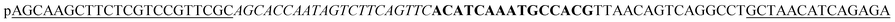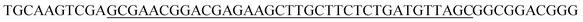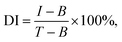Open Access Article
Open Access ArticleHighly sensitive detection of Staphylococcus aureus by a THz metamaterial biosensor based on gold nanoparticles and rolling circle amplification†
Ke Yang,
Wenjing Yu,
Guorong Huang,
Jie Zhou,
Xiang Yang * and
Weiling Fu
* and
Weiling Fu *
*
Department of Laboratory Medicine, Southwest Hospital, Third Military Medical University (Army Medical University), Chongqing 400038, China. E-mail: weiling_fu@126.com; yangxiang@tmmu.edu.cn
First published on 17th July 2020
Abstract
A highly sensitive method for detecting Staphylococcus aureus (S. aureus) is urgently needed to reduce the impact and spread of hospital-acquired infections and food-borne illness. For this purpose, this paper presents a THz metamaterial biosensor based on gold nanoparticles (AuNPs) and rolling circle amplification (RCA). The RCA process amplified the S. aureus DNA fragments and generated copious yields of long single-strand DNA molecules. These molecules were then conjugated with the AuNPs to form complexes that delivered exceptional increases in the refractive indices of the samples, and resulted in corresponding improvements in the THz response of the metamaterial. Under optimal conditions, the shifts in the metamaterial's resonance frequency displayed a linear relationship with concentrations of synthetic S. aureus DNA in the range from 10 fM to 10 pM, with a limit of detection of 2.77 fM. We also tested the practical application of this biosensor in measurements of genomic DNA in clinical bacterial strains, where the sensor showed a detection limit of 0.08 pg μL−1 and a linear range from 0.1 to 5 pg μL−1. It also exhibited reasonable specificity, resisting interference from three other pathogenic bacteria. These findings indicate that the proposed approach offers a cost-effective THz biosensing strategy that can be easily fabricated and conveniently operated to aid the diagnosis of infectious disease and food safety control.
Introduction
Staphylococcus aureus (S. aureus) is among the most common pathogenic bacteria in nosocomial infections and food-borne disease. These bacteria are usually present in open wounds or contaminated food, leading to numerous ailments, from minor skin infections to life-threatening diseases, such as food poisoning, abscesses, meningitis, and septicemia.1 S. aureus detection was conventionally accomplished by bacterial culturing, which is tedious and time-consuming.2 Although nucleic acid-based amplification technologies, including the polymerase chain reaction approach, provide more accurate diagnoses in less time, these strategies require thermostable DNA polymerases and precise control of thermal cycles.3 Matrix-assisted laser desorption ionization-time of flight mass spectrometry can directly detect isolated pure colonies after microbial culture. However, this approach suffers from a relatively poor limit of detection (LOD) of 105–106 colony-forming units (CFU).4 Nucleic acid sequencing also requires expensive instruments and trained personnel, and this method can therefore exceed the budgetary constraints of small clinical laboratories.3,5 Various biosensors have been implemented to make rapid and sensitive detection of S. aureus or other pathogenic bacteria, such as surface plasmon resonance (SPR),6 surface-enhanced Raman scattering (SERS),7 colorimetry,8 electrochemistry9 and terahertz (THz) spectroscopy.10 Most of these approaches had superior features in sample preparation, assay time, cost, portability and complexity, which contributed to satisfy the practical bacterial detection requirements. In this context, exploring biosensing techniques for S. aureus detection would be highly advantageous for various applications.THz spectroscopy has recently been exploited for bacterial detection owing to its rapid, label-free and noninvasive features. The frequency range of THz radiation (0.1–10 THz) corresponds to the low-frequency vibrational energy levels of the hydrogen bonds, van der Waals forces, and nonbonded interactions found in many biomolecules, and the THz range is thus suitable for probing changes in bacterial structure and metabolism.10 Furthermore, water molecules exhibit strong absorption of THz radiation, enabling different bacteria species to be distinguished by their distinct water contents.11,12 This technique has been applied in various bacterial investigations, such as component characterization,13 colony detection,12 spore identification14 and viability assessment.11,12 Moreover, our previous work demonstrated the quantitative THz sensing of bacterial DNA concentrations.10 In that work, rolling circle amplification (RCA) was used to generate many physical copies of the target bacterial DNA, which absorb less THz radiation than the highly THz-absorbing water molecules in a solution.10 As a result, the THz absorption coefficients of the amplified bacterial DNA samples decreased linearly with their increasing concentrations, achieving a sensitivity of 0.05 ng μL−1 for the genomic DNA detection of clinical bacterial strains.10 Nevertheless, this sensitivity requires further improvement to enable measurements of trace bacteria in practical samples.
THz metamaterials are artificially engineered structures composed of subwavelength metallic resonators, which exhibit unique electromagnetic properties that can be leveraged to enhance the THz detection sensitivity.15 Considering the metallic resonator as an inductive–capacitive circuit element, the resonant frequency of THz metamaterials can be expressed as 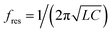 ,15,16 where L and C are the inductance and the capacitance, respectively. The inductance L is mainly related to the structural parameters of the fabricated metallic resonator, whereas the capacitance C is highly associated with the effective dielectric constant of the capacitor. A change in the dielectric constant caused by a foreign substance deposited on the metamaterial's surface can alter the capacitance, which will change the THz resonant frequency. A strong localized electric field and high quality-factor resonances can significantly improve metamaterial's sensitivity to small changes in the electromagnetic environment.17,18 Consequently, trace amounts of a sample deposited over the metamaterial's surface could therefore be probed by monitoring increases in the dielectric constant. Various biological substances, such as bacteria,17,18 cells,19 viruses,20 and biomolecules,21,22 have been detected using THz metamaterials. A maximum sensitivity improvement of 1010 times was reported in a kanamycin sulfate detection, compared with the measurement using THz spectroscopy in the free-space time-domain.22
,15,16 where L and C are the inductance and the capacitance, respectively. The inductance L is mainly related to the structural parameters of the fabricated metallic resonator, whereas the capacitance C is highly associated with the effective dielectric constant of the capacitor. A change in the dielectric constant caused by a foreign substance deposited on the metamaterial's surface can alter the capacitance, which will change the THz resonant frequency. A strong localized electric field and high quality-factor resonances can significantly improve metamaterial's sensitivity to small changes in the electromagnetic environment.17,18 Consequently, trace amounts of a sample deposited over the metamaterial's surface could therefore be probed by monitoring increases in the dielectric constant. Various biological substances, such as bacteria,17,18 cells,19 viruses,20 and biomolecules,21,22 have been detected using THz metamaterials. A maximum sensitivity improvement of 1010 times was reported in a kanamycin sulfate detection, compared with the measurement using THz spectroscopy in the free-space time-domain.22
The THz sensitivity is closely related to the metamaterial design, including the width of the gap area and the refractive index of the substrate.16,17 In particular, a nanometer-scale gap16 and substrates with low refractive indices17,21 exhibit better sensitivity. However, a precise nanometer-scale gap is difficult to fabricate by the conventional photolithography and the better substrates incur high material costs.23 A metamaterial that integrated silver nanowires etched into the network film was recently reported, and with the field enhancement effect at the nanowire tips, this device exhibited a virus LOD as low as approximately 107 CFU mL−1.23 In addition, frequency shifts in metamaterials have been shown to increase with the substrate's refractive index.16,24 This observation can be leveraged by facilitating binding between the detection target and particles with a high refractive index, such as gold nanoparticles (AuNPs), to produce a more robust THz response. This approach was used to improve the LOD of avidin samples by a thousand-fold.24 Thus, nanomaterials have exhibited tremendous potential to improve sensitivity, and offer the advantages of greater accessibility, simpler implementation, and lower cost compared to adjusting the metamaterial design.
Inspired by these previous works, the study presented in this paper developed a THz metamaterial biosensor based on signal enhancement by AuNPs and magnetic bead (MB)-based RCA for highly sensitive detection of S. aureus DNA. In this strategy, species-specific 16S rDNA were first amplified through MB-based RCA reactions and then conjugated with AuNPs to form complexes that significantly enhanced the THz signal. The results show that the AuNPs delivered an exceptional increase in the refractive indices of the samples, with an improved THz sensing capability beyond that achieved simply by RCA in our previous work. Based on this strategy, this THz metamaterial biosensor presented excellent detection sensitivity and specificity for the synthetic S. aureus DNA sequence. In addition, it was also tested in measurements of genomic DNA extracted from clinical bacterial strains, demonstrating the potential of the proposed material for practical applications in the diagnosis of infectious disease and food safety control.
Experimental section
1. Reagents and materials
Table 1 presents the oligonucleotide sequences used in this study. Reagent information is given in the ESI.†The underlined padlock probe (PLP) matches the underlined sequence in the target sequence. The italicized PLP matches the italicized sequence in the capture probe. The bold PLP can generate the complementary sequence in the RCA products, which matches the bold sequence in the AuNP probe. The “p” at the start of the padlock probe sequence represents the phosphate at the 5′ end for the ligation of PLP. A sequence that was not complementary to the PLP sequence was used as a negative control sequence. The capture probe was modified with biotin at the 5′ end to immobilize the streptavidin-coated MBs. The AuNP probe was modified with a thiol group at the 5′ end to immobilize the AuNPs.
2. MB-based RCA for bacterial DNA amplification
2 μL of different concentrations of the target sequence and 1 μL of PLP (100 nM) were added to 5.7 μL of DNA ligase buffer. The solution was denatured at 95 °C for 5 min, cooled to 4 °C, and then incubated at 60 °C for 30 min. After adding 1 μL of 0.05% bovine serum albumin and 0.3 μL of DNA ligase (500 U), the mixture was kept at 37 °C for 45 min to form the circular PLP. In the next step, 1 μL of exonuclease I (400 U), 1 μL of exonuclease III (400 U), and 8 μL of exonuclease buffer were added to the ligation mixture to remove the excess target sequences and PLPs that had not been ligated. The ligation mixture was then incubated at 37 °C for 30 min, and terminated by heating at 95 °C for 5 min. After the ligation process, 2 μL of capture probe-modified MBs (CP-MBs, 1 μM, prepared as described in the ESI†) and 10 μL of Phi29 DNA polymerase buffer were added to 40 μL of the ligation mixture to hybridize the CP-MBs and circular PLP at 37 °C for 30 min. Then, 2 μL of Phi29 DNA polymerase (300 U) and 6 μL of deoxyribonucleosides (100 mM) were added to the mixture and incubated at 37 °C for 60 min for the RCA reaction. Finally, the MBs with the RCA products (RCA–MBs) were washed twice and suspended in 60 μL of deionized water.3. Conjugation of RCA products with AuNPs
First, AuNPs with an average diameter of 60 nm were modified with the AuNP probe presented in Table 1 to form the nano probes according to the protocol listed in the ESI.† 100 μL of the nano probe (1 nM) was added to 50 μL of the RCA–MBs solution. The mixture was incubated at 37 °C for 30 min to conjugate the AuNPs with the RCA products and thereby form the AuNP–RCA–MBs. Excess nano probes were removed by magnetic separation for 5 min. The AuNP–RCA product complexes were separated from the MBs by adding 50 μL of de-hybridization solution and heating the mixture at 90 °C for 10 min. After removing all MBs, the solution containing the AuNP–RCA product complexes was used in the subsequent THz metamaterial measurements.4. Bacterial genomic DNA extraction
Four clinical bacterial strains—S. aureus, Escherichia coli (E. coli), Acinetobacter baumannii (A. baumannii), and Pseudomonas aeruginosa (P. aeruginosa)—were obtained from the Department of Laboratory Medicine, Southwest Hospital (Chongqing, China). The strains were inoculated on blood agar plates overnight at 37 °C in an atmosphere containing 5% CO2. Following incubation, single colonies were lifted off the plates and suspended in sterilized saline water to an optical density of 1.0 at 600 nm. A TaKaRa Mini BEST Bacteria Genomic DNA Extraction Kit was used to extract the bacterial genomic DNA, which were then fragmented by digestion using QuickCut BamHI, EcoRI, and HindIII in 50 μL of reaction mixture at 37 °C for 10 min. Finally, the genomic DNA were diluted to the desired concentration with deionized water. An approximate range from 3–8 μg of genomic DNA can be extracted from 2.0 × 109 of S. aureus cells according to the product manual.5. Fabrication of THz metamaterial
THz metamaterials composed of periodically arranged golden split ring resonators were fabricated on a silicon substrate by conventional photolithography. The manufacturing process is described in the ESI, which also presents a microscopic image of the THz metamaterial pattern (Fig. S1).†6. THz metamaterial measurements
THz metamaterial measurements were conducted with a commercial THz time-domain spectroscopy system in transmission mode (TAS7500SP, Advantest Co., Tokyo, Japan), according to the following protocol. First, a high-resistivity silicon substrate was measured to obtain a reference to which subsequent sample measurements could be compared. Second, the THz signal transmitted from a bare metamaterial was measured, before the deposition of 10 μL of the solution containing the AuNP–RCA product complexes. Third, the metamaterials coated with the complexes was completely dried on a hotplate at 50 °C for 10 min, and the THz signals from the coated metamaterial were measured. After each measurement, the coated metamaterials were rinsed thoroughly with distilled water followed by nitrogen blow drying for the next sample loading. Every sample was measured three times. By calculating the differences in the resonance peaks of the transmission spectra from the coated and bare metamaterials, the metamaterial frequency shifts (Δf) of the various samples could be compared. The blank control was obtained according to the same aforementioned protocol but replacing the bacterial DNA with deionized water.Results and discussion
1. Principle of THz metamaterial biosensor for bacterial DNA detection
As illustrated in Fig. 1, the THz metamaterial's detection process as a biosensor includes the following three steps. (i) MB-based RCA procedures for bacterial DNA amplification. The bacterial DNA sequence was initially hybridized with the binding region of a linear PLP and DNA ligase was added to form a complete circle with the hybridized DNA. Circular PLPs were then captured by capture probes on MBs via their complementary sequences. With the DNA polymerase and deoxyribonucleosides, the RCA reaction was triggered on the surface of MBs, generating multiple long single-strand DNA containing repetitive secondary target sequences, hereafter referred to as the RCA products. (ii) Conjugation of RCA products with AuNPs. AuNPs were first modified with the AuNP probe to form nano probes according to the aforementioned protocol. The nano probes were then hybridized with the complementary parts of the RCA products, assembling AuNP–RCA product complexes. (iii) THz metamaterial measurement. After magnetic separation and de-hybridization, the pure AuNP–RCA product complexes were separated from the MBs and measured using THz metamaterials. Because the bacterial DNA concentration is highly correlated with the number of complexes present, the concentrations can be determined by calculating the relative changes in the frequency shifts (Δf) of the coated metamaterial.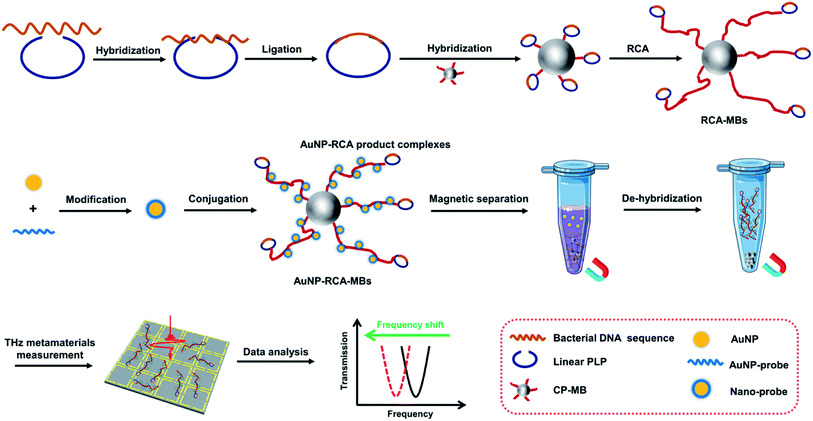 | ||
| Fig. 1 Schematic of the fabrication and operating principle of the proposed THz metamaterial biosensor for bacterial DNA detection. | ||
2. Feasibility of THz metamaterial biosensing strategy
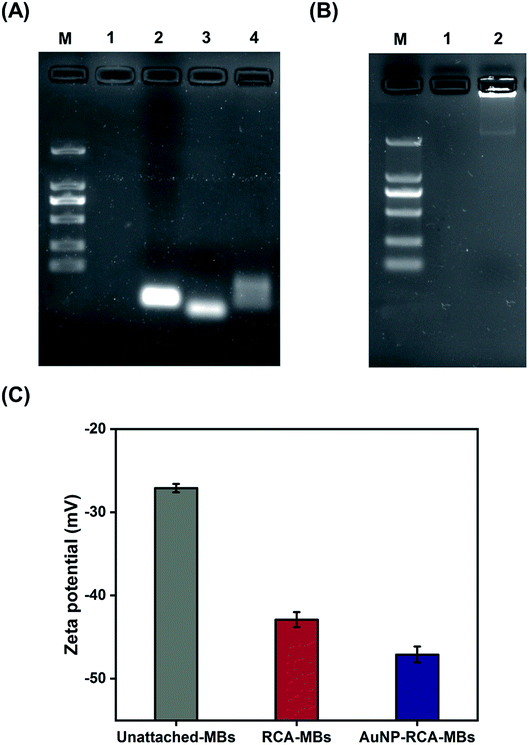
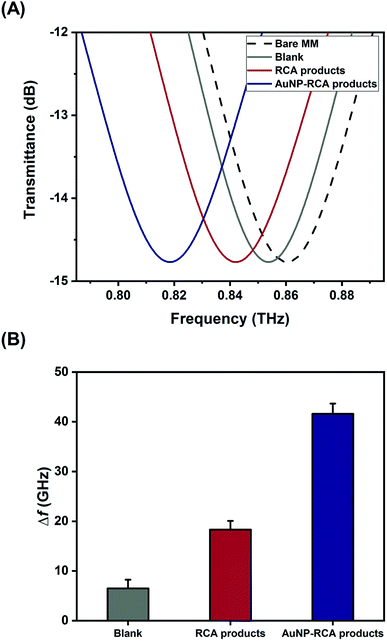
Fig. 2A and B show the gel electrophoresis results of S. aureus DNA ligation products and RCA products, respectively. The circular PLPs migrated at slower rates than the linear PLPs and the S. aureus DNA sequences, indicating effective hybridization and ligation of bacterial DNA to linear PLPs. In addition, Fig. 2B shows that the RCA products were too large to enter the gel, and this result confirmed that the generated RCA products were long single-strand DNA molecules with high molecular masses. To further validate that the RCA occurred on the MB surface, zeta potential measurements were taken to characterize the RCA–MBs and AuNP–RCA–MBs. As shown in Fig. 2C, RCA–MBs (−42.90 ± 0.90 mV) exhibited a more negative zeta potential than unattached-MBs (−27.10 ± 0.51 mV). These results suggested that MBs were loaded with negatively charged molecules such as RCA products. With the introduction of the AuNPs, the zeta potential further decreased (−47.10 ± 0.96 mV), suggesting that the conjugation of the AuNPs with the RCA products enhanced the negative charge. These results are consistent with a previous zeta potential measurement from the amplified DNA of Leishmania, which also showed a more negative zeta potential after labeling with AuNPs.25 Therefore, these results demonstrate the immobilization of AuNP–RCA product complexes on the MBs.Fig. 3A displays the THz transmission spectra of the RCA products and AuNP–RCA product complexes. The resonant frequency of the bare metamaterial was found to be approximately 0.86 THz. After the RCA process and AuNP conjugation, the resonant frequency shifted towards a lower frequency. By calculating the relative changes in the resonance peaks of the transmission spectra between the sample coated metamaterials and the bare metamaterials, the sample's frequency shifts (Δf) were obtained. As shown in Fig. 3B, the Δf of the AuNP–RCA product complexes (41.58 ± 2.07 GHz) was much larger than that of the pure RCA products (18.31 ± 1.75 GHz). This finding was consistent with a previous detection of avidin using a THz metamaterial, which also showed an increased Δf after conjugation with AuNPs.24 The signal enhancement was mainly attributed to the higher refractive index resulting from the AuNP conjugation because the Δf of metamaterials has been demonstrated to be proportional to the refractive indices of the samples.24 Under most circumstances, the refractive index of the metal far outweighs that of an organic compound, such as a DNA molecule.24,26 Therefore, the detection sensitivity of THz metamaterials can be improved by introducing AuNPs. Compared to that of the blank control, the pure RCA products generated a larger Δf, which could be explained by the more copious yields of amplified DNA molecules. This result verified the complementary THz response improvement conferred by the RCA process.
3. Optimization of experimental conditions
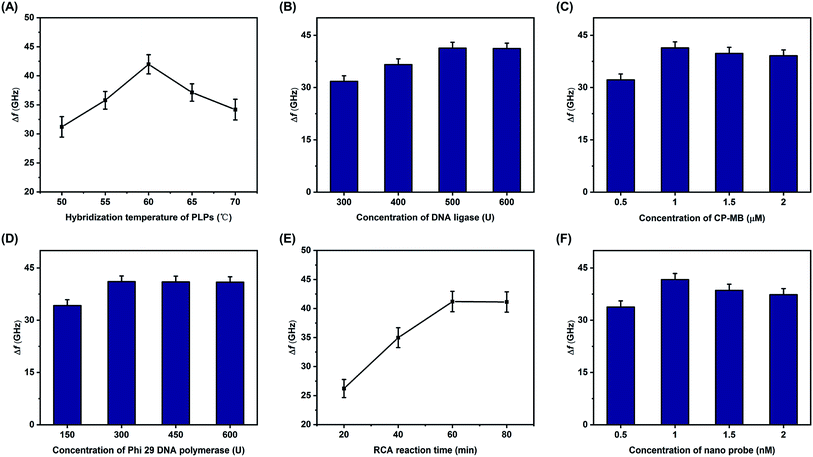
Several experimental conditions were optimized, including the hybridization temperature of the PLPs, the concentration of the DNA ligase, the concentration of the CP-MB, the concentration of Phi 29 DNA polymerase, the RCA reaction time, and the concentration of nano probe. One pM of S. aureus DNA sequences was used in all of the optimization studies, and all of the optimal concentrations are reported in terms of the initial reagent concentrations. As shown in Fig. 4A, the Δf increased with increases in the PLP hybridization temperature, with the maximum Δf observed at 60 °C. Therefore, 60 °C was selected as the optimal hybridization temperature. The DNA ligase concentration was also considered to be essential for the RCA reaction, and as displayed in Fig. 4B, the Δf increased with increasing concentrations of DNA ligase until the maximum at 500 U, remaining relatively stable thereafter. Therefore, 500 U was chosen as the optimal DNA ligase concentration. Using the same method, the effects of the other key conditions on Δf were also investigated, as detailed in Fig. 4C through 4F. The optimal conditions were as follows: the optimal concentrations of the CP-MB, nano probe, and Phi 29 DNA polymerase were 1 μM, 1 nM, and 300 U, respectively; the optimal RCA reaction time was 60 min.4. THz metamaterial biosensor for synthetic S. aureus DNA detection
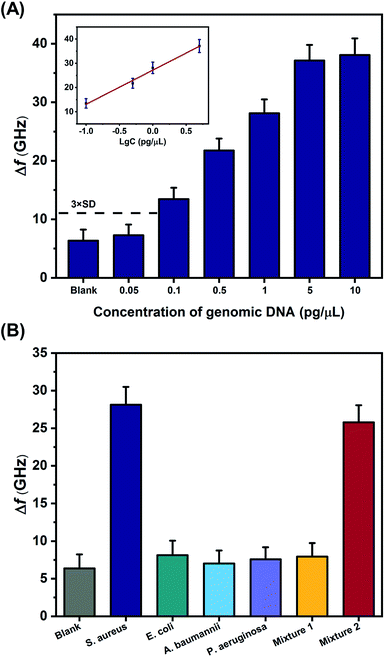
Under the optimal experimental conditions, different concentrations of synthetic S. aureus DNA were analyzed with the THz metamaterial biosensor. 100 nM of PLPs were used in the S. aureus DNA detection. As illustrated in Fig. 5A, the Δf increased with increasing concentrations of S. aureus DNA from 1 fM to 100 pM. The inset in Fig. 5A shows the clear linear correlation between the Δf and the logarithm of the S. aureus DNA concentration over the range from 10 fM to 10 pM, with a correlation coefficient (R2) of 0.9878. The linear regression equation was expressed as Δf = 10.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c + 168.46. According to this equation, the LOD was determined to be 2.77 fM, which was calculated from the average Δf of the blank control plus three times the standard deviation. Compare to our previous work on E. coli DNA detection, which showed a LOD of 60 pM,10 the sensitivity of the THz metamaterial biosensor presented here achieved an approximately 104-fold improvement. This significant enhancement was attributed mainly to the use of the AuNPs in complexes with the target DNA. The introduced AuNPs enhance the THz responses of the samples on the metamaterials because of the high refractive index of the metal. Several other strategies for improving the sensitivity of THz detection have also been developed, such as the utilization of metallic mesh structures27 and plasmonic antennae.28 Compared to those methods, the AuNP-enhanced metamaterials presented here have significant advantages of easy accessibility, simple implementation, and low cost, and these conditions favor the development of THz spectroscopy into a practical tool for quantifying bacterial DNA.
c + 168.46. According to this equation, the LOD was determined to be 2.77 fM, which was calculated from the average Δf of the blank control plus three times the standard deviation. Compare to our previous work on E. coli DNA detection, which showed a LOD of 60 pM,10 the sensitivity of the THz metamaterial biosensor presented here achieved an approximately 104-fold improvement. This significant enhancement was attributed mainly to the use of the AuNPs in complexes with the target DNA. The introduced AuNPs enhance the THz responses of the samples on the metamaterials because of the high refractive index of the metal. Several other strategies for improving the sensitivity of THz detection have also been developed, such as the utilization of metallic mesh structures27 and plasmonic antennae.28 Compared to those methods, the AuNP-enhanced metamaterials presented here have significant advantages of easy accessibility, simple implementation, and low cost, and these conditions favor the development of THz spectroscopy into a practical tool for quantifying bacterial DNA.
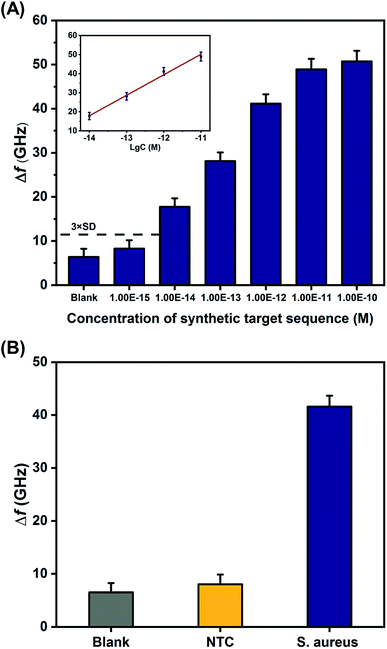 | ||
| Fig. 5 Detection of synthetic S. aureus DNA by the proposed THz metamaterial biosensor. (A) Evaluation of the sensitivity of synthetic S. aureus DNA detection, based on metamaterial frequency shifts (Δf) resulting from the variation of DNA concentrations ranging from 1 fM to 100 pM. The inset shows the linear relationship between Δf and the logarithm of S. aureus DNA concentration. (B) Evaluation of the specificity of synthetic S. aureus DNA detection. A sequence that was not complementary to the PLP (shown in Table 1) served as the negative control (NTC). Error bars indicate the standard deviation (n = 3 in all cases). | ||
To investigate the specificity of synthetic S. aureus DNA detection with the proposed metamaterial biosensor, the frequency shifts observed from samples containing the S. aureus DNA were compared with samples containing a sequence that was not complementary to the PLP (negative control, NTC) and samples containing deionized water (blank control). The concentrations of S. aureus DNA and NTC were both 1 pM. As shown in Fig. 5B, S. aureus DNA generated a larger Δf than either the NTC or the blank control, indicating that the THz metamaterial biosensor could specifically discriminate synthetic S. aureus DNA. In addition, the reproducibility of the results from this biosensor was also evaluated from the relative standard deviation obtained from five replicate measurements of the 1 pM of synthetic S. aureus DNA. The relative standard deviation was calculated to be 3.22%, suggesting acceptable reproducibility.
5. THz metamaterial biosensor for S. aureus genomic DNA detection
The proposed THz metamaterial biosensor was tested in a practical application by measuring the genomic DNA extracted from clinical bacterial strains, using 100 nM of PLPs. As in the tests on the synthetic S. aureus DNA, the Δf of the genomic DNA samples displayed a linear relationship with the logarithm of the DNA concentration over the range of 0.1 to 5 pg μL−1 (Δf = 14.08![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c + 27.17, R2 = 0.9856) (Fig. 6A). The LOD for the genomic DNA was calculated to be 0.08 pg μL−1 (2–5.3 × 104 bacterial cell per ml according to the genomic DNA extraction protocol), which is less than or equal to the detection limits of other reported bacterial DNA detection methods, such as SPR, colorimetric, and electrochemical biosensors, as shown in Table 2. These results suggested that the THz metamaterial biosensor offers significant potential for detecting bacterial DNA in clinical assays.
c + 27.17, R2 = 0.9856) (Fig. 6A). The LOD for the genomic DNA was calculated to be 0.08 pg μL−1 (2–5.3 × 104 bacterial cell per ml according to the genomic DNA extraction protocol), which is less than or equal to the detection limits of other reported bacterial DNA detection methods, such as SPR, colorimetric, and electrochemical biosensors, as shown in Table 2. These results suggested that the THz metamaterial biosensor offers significant potential for detecting bacterial DNA in clinical assays.
| Detection methods | Target species | Linear range | Detection limit | Reference |
|---|---|---|---|---|
| SPR | S. aureus | 1–5000 pg μL−1 | 0.5 pg μL−1 | 6 |
| SPR | M. tuberculosis | 104–108 CFU mL−1 | 5 pg μL−1 | 29 |
| SPR | M. avium | 104–108 CFU mL−1 | 2 pg μL−1 | 29 |
| Colorimetry | P. aeruginosa | 10–50 pg μL−1 | 9.899 pg μL−1 | 8 |
| Colorimetry | E. coli | — | 0.03 pg μL−1 | 30 |
| Electrochemistry | M. tuberculosis | 10–104 pg μL−1 | 10 pg μL−1 | 9 |
| Electrochemistry | M. tuberculosis | — | 40 pg μL−1 | 31 |
| Electrochemistry | V. cholerae | 100–500 ng μL−1 | 31.5 ng μL−1 | 32 |
| Electrochemistry | E. coli | 1–1000 pg μL−1 | 1 pg μL−1 | 33 |
| Electrochemistry | S. pneumoniae | 0–8 pg μL−1 | 0.218 pg μL−1 | 34 |
| THz metamaterials | S. aureus | 0.1–5 pg μL−1 | 0.08 pg μL−1 | This work |
To confirm whether this biosensor could specifically differentiate the genomic DNA of S. aureus, THz signals from the target bacteria (S. aureus) and three interfering pathogenic bacteria (E. coli, A. baumannii, and P. aeruginosa) were compared. In addition, signals from a mixture of the three interfering pathogenic bacteria (Mixture 1) and a mixture of the target bacteria and three interfering pathogenic bacteria (Mixture 2) were also measured. The concentrations of all bacterial samples were 1 pg μL−1. As shown in Fig. 6B, the Δf of S. aureus was significantly higher than those of the three interfering pathogenic bacteria and that of Mixture 1. Moreover, the Δf of Mixture 2 was almost the same as that of S. aureus, indicating that the THz signal of Mixture 2 mostly came from S. aureus. These results suggested that the THz metamaterial biosensor could specifically discriminate S. aureus from the mixture samples. Because numerous interfering signals are often present in clinical samples, we also calculated the degree of interference (DI) of bacterial samples using the following equation:
Compared to conventional bacterial identification methods, the proposed THz metamaterial sensing platform, complete with AuNPs preparation and RCA processing, can be easily fabricated and operated, and the biosensor is thus cost-effective even for small clinical laboratories in resource-limited settings. Unlike the polymerase chain reaction approach, which requires that samples be tagged with fluorescent probes or staining reagents, the THz biosensor enables nondestructive and environmentally safe measurements, avoiding biological hazards and carcinogenic risks from chemicals used in other approaches. In the future, the specific capture of bacterial DNA and the RCA process could be simultaneously conducted by directly modifying the probes to the metamaterial's surface. Integrating the proposed biosensor with microfluidic technology would also enable its use for high-throughput detection of pathogenic bacteria.
Conclusion
In summary, a THz metamaterial biosensor was developed for the highly sensitive detection of S. aureus based on signal enhancement by RCA and AuNPs. With the cyclic RCA process, trace S. aureus DNA can be used to generate many long single-strand DNA, which are then hybridized with AuNPs to form AuNP–RCA product complexes. These complexes produced remarkable shifts in the resonance frequency of the THz metamaterials because of the considerable variation in the refractive index induced by the conjugation with AuNPs. These dual signal enhancement strategies resulted in a THz metamaterial biosensor that exhibited favorable capability for both synthetic and clinical S. aureus DNA detection. In measurements of clinical bacterial strains of S. aureus, the biosensor showed an excellent detection sensitivity with a detection limit of 0.08 pg μL−1 and a linear range from 0.1 to 5 pg μL−1. The biosensor also exhibited excellent specificity, resisting interferences from three other pathogenic bacteria. These findings demonstrate that the proposed THz biosensor is capable of highly sensitive S. aureus detection, and is simple to fabricate and easy to operate, all of which are advantages in practical applications.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (81920108024, 81802118), the Military logistics scientific research project (AWS17J010), the Joint Medical Scientific Research Project of Chongqing (2018QNXM007) and the Southwest Hospital Research Project (SWH2019BJKJ-07).References
- G. A. R. Y. Suaifan, S. Alhogail and M. Zourob, Biosens. Bioelectron., 2017, 90, 230–237 CrossRef CAS PubMed.
- O. Opota, A. Croxatto, G. Prod'hom and G. Greub, Clin. Microbiol. Infect., 2015, 21, 313–322 CrossRef CAS PubMed.
- S. H. Lee, S.-m. Park, B. N. Kim, O. S. Kwon, W.-Y. Rho and B. H. Jun, Biosens. Bioelectron., 2019, 141, 111448–111465 CrossRef CAS PubMed.
- M. Narayanan, Arch. Dis. Child., 2019, 104, 513–517 CrossRef PubMed.
- S. Ardui, A. Ameur, J. R. Vermeesch and M. S. Hestand, Nucleic Acids Res., 2018, 46, 2159–2168 CrossRef CAS PubMed.
- D. Shi, J. Huang, Z. Chuai, D. Chen, X. Zhu, H. Wang, J. Peng, H. Wu, Q. Huang and W. Fu, Biosens. Bioelectron., 2014, 62, 280–287 CrossRef CAS PubMed.
- X. Chen, M. Tang, Y. Liu, J. Huang, Z. Liu, H. Tian, Y. Zheng, M. L. de la Chapelle, Y. Zhang and W. Fu, Microchim. Acta, 2019, 186, 102–109 CrossRef PubMed.
- B. Amini, M. Kamali, M. Salouti and P. Yaghmaei, Spectrochim. Acta, Part A, 2018, 199, 421–429 CrossRef CAS PubMed.
- E. Torres-Chavolla and E. C. Alocilja, Biosens. Bioelectron., 2011, 26, 4614–4618 CrossRef CAS PubMed.
- X. Yang, K. Yang, X. Zhao, Z. Lin, Z. Liu, S. Luo, Y. Zhang, Y. Wang and W. Fu, Analyst, 2017, 142, 4661–4669 RSC.
- Y. Xiang, W. Dongshan, Y. Shihan, L. Yueping, Y. Shu, Z. Mingkun, Y. Zhongbo, Z. Xiaoyan, H. Qing, C. Hong-Liang and F. Weiling, J. Biophotonics, 2016, 9, 1050–1058 CrossRef PubMed.
- X. Yang, J. Shi, Y. Wang, K. Yang, X. Zhao, G. Wang, D. Xu, Y. Wang, J. Yao and W. Fu, J. Biophotonics, 2018, 11, e201700386 CrossRef PubMed.
- C. Wang, J. Gong, Q. Xing, Y. Li, F. Liu, X. Zhao, L. Chai, C. Wang and A. M. Zheltikov, J. Biophotonics, 2010, 3, 641–645 CrossRef CAS PubMed.
- W. Zhang, E. R. Brown, L. Viveros, K. P. Burris and C. N. Stewart Jr, J. Biophotonics, 2014, 7, 818–824 CrossRef CAS PubMed.
- S. RoyChoudhury, V. Rawat, A. H. Jalal, S. N. Kale and S. Bhansali, Biosens. Bioelectron., 2016, 86, 595–608 CrossRef CAS PubMed.
- W. Xu, L. Xie and Y. Ying, Nanoscale, 2017, 9, 13864–13878 RSC.
- S. J. Park, B. H. Son, S. J. Choi, H. S. Kim and Y. H. Ahn, Opt. Express, 2014, 22, 30467–30472 CrossRef CAS PubMed.
- S. J. Park, J. T. Hong, S. J. Choi, H. S. Kim, W. K. Park, S. T. Han, J. Y. Park, S. Lee, D. S. Kim and Y. H. Ahn, Sci. Rep., 2014, 4, 4988 CrossRef CAS PubMed.
- C. Zhang, L. Liang, L. Ding, B. Jin, Y. Hou, C. Li, L. Jiang, W. Liu, W. Hu and Y. Lu, Appl. Phys. Lett., 2016, 108, 241105 CrossRef.
- S. J. Park, S. H. Cha, G. A. Shin and Y. H. Ahn, Biomed. Opt. Express, 2017, 8, 3551–3558 CrossRef CAS PubMed.
- D.-K. Lee, J.-H. Kang, J.-S. Lee, H.-S. Kim, C. Kim, J. H. Kim, T. Lee, J.-H. Son, Q. H. Park and M. Seo, Sci. Rep., 2015, 5, 15459 CrossRef CAS PubMed.
- L. Xie, W. Gao, J. Shu, Y. Ying and J. Kono, Sci. Rep., 2015, 5, 8671 CrossRef PubMed.
- J. T. Hong, S. W. Jun, S. H. Cha, J. Y. Park, S. Lee, G. A. Shin and Y. H. Ahn, Sci. Rep., 2018, 8, 15536 CrossRef CAS PubMed.
- W. Xu, L. Xie, J. Zhu, X. Xu, Z. Ye, C. Wang, Y. Ma and Y. Ying, ACS Photonics, 2016, 3, 2308–2314 CrossRef CAS.
- A. D. L. Escosura-Muñiz, L. Baptista-Pires, L. Serrano, L. Altet and A. Merkoçi, Small, 2015, 12, 205–213 CrossRef PubMed.
- K. Zeng, H. Li and Y. Peng, Microchim. Acta, 2017, 184, 2637–2644 CrossRef CAS.
- T. Hasebe, S. Kawabe, H. Matsui and H. Tabata, J. Appl. Phys., 2012, 112, 094702 CrossRef.
- L. Masini, S. Meucci, J. Xu, R. Degl'Innocenti, F. Castellano, H. E. Beere, D. Ritchie, D. Balduzzi, R. Puglisi and A. Galli, Laser Photonics Rev., 2015, 8, 734–742 CrossRef.
- Y. Xiang, X. Zhu, Q. Huang, J. Zheng and W. Fu, Biosens. Bioelectron., 2015, 66, 512–519 CrossRef CAS PubMed.
- A. Sayad, F. Ibrahim, S. M. Uddin, J. Cho, M. Madou and K. L. Thong, Biosens. Bioelectron., 2018, 100, 96–104 CrossRef CAS PubMed.
- M.-N. Tsaloglou, A. Nemiroski, G. Camci-Unal, D. C. Christodouleas, L. P. Murray, J. T. Connelly and G. M. Whitesides, Anal. Biochem., 2018, 543, 116–121 CrossRef CAS PubMed.
- M. K. Patel, J. Singh, M. K. Singh, V. V. Agrawal, S. G. Ansari and B. D. Malhotra, J. Nanosci. Nanotechnol., 2013, 13, 1671–1678 CrossRef CAS PubMed.
- D. Jiang, F. Liu, C. Liu, L. Liu and X. Pu, Int. J. Electrochem. Sci., 2013, 8, 9390–9398 CAS.
- J. Wang, M. C. Leong, E. Z. W. Leong, W. S. Kuan and D. T. Leong, Anal. Chem., 2017, 89, 6900–6906 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methodology, as well as an image of the THz metamaterial (Fig. S1). See DOI: 10.1039/d0ra03116j |
| This journal is © The Royal Society of Chemistry 2020 |