. Ishida and T. Imanishi, Tetrahedron Lett., 1997, 38, 8735 CrossRef CAS; (b) S. Obika, D. Nanbu, Y. Hari, J. Andoh, K. Morio, T. Doi and T. Imanishi, Tetrahedron Lett., 1998, 39, 5401 CrossRef CAS.
- A. Maciaszek, A. Krakowiak, M. Janicka, A. Tomaszewska, M. Sobczak, B. Mikołajczyk and P. Guga, Org. Biomol. Chem., 2015, 13, 2375 RSC.
- K. Jastrzębska, A. Maciaszek, R. Dolot, G. Bujacz and P. Guga, Org. Biomol. Chem., 2015, 13, 10032 RSC.
- R. Kumar, S. K. Singh, A. A. Koshkin, V. K. Rajwanshi, M. Meldgaard and J. Wengel, Bioorg. Med. Chem. Lett., 1998, 8, 2219 CrossRef CAS PubMed.
- A. Maciaszek, K. Jastrzębska and P. Guga, Org. Biomol. Chem., 2019, 17, 4611 RSC.
- T. Brown, C. E. Pritchard, G. Turner and S. A. Salisbury, J. Chem. Soc., Chem. Commun., 1989, 891 RSC.
- Y. Zhou, E. Kierzek, Z. P. Loo, M. Antonio, Y. H. Yau, Y. W. Chuah, S. Geifman-Shochat, R. Kierzek and G. Chen, Nucleic Acids Res., 2013, 41, 6664 CrossRef CAS PubMed.
- L. A. H. van Bergen, M. Alonso, A. Palló, L. Nilsson, F. De Proft and J. Messens, Sci. Rep., 2016, 6, 30369 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03934a |
| ‡ Dedicated to Professor Wojciech J. Stec on the occasion of his 80th birthday. |
§ The mechanism of stabilization (proposed by C. Hélène and also discussed by others12,42), based on the hydrogen bonding between the 2′-OH group of ribose in the Hp-strand and the pro-RP oxygen atom (or the RP sulfur atom in our work) cannot operate because parallel triplexes and parallel duplexes formed with Hp-mRNAs are thermally more stable. Also hydrogen bonding pyrimidine–C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–S–P(O)(OR)2, analogous to Cys–S–H⋯O O⋯H–S–P(O)(OR)2, analogous to Cys–S–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C in proteins,43 is less likely because at neutral pH phosphorothioate diesters are fully ionized. C in proteins,43 is less likely because at neutral pH phosphorothioate diesters are fully ionized. |
| ¶ We assumed high melting temperatures of the investigated complexes, therefore, to avoid degradation of RNA oligomers at elevated temperatures we decided to use (2′-OMe)-RNA oligomers. |
| This journal is © The Royal Society of Chemistry 2020 |