Open Access Article
Open Access ArticleCytotoxic effect of green silver nanoparticles against ampicillin-resistant Klebsiella pneumoniae
Reham Samir Hamida *a,
Mohamed Abdelaal Alib,
Doaa A. Godac,
Mahmoud Ibrahim Khalilad and
Alya Redhwan*e
*a,
Mohamed Abdelaal Alib,
Doaa A. Godac,
Mahmoud Ibrahim Khalilad and
Alya Redhwan*e
aMolecular Biology Unit, Department of Zoology, Faculty of Science, Alexandria University, Egypt. E-mail: reham.hussein@alexu.edu.eg; Tel: +201156298937
bBiotechnology Unit, Department of Plant Production, College of Food and Agriculture Science, King Saud University, Riyadh, Saudi Arabia
cBioprocess Development Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), Alexandria, Egypt
dDepartment of Biological Sciences, Faculty of Science, Beirut Arab University, Lebanon
eDepartment of Health, College of Health and Rehabilitation Sciences, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. E-mail: AMRedhwan@pnu.edu.sa; Tel: +966555237223
First published on 3rd June 2020
Abstract
Considering the harmful effects and high spread of drug-resistant Klebsiella pneumoniae, many researchers have been trying to produce new antibacterial agents to combat the emergence of multidrug-resistant (MDR) strains of this bacterium. Recent progress in the nanomedicine field has provided opportunities for synthesizing unique nanoagents to battle MDR bacteria by targeting virulence and resistance signalling. The biocidal effects of 14.9 nm silver nanoparticles fabricated using Nostoc sp. Bahar M (N-SNPs) and AgNO3 were examined against drug-resistant K. pneumoniae using the agar well diffusion method. Transmission electron microscopy (TEM) was used to detect the ultrastructural changes caused by N-SNPs and AgNO3. To address the mode of action of N-SNPs and AgNO3, CAT, GPx, LDH and ATPase levels were assessed. The toxicity of N-SNPs and AgNO3 was evaluated against the mfD, flu, hly, 23S, hns, hcp-1, VgrG-1 and VgrG-3 genes as well as cellular proteins. N-SNPs showed the greatest inhibitory activity against K. pneumoniae, with MIC and MBC values of 0.9 and 1.2 mg mL−1, respectively. Furthermore, N-SNPs and AgNO3 induced apoptotic features, including cell shrinkage and cell atrophy. N-SNPs were more potent bactericidal compounds than AgNO3, causing increased leakage of LDH and GPx activities and depletion of ATPase and CAT activities, resulting in induced oxidative stress and metabolic toxicity. Compared to AgNO3, N-SNPs exhibited the highest toxicity towards the selected genes and the greatest damage to bacterial proteins. N-SNPs were the most potent agents that induced bacterial membrane damage, oxidative stress and disruption of biomolecules such as DNA and proteins. N-SNPs may be used as effective nanodrugs against MDR bacteria.
1 Introduction
Worldwide, humans face a potentially life-threatening risk from antimicrobial resistance (AMR), regardless of gender, age, or socioeconomic background. The mortality rate due to AMR is rising and is expected to reach 50 million by 2050.1 Despite this, the discovery rate of new antibiotics has been decreasing over time. Clinically important Gram-positive and Gram-negative bacteria are rapidly mounting resistance to the available antimicrobial agents.2 In particular, Klebsiella sp. that is the second most common disease-causing Gram-negative bacterium, resulting in invasive infections such as pneumonia and meningitis.3Antimicrobial-resistant bacteria, known as superbugs, tend to enhance their levels of defence against antibiotics by modifying their mechanisms of defence over time.4 For example, multidrug-resistant (MDR) bacteria can form biofilms as protective barriers against antibacterial agents.5,6 Moreover, bacterial fitness during infection is mainly related to the acquisition of virulence factors.7 Virulence factors, including adherence and invasion factors, capsules, endo- and exotoxins, and secretory system components, are normally used by bacteria to establish and cause disease or infection and to evade host defence strategies.8,9 Currently, there is an urgent need for alternative treatments to tackle the AMR threat, resulting from the dramatic increase in antimicrobial-resistant microbial strains worldwide coupled with the limited available treatments.10 One of these strategies is nanotechnology-based medicine.11 Emerging nanoscience allows the synthesis of different nanoparticles (NPs) from bulk materials. These NPs can be distinguished from their precursor materials based on their unique chemical, physical and biological properties.12 NPs can be fabricated by different routes, including physical, chemical and biofabrication methods.11 The green synthesis method is based on using natural sources, including plants, algae and microorganisms, such as bacteria, cyanobacteria and fungi.13 Cyanobacteria are a diverse group of photoautotrophic prokaryotes that exist in a wide range of ecosystems.14 These microorganisms contain a variety of biochemical components that have antimicrobial and anticancer activities.15,16 Moreover, recent publications have reported the ability of cyanobacteria such as Desertifilum sp. and Nostoc sp. Bahar M to synthesize NPs that can be distinguished by different biological applications.13,17 Since ancient times, silver ions have been used as broad-spectrum antimicrobial agents against different microorganisms.18 Currently, silver ions are used in many nanoproducts that are applied in many fields, such as industry and medicine.19 Silver nanoparticles (SNPs) is one of the most potent antimicrobial agents due to their large surface area and small size, facilitating the interaction between SNPs and their targets, such as bacterial cells.20 Based on the fact of bacterial virulence factors could be useful targets for the prevention and supplementary treatment of bacterial infections.21,22 Recent studies have demonstrated the relation between NPs and some virulence- and biofilm formation-related genes.21,23 However, there is a lack of knowledge regarding the link between bacterial virulence factors, including secretion systems, and NPs. Until now, the exact inhibitory mechanism of NPs against bacterial cells is unclear, however, there are a few proposed mechanisms of action of NPs against bacteria. One of these considerations is that NPs induce ROS formation, causing oxidative stress that results in an imbalance in bacterial activities.24 Another possible mechanism is the interaction of NPs with biomolecules such as proteins, including enzymes, and genes.25 Thus, this study aims to study the inhibitor activity of N-SNPs synthesized by Nostoc Bahar M for first time against Klebsiella pneumoniae in comparing to silver nitrate (bulk material). Moreover, explain the killing mechanism of N-SNPs and silver nitrate against K. pneumoniae to fill the gap in knowledge regarding this association, which might lead to drug discovery or to the development of methods for diagnosis or prevention and intervention to tackle infections caused by drug-resistant K. pneumoniae.
2 Experimental
2.1. Materials
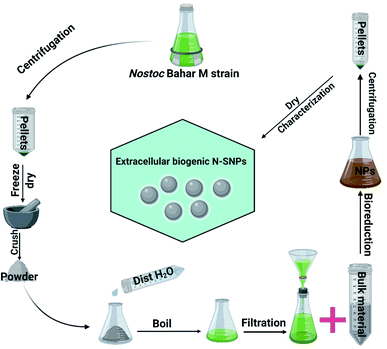
Silver nitrate (AgNO3), bacterial culture materials, and the enzymes catalase (CAT) and glutathione peroxidase (GPx) were purchased from Sigma-Aldrich (St. Louis, MO, USA); the LDH Assay Kit (colorimetric) was purchased from Abcam (Cambridge, UK); and PiBind resin was purchased from Expedeon (San Diego, USA). TRIzol reagent was purchased from Life Technologies (California, USA), and Maximas SYBR Green/Fluorescein qPCR Master Mix and the QuantiTects Reverse Transcription Kit were purchased from Qiagen (Germantown, USA). TriFast was purchased from Peqlab VWR (Pennsylvania, USA). Spherical (8.5 to 26.44 nm; mean size 14.9 nm) biogenic SNPs were extracellularly synthesized using Nostoc Bahar M (N-SNPs), characterized and published previously (Fig. 1).132.2. Bacterial culture
K. pneumoniae (clinical isolate) bacterial cells were obtained from Almery University Hospital (Alexandria, Egypt). Ten microlitres was taken from glyceride bacterial stocks, seeded into Luria–Bertani (LB) broth, and grown for 24 h at 37 °C. Fresh bacterial cultures were obtained at a concentration of 0.5 on the McFarland scale (104 CFU mL−1). Then, 50 μL of each bacterial strain was poured and gently spread onto freshly prepared LB agar plates. After spreading, four 8 mm wells were made using cork borer in all the plates for further experiments with the treatments.2.3. Agar well diffusion assay
To assess the biocidal activity of N-SNPs and AgNO3 against K. pneumoniae, 100 μL of N-SNPs, AgNO3 (1.54 mg mL−1), distilled water as a negative control and the antibiotic ampicillin as a positive control were added to each agar plate well. The treated plates were incubated at 37 °C for 24 h. At the end of incubation, the diameter of each inhibition zone (IZ) was measured using a standard metric ruler and the values recorded in mm. Data collected from independent experiments performed in at least triplicate and are presented as the mean ± SEM.2.4. Minimum inhibition concentration and minimum bactericidal concentration
The minimum inhibitory concentration (MIC) refers to the lowest concentration of antimicrobial agent at which bacterial growth is inhibited. However, the minimum bactericidal concentration (MBC) is the lowest concentration of antimicrobial drug that kills bacteria. To determine the MIC and MBC values, different concentrations of N-SNPs and AgNO3 were prepared (1.8, 1.5, 1.2, 0.9, 0.6 and 0.3 mg mL−1). In 96-well plates, 100 μL per well bacterial suspension (104 CFU mL−1) was treated with 100 μL of different concentrations of both N-SNPs and AgNO3 and then incubated for 24 h at 37 °C. After 24 h, the bacterial turbidity was checked with the naked eye and compared with a 0.5 McFarland standard medium. To verify the result of the previous step, 10 μL of each antimicrobial agent at the determined MIC and MBC values was tested against the bacterial strain using the agar well diffusion method.2.5. LDH assay
According to Yuan et al. (2017), 6 mL of 104 CFU mL−1 bacterial samples before and after being treated with 1.54 mg mL−1 N-SNPs or silver nitrate for 24 h at 37 °C was centrifuged at 5000 rpm for 10 min at 4 °C. The pellets were washed twice and treated with LDH reaction solution, followed by gentle shaking for 30 min at ambient temperature. After the incubation period, the OD of the samples was detected at 490 nm.262.6. Measurement of ATPase activity
The influence of N-SNPs and AgNO3 (1.54 mg mL−1) on the ATPase activity of the selected bacteria was determined using a colorimetric ATPase assay according to the method described by Andrés et al.27 Ten microlitres of PiBind resin was added to 10 μL of each culture supernatant to remove the free inorganic phosphate (Pi). The amount of Pi released was determined by spectrophotometry (UV 2505 spectrophotometer, Thomas Scientific, New Jersey, USA) at A650. For all experiments, calibration was performed using a standard range of Pi concentrations, and data were determined from a minimum of three independent assays.2.7. Estimation of antioxidant activity
For determination of CAT and GPx activities, the cells before and after treatment with N-SNPs or AgNO3 (1.54 mg mL−1) were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min at 4 °C. Then, the pellets were washed with PBS and lysed using a sonicator (IKAT10 basic sonicator, Cole-Parmer, Illinois, USA). The oxidative marker activities were measured using reagents from various kits according to the corresponding instructions.28
000 rpm for 5 min at 4 °C. Then, the pellets were washed with PBS and lysed using a sonicator (IKAT10 basic sonicator, Cole-Parmer, Illinois, USA). The oxidative marker activities were measured using reagents from various kits according to the corresponding instructions.28
2.8. Specimen preparation for TEM
Bacterial specimens before and after treatment with 1.54 mg mL−1 N-SNPs or AgNO3 for 24 h at 37 °C were centrifuged at 3500 rpm for 10 min. The pellets were resuspended in PBS and centrifuged again for 10 min for washing. The washed pellets were fixed by immersing the cells immediately in ice-cold 4F1G (4% formaldehyde and 1% glutaraldehyde) in 0.1 M PBS for 2 h at 4 °C. Then, 1% osmium tetroxide (OsO4) was added for 2 h at 4 °C, and the stained specimens were washed with 0.1 M PBS several times for 10 min. The samples were serially dehydrated with graded ethanol (25, 50, 75, 95 and 100%). Then, the samples were infiltrated in propylene oxide and embedded in an Araldite Epon mixture. Ultrathin (70 nm) sections from the selected area were cut with a glass knife on an LKB Ultramicrotome. The ultrathin sections after double staining with 2% uranyl acetate and lead citrate were placed on 200-mesh copper grids and then examined under a JEOL 100 CX electron microscope (JEOL, Tokyo, Japan) operating at 80 kV.292.9. Estimation of the frequency distribution of NPs size
The diameter of NPs distributed inside, and outside bacterial cells was measured using Image J software based on TEM micrographs.2.10. Gene expression analysis
qRT-PCR was applied to evaluate the change in the expression levels of transcription-repair coupling factor (mfD), α-haemolysin (hly), Ag43 phase-variable biofilm formation autotransporter CP4-44 prophage (flu), 23 rRNA (23S), histone-like nucleotide structure protein (hns), haemolysin coregulated protein (hcp-1) and valine glycine repeat proteins G1 and G3 (VgrG-1 and VgrG-3) in K. pneumoniae before and after treatment with the two silver species (1.54 mg mL−1) for 24 h at 37 °C. In brief, total RNA was extracted from the samples using TRIzol reagent. The yield and quality of the total RNA were determined spectrophotometrically based on the absorbance at 260 nm and the 260/280 nm ratio, respectively. The mRNA of the tested genes was evaluated using Maximas SYBR Green/Fluorescein qPCR Master Mix by a Rotor-Gene Q instrument. A reverse transcription step was applied to the total RNA using the QuantiTects Reverse Transcription Kit with a random primer hexamer in a two-step RT-PCR in which any genomic DNA (gDNA) contamination was eliminated using gDNA Wipeout Buffer. Total cDNA (30 ng) was used as a template for amplification with the specific primer pair related to the eight selected genes (Table 1) at a final concentration of 300 nM. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized as a housekeeping gene. The Rotor-Gene Q automatically collected the data and analysed the value of the threshold cycle (Ct), which was normalized to an average Ct value of the housekeeping gene (ΔCt), and the relative expression of each representative was calculated as 2 − ΔCt.| Gene | Primers | Reference |
|---|---|---|
| mfD | F: TCAGGAAGCTGGAAGGTAATG | 43 |
| R: GGACCATCAAGGCGGTAAT | ||
| flu | F: CACAGATACGTACAGAAAGACATTCAGG | 53 |
| R: GGCTGTGGGAGTTTCTGAATTG | ||
| hly | F: TGAATCCTGTCGCTAATG | 21 |
| R: TATCATCCGACCTTTCACT | ||
| 23S | F: AGGCGATGAAGGACGTGCTA | 54 |
| R: A TTCGGACATCGCCGGTTATA | ||
| hns | F: GAAATTGAAGAGCGTACGCGTAA | |
| R: CAATACCGTCAGCAATCAGCAT | ||
| hcp-1 | F: CAAAAACCCCGCCTACGA | |
| R: TCGTAGCGCAGCTCAATCTG | ||
| VgrG-1 | F: CCCCGGCCAACAACAAA | |
| R: GTCGAGACCTTGATATGCTCTTTTC | ||
| VgrG-3 | F: TCCTCGCCGCGCTTATC | |
| R: CCCGCAATGTGATTCGTACTC |
2.11. SDS-PAGE
To detect the toxic effect of N-SNPs and silver nitrate, total cellular proteins of the selected bacteria were extracted before and after treatment with 1.54 mg mL−1 N-SNPs or silver nitrate for 24 h at 37 °C and purified using TriFast. Ten micrograms of the purified protein from each sample was loaded into an OmniPAGE Mini vertical electrophoresis unit with a Power Pro 5 as the power supply (Cleaver Scientific, Warwickshire, UK) on a SERVAGel™ TG PRiME™ 10% (SERVA, Heidelberg, Germany). Then, the gel was stained using 0.1% Coomassie blue R-250 for 2 h and destained with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 solution of glacial acetic acid
6 solution of glacial acetic acid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water. For data analysis, a gel documentation system and TotalLab analysis software version 1.0.1 (Newcastle-Upon-Tyne, UK) (Geldoc-it, UVP, England) were applied.20
water. For data analysis, a gel documentation system and TotalLab analysis software version 1.0.1 (Newcastle-Upon-Tyne, UK) (Geldoc-it, UVP, England) were applied.20
2.12. Statistical analysis
All experiments were repeated at least three times, and the data are presented as the mean ± SEM. Prism 8.3 software (GraphPad Software Inc., La Jolla, CA, USA) was used to evaluate the statistically significant difference between the treatments and the control through one-way analysis of variance (ANOVA). The significance of the data is represented at P < 0.0001, P < 0.001 and P < 0.01. For SNP size analysis, Image J (National Institutes of Health, Bethesda, MD, USA) was used.3 Results
3.1. Antibacterial activity
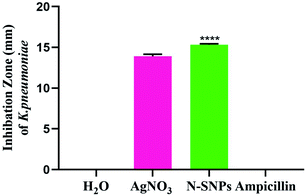
The inhibitory effect of biogenic N-SNPs and AgNO3 against drug-resistant K. pneumoniae was evaluated using an agar well diffusion assay. As shown in Fig. 2, N-SNPs and AgNO3 were able to suppress K. pneumoniae growth; however, the same bacteria resisted the antibiotic ampicillin. In addition, N-SNP was the antibacterial agent that showed the greatest inhibition of K. pneumoniae, with an IZ diameter of 15.33 ± 0.1, followed by silver nitrate, with an IZ diameter of 13.9 ± 0.3 (Table 2).| Bacteria | Measurements | Treatments | ||
|---|---|---|---|---|
| AgNO3 | N-SNPs | Ampicillin | ||
| a NA: not applicable. | ||||
| K. pneumoniae | IZ (mm) | 13.9 ± 0.3 | 15.33 ± 0.1 | NA |
| MIC (mg mL−1) | 1.2 | 0.9 | NA | |
| MBC (mg mL−1) | 1.5 | 1.2 | NA | |
| MIC/MBC | 0.8 | 0.8 | NA | |
Based on data from the serial dilution and agar well diffusion methods, K. pneumoniae was highly responsive to N-SNPs, with an MIC of 0.9 mg mL−1, compared to AgNO3, with an MIC of 1.2 mg mL−1. Additionally, the results indicated that the MBC value of N-SNPs was 1.2 mg mL−1, while the MBC value of AgNO3 was 1.5 mg mL−1 (Table 2). The results showed that N-SNPs had a stronger bactericidal effect than silver nitrate, with low MIC and MBC values against K. pneumoniae.
3.2. Ultrastructural changes caused by N-SNPs and silver nitrate
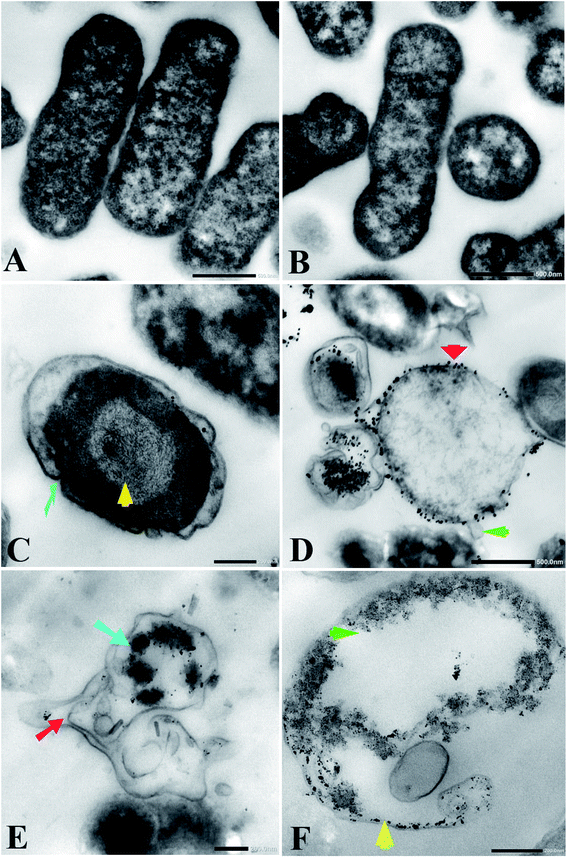
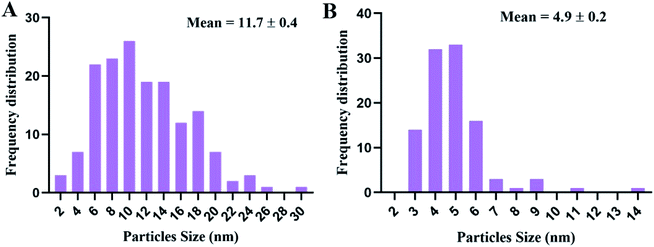
Transmission electron microscopy (TEM) examination of untreated K. pneumoniae showed that the cells retained their rod shape and exhibited intact morphology with an evenly distributed nucleoplasm (Fig. 3A and B). On the other hand, K. pneumoniae treated with silver nitrate exhibited an aberrant morphology, including shrinking cells, thin cell walls with pore and fold formation, and a decreased electron-dense cytoplasm with vacuole formation (Fig. 3C and D). Additionally, Fig. 3D shows the distribution of electron-dense nanosized particles ranging from 2 to 30 nm (Fig. 4A) in the cell wall and cytoplasm. TEM showed obvious ultrastructural changes in K. pneumoniae treated with N-SNPs, where deformation of the bacteria, shrinkage of cells and thinning of cell membranes were observed. In addition, complete separation of the plasma membrane from the bacterial cell wall and the appearance of atrophied cells were accompanied by folded and ruptured membranes. Moreover, agglutination of the nucleoplasm, cytoplasmic lashing, liberation of the cytoplasmic contents to the surrounding environment, vacuole formation and cell debris were observed (Fig. 3C and E). The cells seemed to undergo lysis, and the N-SNPs were concentrated inside the bacterial cells (Fig. 3F). The analysis data obtained from Image J software exhibited that the N-SNPs distributed inside the bacterial cells have a size range from 3 to 14 nm. This size range (after subjection) was found to be less than the original size range of the SNPs (8.5 to 26.44 nm) synthesized by Nostoc Bahar M before subjection to bacteria (Fig. 4B).3.3. Enzymes and antioxidants
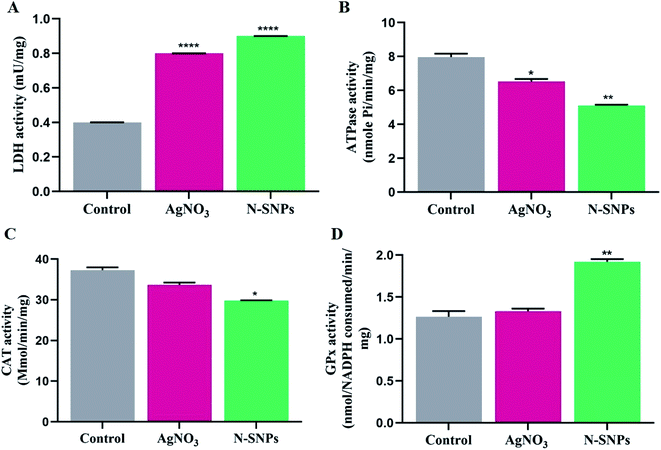
LDH activity was investigated to assess the lethal effects of N-SNPs and AgNO3 against K. pneumoniae. LDH activity increased after treatment with N-SNPs and silver nitrate (Fig. 5A). However, the most significant increase in LDH level was observed in K. pneumoniae treated with N-SNPs. The role of N-SNPs and AgNO3 in metabolic toxicity was assessed by measuring ATPase activity. As shown in Fig. 5B, N-SNPs were the most potent treatment, leading to significant inhibition of ATPase activity compared with silver nitrate treatment and no treatment. N-SNP and silver nitrate treatment resulted in a significant decrease in CAT levels (Fig. 5C). In contrast, the GPx level increased after exposure of the bacteria to N-SNPs and silver nitrate for 24 h (Fig. 5D). We noted that silver nitrate and biogenic N-SNPs have the same action towards the selected antioxidants and enzymes of the bacterial cells; however, N-SNPs were the most potent treatment and caused an imbalance in the activities of these biomolecules.3.4. Influence of N-SNPs and AgNO3 on gene expression
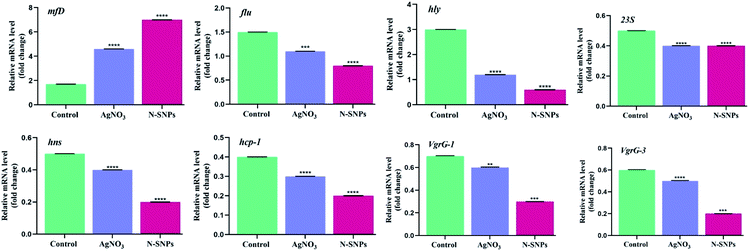
The changes in the expression of the mfD, hly, flu, 23S, hns, hcp-1, VgrG-1 and VgrG-3 genes were evaluated using qRT-PCR before and after subjecting K. pneumoniae to N-SNP and silver nitrate treatment for 24 h. Total RNA was extracted, cDNA was generated, quantitative real-time PCR was performed using specific primers, and GAPDH was used as a housekeeping gene control. The data showed that the exposure of K. pneumoniae to N-SNPs and silver nitrate caused significant upregulation of mfD gene expression and significant downregulation of hly, flu, 23S, hns, hcp-1, VgrG-1 and VgrG-3 mRNA expression (Fig. 6). However, the data showed that N-SNPs exhibited greater toxicity against the selected genes than silver nitrate.3.5. SDS-PAGE
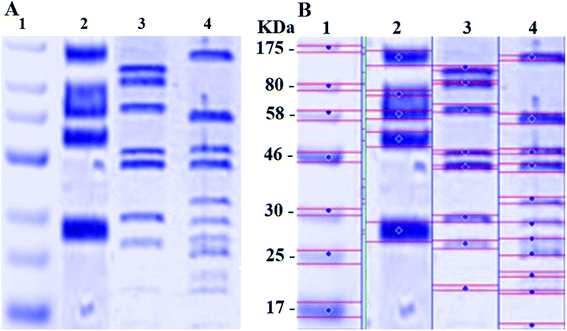
The SDS-PAGE result for the proteins extracted from K. pneumoniae before and after treatment with N-SNPs and silver nitrate is shown in Fig. 7A and B. Compared with the control, the gel pattern showed that the bands of the proteins of K. pneumoniae treated with N-SNPs were very thin, and new protein bands appeared. Additionally, in comparison with untreated bacteria, the high-molecular-weight protein bands were generally prevented from entering the electrophoresis gel. For K. pneumoniae treated with silver nitrate, high-molecular-weight bands and protein bands slightly heavier than those from the N-SNP treatment were observed. Additionally, 8 protein bands were observed after treatment with silver nitrate, while 11 bands were observed after N-SNP treatment, and 5 were observed in the control (Fig. 7B), indicating that the N-SNPs may have caused protein fragmentation and denaturation.4 Discussion
In comparison with silver nitrate and ampicillin, we have shown that N-SNPs have the strongest bactericidal activity against K. pneumoniae, and the largest IZ (15.33 ± 0.1) was obtained when using N-SNPs. A recent study reported that SNPs synthesized using tea leaf extracts had antibacterial activity against K. pneumoniae with an IZ diameter of 10 mm.30 The MICs and MBCs against bacteria of N-SNPs were found to be lower than those of silver nitrate. The data indicated that a lower concentration of N-SNPs than of AgNO3 could inhibit bacterial growth. This is similar to the data reported by Hamida et al., which showed that the MIC and MBC of SNPs and silver nitrate against bacteria at 104 CFU mL−1 were 1.2 and 1.5 mg mL−1, respectively.20 The slight variance in the MIC and MBC values of N-SNPs and silver nitrate reported in this study may be due to particle size, the nature of the material coating the NPs and the response of the bacteria to treatment.29,31TEM examination revealed that both silver nitrate and N-SNPs caused observable morphological changes and membrane disruption; however, N-SNP treatment resulted in the most acute ultrastructural damage. Additionally, N-SNPs were found to be concentrated inside the bacterial cells, and their size ranged between 3 and 14 nm. We suggested that N-SNPs were initially adsorbed to the surface of bacterial cell walls through electrostatic forces and then entered through the cell membrane to spread into the cytoplasm and interact with different biomolecules, such as antioxidants, proteins, DNA and enzymes, either directly or indirectly, leading to DNA degradation, protein denaturation, release of cellular contents and eventually cell death.32 El Din et al. reported that P. aeruginosa treated with SNPs showed membrane and genetic material disruption.33 The decreased size of the biogenic N-SNPs inside K. pneumoniae compared with their original size (8.5 to 26.44 nm) may be due to the aggregated NPs being trapped outside the cells.13,29 This indicates that the size of the NPs is an important factor for their activity.18 In addition to ultrastructural changes in K. pneumoniae treated with silver nitrate, electron-dense granules were found to be distributed both outside and inside cell membranes (Fig. 3B and C). These dark granules have nanosizes ranging from 2 to 30 nm. These data indicated that K. pneumoniae is possibly able to convert silver nitrate at a high concentration (10 mM AgNO3) to SNPs as a defence response against heavy metals. Based on these data, it is possible that these synthesized NPs by K. pneumoniae are the cause of the changes in K. pneumoniae morphology not the silver nitrate. Many studies have stated that K. pneumoniae has the ability to synthesize SNPs.34,35 Additionally, the result is consistent with the observations by Jung et al. about the existence of dark electron-dense particles or precipitates around damaged bacterial cells.19
To assess the influence of N-SNPs and silver nitrate on bacterial membrane, an LDH assay was employed, and the results showed that the level of LDH increased after 24 h of treatment of K. pneumoniae with N-SNPs compared with that observed after treatment with silver nitrate. The higher level of LDH resulting from N-SNP treatment than from silver nitrate treatment indicated the greater toxicity of N-SNPs against the bacteria than that of AgNO3. The results are in accordance with those of Gurunathan et al., who reported that SNPs synthesized using the biomolecule apigenin resulted in an increase in LDH levels in Prevotella melaninogenica and Arcanobacterium pyogenes.28
ATPase enzymes are important cellular enzymes that aid cells in obtaining energy to exert their biological functions.36 The current results showed that ATPase activity decreased after treatment of K. pneumoniae with the two sliver species, but N-SNPs caused a greater decrease in ATPase levels than silver nitrate. The decrease in ATPase levels could be an indicator of failure of cellular biological functions.37 Moreover, these data indicated that the biogenic SNPs may affect the metabolic activity of K. pneumoniae by interacting with enzymes. Recent publications have shown that gold NPs cause metabolic toxicity by interfering with the ATPase enzyme.38
One mode of action of SNPs against bacteria is to increase oxidative stress by influencing antioxidant activities.26 The antioxidant enzymes GPx and CAT are involved in scavenging ROS, and they are important enzymes that act as a barrier against oxidative stress.39 The current findings showed that N-SNPs were the most potent agents that caused an imbalance in GPx and CAT antioxidant activities, resulting in a decrease in the CAT level and an increase in the GPx level. The disruption in CAT and GPx activities reflects the ability of N-SNPs to induce ROS formation, leading to intense oxidative stress. Different studies explained that one mode of action of SNPs against bacteria is the induction of ROS formation, leading to the oxidation of different biomolecules, such as enzymes, non-enzyme proteins and nucleic acids, resulting in cell death.24,40 The action of silver nitrate against LDH, ATPase, GPx and CAT was similar to that of Nostoc-mediated SNPs. However, the potentiality of N-SNPs against these biomolecules was greater than that of silver ions, which may be due to the small size and large surface area, charge, and nature of the biocoat surrounding the SNPs.41
To better understand the effects of N-SNPs and silver nitrate against K. pneumoniae pathogenicity, we selected a number of genes associated with DNA damage, biofilm formation, protein synthesis and virulence activity, especially those related to invasion and the type VI secretion system. The data demonstrated that the two silver species caused an imbalance in the mRNA expression levels of the mfD, flu, hly, 23S, hns, hcp-1, VgrG-1 and VgrG-3 genes; however, N-SNPs were the most potent agents that caused a significant imbalance in gene expression levels. This may be due to the surface chemistry of the N-SNPs and their size and charge.41 Our data showed that N-SNPs caused a significant increase in the mfD mRNA expression level, indicating that N-SNPs may cause DNA damage through direct effects by attaching to nucleic acids and/or through indirect influences by causing oxidative stress that leads to oxidative lesions on DNA.42 A recent report showed that polymer-coated SNPs caused a significant increase in mfD mRNA expression levels in E. coli.43 Similarly, the significant decrease in flu and hly gene expression levels indicated that SNPs caused genotoxicity through direct and/or indirect interactions with these genes.42,44 Furthermore, cellular biological functions, such as the ability of bacteria to form biofilms and invade hosts, are disrupted as a result of direct interactions of N-SNPs with DNA molecules and/or proteins, including enzymes, or indirectly influence through increasing oxidative stress in bacterial cells.45 Recent publications showed that zinc oxide and silver nanoparticles caused a reduction in the mRNA expression level of the flu and hly genes, respectively, in E. coli.21,23 Meanwhile, the downregulation of 23S rRNA after treatment of K. pneumoniae with N-SNPs and AgNO3 may be due to the ability of N-SNPs and silver ions to reduce the translation ability, leading to disruption of the protein synthesis process. Liiv et al. reported that mutation in the 23S gene strongly suppresses protein translation.46 Liao et al. showed that SNPs caused a decrease in DNA, RNA and ribosomal protein levels in multidrug-resistant P. aeruginosa.47 Secretion system VI is a bacterial survival mechanism that allows bacteria to battle their hosts.48 Although silver nitrate and N-SNPs caused downregulation of the expression level of secretion system VI genes, N-SNPs were the most significant agents causing an imbalance in the expression of these genes, including hns, hcp-1, VgrG-1 and VgrG-3. The downregulation of these four genes, proving the relationship between N-SNPs and the virulence activity of K. pneumoniae. We hypothesized that the downregulation of these four genes observed after subjecting K. pneumoniae to N-SNPs may be due to a direct interaction between N-SNPs and the genes and/or due to indirect effects through the interaction of N-SNPs with proteins, including enzymes, related to the activation signals of these genes.45 This finding may provide an opportunity to use biogenic SNPs as significant anti-virulence agents to disrupt the interaction between MDR bacteria and their hosts.
The SDS-PAGE pattern of bacteria treated with N-SNPs showed that the band intensity of the proteins was very low, and new protein bands appeared. The number of protein bands resulting from treatment with N-SNPs was greater than that resulting from silver nitrate treatment and no treatment. These findings indicate that N-SNPs were able to cause more damage to protein structures than AgNO3. The higher toxicity of N-SNPs may be due to their physicochemical characteristics. These unique properties may aid in inducing intense oxidative stress, causing unfolding of the protein chain, which might cause cellular protein modification and degradation of these proteins.49,50 Moreover, the appearance of new protein bands observed after subjecting K. pneumoniae to N-SNPs may have been the result of a stress response and/or prevention of the synthesis of other proteins by inhibition of many translation factors.51,52
5 Conclusions
The effects of silver nitrate and novel SNPs synthesized by Nostoc sp. Bahar M on bacterial growth, morphology, antioxidants, enzymes, total protein content and genes related to DNA damage, biofilm formation and virulence were examined. It is possible to conclude that N-SNPs were more potent than silver nitrate, given the increased leakage of LDH and GPx activities and depletion of ATPase and CAT activities, resulting in induced oxidative stress and metabolic toxicity. Moreover, N-SNPs caused an imbalance in the gene expression of mfD, flu, hly and 23S. Additionally, this investigation demonstrated for the first time the relationship between biogenic N-SNPs and the genes of the type VI secretion system. N-SNPs significantly reduced hns, hcp-1, VgrG-1 and VgrG-3 gene expression. In addition, N-SNPs caused cellular protein degradation. Furthermore, insight into how novel N-SNPs impact bacterial biological functions such as metabolism, biofilm formation and virulence activity could provide key information on the mechanisms of action, resulting in new therapeutic methodologies for the development of better antibacterial agents. The present investigation demonstrates the possible cell-killing mechanism of N-SNPs against K. pneumoniae, including (a) direct influences of N-SNPs through interactions with cellular membranes and biomolecules such as proteins (including enzymes), antioxidants and DNA and (b) indirect influences through the induction of ROS formation and oxidative stress, causing oxidation of cellular biomolecules (Fig. 8). This work demonstrates that green N-SNPs are efficient antibacterial agents that may be alternatives to antibiotics. Other studies are needed to complete the study of the effects of N-SNPs against different bacteria and fungi and to target other genes to determine the specific mechanism of action of SNPs in microbial pathogenicity.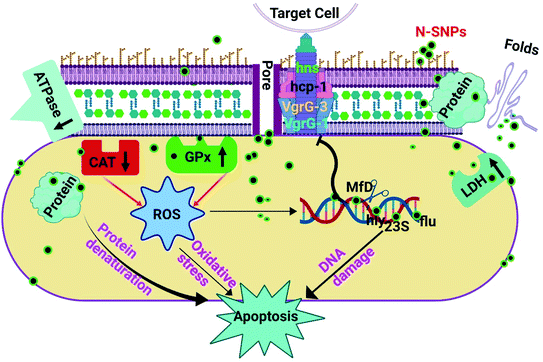 | ||
| Fig. 8 The potential mechanism of action of SNPs synthesized by Nostoc Bahar M sp. (N-SNPs) against K. pneumoniae. | ||
Ethics statement
The current study was performed in accordance with the Research Ethical Committee guidelines published by the National Health and Medical Research Council and the Ministry of Health and Population in Egypt. The Department of Microbiology, Medicine Faculty, Alexandria University, Alexandria, Egypt, granted permission for this work.Author contributions
RH, MA, MK and AR contributed conception and design of the study. RH, DG and MA downloaded and organized datasets. RH performed the statistical and result analysis and wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.Data availability
The data supporting this article are available in Fig. 1–8 and Tables 1 and 2. The data sets analysed in the present study are available from the corresponding author upon reasonable request.Funding
Not applicable.Abbreviations
| NPs | Nanoparticles |
| N-SNPs | Silver nanoparticles synthesized by Nostoc sp. Bahar M |
| SNPs | Silver nanoparticles |
| AgNO3 | Silver nitrate |
| PCR | Polymerase chain reaction |
| Nm | Nanometre |
| Min | Minute |
| μL | Microlitre |
| mg | Milligram |
| mL | Millilitre |
| μg | Microgram |
| mM | Millimolar |
| h | Hour |
| rpm | Revolutions per minute |
| TEM | Transmission electron microscopy |
| kV | Kilovolt |
| kb | Kilobase |
| IZ | Inhibition zone |
Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. This research was included in a request for patent 120410494 from King Abdulaziz City for Science and Technology, Saudi Arabia, submitted on 09/02/2020, titled “Green silver nanoparticles and their antibacterial activities” https://www.epatentsso.saip.gov.sa/.Acknowledgements
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.Notes and references
- J. O'Neill, Tackling Drug-resistant Infections Globally: Final Report and Recommendations–The Review on Antimicrobial Resistance Chaired by Jim O'Neill, Wellcome Trust and HM Government, London, 2016 Search PubMed.
- T. C. Dakal, A. Kumar, R. S. Majumdar and V. Yadav, Front. Microbiol., 2016, 7, 1831–1848 CrossRef PubMed.
- M. Frieri, K. Kumar and A. Boutin, J. Infect. Public Heal., 2017, 10, 369–378 CrossRef PubMed.
- J. M. Blair, M. A. Webber, A. J. Baylay, D. O. Ogbolu and L. J. Piddock, Nat. Rev. Microbiol., 2015, 13, 42–51 CrossRef CAS PubMed.
- B. Li, Y. Zhao, C. Liu, Z. Chen and D. Zhou, Future Microbiol., 2014, 9, 1071–1081 CrossRef CAS PubMed.
- C. N. Murphy and S. Clegg, Future Microbiol., 2012, 7, 991–1002 CrossRef CAS PubMed.
- A. Beceiro, M. Tomas and G. Bou, Clin. Microbiol. Rev., 2013, 26, 185–230 CrossRef CAS PubMed.
- T. Guillard, S. Pons, D. Roux, G. B. Pier and D. Skurnik, Bioessays, 2016, 38, 682–693 CrossRef PubMed.
- D. A. Rasko and V. Sperandio, Clin. Microbiol. Rev., 2010, 9, 117–135 CAS.
- M. K. Rai, S. D. Deshmukh, A. P. Ingle and A. K. Gade, J. Appl. Microbiol., 2012, 112, 841–852 CrossRef CAS PubMed.
- A. Gour and N. K. Jain, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 844–851 CrossRef CAS PubMed.
- H. Dong, Y. Gao, P. J. Sinko, Z. Wu, J. Xu and L. Jia, Nano Today, 2016, 11, 7–12 CrossRef CAS.
- M. M. Bin-Meferij and R. S. Hamida, Int. J. Nanomed., 2019, 14, 9019–9029 CrossRef PubMed.
- R. Rippka, J. Deruelles, J. B. Waterbury, M. Herdman and R. Y. Stanier, Microbiology, 1979, 111, 1–61 Search PubMed.
- R. Tyagi, B. Kaushik and J. Kumar, in Microbial Diversity and Biotechnology in Food Security, Springer, 2014, pp. 463–470 Search PubMed.
- S. Mukund and V. Sivasubramanian, Int. j. appl. biol. pharm. technol., 2014, 5, 34–45 Search PubMed.
- R. S. Hamida, N. E. Abdelmeguid, M. A. Ali, M. M. Bin-Meferij and M. I. Khalil, Int. J. Nanomed., 2020, 15, 49–63 CrossRef PubMed.
- A. Kedziora, M. Speruda, E. Krzyzewska, J. Rybka, A. Lukowiak and G. Bugla-Ploskonska, Int. J. Mol. Sci., 2018, 19, 444–461 CrossRef PubMed.
- W. K. Jung, H. C. Koo, K. W. Kim, S. Shin, S. H. Kim and Y. H. Park, Appl. Environ. Microbiol., 2008, 74, 2171–2179 CrossRef CAS PubMed.
- R. S. Hamida, M. A. Ali, D. A. Goda, M. I. Khalil and M. I. Al-Zaban, Front. Bioeng. Biotech., 2020, 8, 433 CrossRef.
- A. Tanhaeian and F. S. Ahmadi, Zahedan J. Res. Med. Sci., 2018, 20, 1–6 Search PubMed.
- A. E. Clatworthy, E. Pierson and D. T. Hung, Nat. Chem. Biol., 2007, 3, 541–549 CrossRef CAS PubMed.
- A. Shakerimoghaddam, E. A. Ghaemi and A. Jamalli, Iran. J. Basic Med. Sci., 2017, 20, 451–456 Search PubMed.
- S. Sutterlin, E. Tano, A. Bergsten, A. B. Tallberg and A. Melhus, Acta Derm.-Venereol., 2012, 92, 34–45 CrossRef CAS PubMed.
- M. Konop, T. Damps, A. Misicka and L. Rudnicka, J. Nanomater., 2016, 2016, 1–10 CrossRef.
- Y.-G. Yuan, Q.-L. Peng and S. Gurunathan, Int. J. Mol. Sci., 2017, 18, 569–591 CrossRef PubMed.
- M. T. Andrés and J. F. Fierro, Antimicrob. Agents Chemother., 2010, 54(10), 4335–4342 CrossRef PubMed.
- S. Gurunathan, Y. J. Choi and J. H. Kim, Int. J. Mol. Sci., 2018, 19, 1210–1230 CrossRef PubMed.
- D. G. Romero-Urbina, H. H. Lara, J. J. Velazquez-Salazar, M. J. Arellano-Jimenez, E. Larios, A. Srinivasan, J. L. Lopez-Ribot and M. J. Yacaman, Beilstein J. Nanotechnol., 2015, 6, 2396–2405 CrossRef CAS PubMed.
- Y. Y. Loo, Y. Rukayadi, M. A. Nor-Khaizura, C. H. Kuan, B. W. Chieng, M. Nishibuchi and S. Radu, Front. Microbiol., 2018, 9, 1555–1562 CrossRef PubMed.
- Y. He, S. Ingudam, S. Reed, A. Gehring, T. P. Strobaugh Jr and P. Irwin, J. Nanobiotechnol., 2016, 14, 54–63 CrossRef PubMed.
- C. Marambio-Jones and E. M. Hoek, J. Nanopart. Res., 2010, 12, 1531–1551 CrossRef CAS.
- S. N. El Din, T. A. El-Tayeb, K. Abou-Aisha and M. El-Azizi, Int. J. Nanomed., 2016, 11, 1749–1758 CAS.
- D. Kalpana and Y. S. Lee, Enzyme Microb. Technol., 2013, 52, 151–156 CrossRef CAS PubMed.
- A. Ratan, E. Gupta and R. Ragunathan, Synthesis, 2012, 1, 1–7 Search PubMed.
- M. Riley and M. Peters, Biochim. Biophys. Acta, 1981, 644, 251–256 CrossRef CAS.
- C.-N. Lok, C.-M. Ho, R. Chen, Q.-Y. He, W.-Y. Yu, H. Sun, P. K.-H. Tam, J.-F. Chiu and C.-M. Che, J. Proteome Res., 2006, 5, 916–924 CrossRef CAS PubMed.
- Y. Cui, Y. Zhao, Y. Tian, W. Zhang, X. Lü and X. Jiang, Biomaterials, 2012, 33, 2327–2333 CrossRef CAS PubMed.
- P. Chelikani, I. Fita and P. C. Loewen, Cell. Mol. Life Sci., 2004, 61, 192–208 CrossRef CAS PubMed.
- K. Muthukumar, S. Rajakumar, M. N. Sarkar and V. Nachiappan, Antonie Leeuwenhoek, 2011, 99, 761–771 CrossRef CAS PubMed.
- S. Tang and J. Zheng, Adv. Healthcare Mater., 2018, 7, 1701503–1701513 CrossRef PubMed.
- M. A. Radzig, V. A. Nadtochenko, O. A. Koksharova, J. Kiwi, V. A. Lipasova and I. A. Khmel, Colloids Surf., B, 2013, 102, 300–306 CrossRef CAS PubMed.
- D. A. Ashmore, A. Chaudhari, B. Barlow, B. Barlow, T. Harper, K. Vig, M. Miller, S. Singh, E. Nelson and S. Pillai, Rev. do Inst. Med. Trop. São Paulo, 2018, 60, 1–11 Search PubMed.
- S. Vishnupriya, K. Chaudhari, R. Jagannathan and T. Pradeep, Part. Part. Syst. Char., 2013, 30, 1056–1062 CrossRef CAS.
- S. Pal, Y. K. Tak and J. M. Song, Appl. Environ. Microbiol., 2007, 73, 1712–1720 CrossRef CAS PubMed.
- A. Liiv, D. Karitkina, U. Maivali and J. Remme, BMC Mol. Biol., 2005, 6, 18–27 CrossRef PubMed.
- S. Liao, Y. Zhang, X. Pan, F. Zhu, C. Jiang, Q. Liu, Z. Cheng, G. Dai, G. Wu and L. Wang, Int. J. Nanomed., 2019, 14, 1469–1487 CrossRef CAS PubMed.
- M. Gallique, M. Bouteiller and A. Merieau, Front. Microbiol., 2017, 8, 1454–1463 CrossRef PubMed.
- Y. Wang, S. D. Jett, J. Crum, K. S. Schanze, E. Y. Chi and D. G. Whitten, Langmuir, 2013, 29, 781–792 CrossRef CAS PubMed.
- H. Soliman, A. Elsayed and A. Dyaa, Egypt. j. basic appl. sci., 2018, 5, 228–233 CrossRef.
- S. K. Gogoi, P. Gopinath, A. Paul, A. Ramesh, S. S. Ghosh and A. Chattopadhyay, Langmuir, 2006, 22, 9322–9330 CrossRef CAS PubMed.
- P. Velusamy, G. V. Kumar, V. Jeyanthi, J. Das and R. Pachaiappan, Toxicol. Res., 2016, 32, 95–102 CrossRef CAS PubMed.
- A. Shakerimoghaddam, E. A. Ghaemi and A. Jamalli, Iran. J. Basic Med. Sci., 2017, 20, 451 Search PubMed.
- P.-F. Hsieh, Y.-R. Lu, T.-L. Lin, L.-Y. Lai and J.-T. Wang, J. Infect. Dis., 2019, 219, 637–647 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |