Open Access Article
Open Access ArticleHigh-sensitivity SERS based sensing on the labeling side of glass slides using low branched gold nanoparticles prepared with surfactant-free synthesis†
Tuğba
Tezcan
 a and
Chia-Hsien
Hsu
a and
Chia-Hsien
Hsu
 *abc
*abc
aInstitutes of Biomedical Engineering and Nanomedicine, National Health Research Institutes, Zhunan, Taiwan. E-mail: ttezcan@nhri.edu.tw
bInstitute of Nano Engineering and MicroSystems, National Tsing Hua University, Hsinchu, Taiwan. E-mail: chsu@nhri.org.tw
cPh.D. Program in Tissue Engineering and Regenerative Medicine, National Chung Hsing University, Taichung, Taiwan
First published on 18th September 2020
Abstract
Surface-enhanced Raman scattering (SERS) has become a more attractive tool for biological and chemical sensing due to having a great detection potential to extremely low concentrations of analyte. Here, we report high-sensitivity SERS detection of low branched gold nanoparticles which are produced by a surfactant-free synthesis method. The effects of the size and branches of nanoparticles on the SERS signal intensity were also investigated. Among the prepared nanoparticles, a new type of nanoparticle with small protrusions produced by using a very low concentration of silver ions (2 μM in final solution) achieved the best enhancement factor of ∼4 × 105 for DTNB used as a probe molecule. SERS measurements were performed on the labeling side of microscope glass slides for the first time. The substrate exhibited a good reproducible SERS signal with a relative standard deviation (RSD) of 1.7%. SERS signal intensity obtained using the labelling side was three times larger compared to that obtained using bare glass. To validate the sensing platform, dopamine, an important modulatory neurotransmitter in the brain, was tested. The reported platform was able to achieve label-free detection of dopamine at picomolar and nanomolar concentration level in aqueous and fetal bovine serum (FBS) solution at pH 8.5 respectively. Due to its surfactant-free preparation and enhanced SERS-based sensing features, our reported platform represents a strong alternative to be used in SERS-based sensing applications.
Introduction
SERS is a powerful non-destructive analytical technique with the potential to achieve single molecule sensitivity under appropriate conditions. It can reveal information about molecular species in chemical and biological molecules.1,2 Two mechanisms, electromagnetic (EM) and chemical mechanisms, are used to describe the enhancement of Raman signals in SERS. EM enhancement can be attributed to the localized EM field at the nanostructured metal surface, while chemical enhancement depends on the charge transfer between the noble metal and the adsorbed analyte. EM enhancement is the dominant mechanism, because of its high enhancement factor, which ranges between 104 to 108 compared to chemical enhancement, which has a factor ranging between 10 and 102.3–7 Several studies have recently investigated the use of nanoparticles to improve SERS signals. The aggregation of nanoparticles creates “hotspots” that generate a highly enhanced EM field and offer a very low detection limit. However, this aggregation decreases reproducibility.8,9 Many researchers have investigated the formation of ordered metal nanostructures on solid substrates to enhance the signal without compromising the reproducibility.10–12 However, these efforts faced some challenges such as process complexity and relatively high cost. An alternative approach utilized hydrophobic surfaces to obtain a reproducible and enhanced SERS signal. The process involved evaporation of a droplet of the sample on the hydrophobic substrate. This technique resulted in a highly uniform distribution of nanoparticles, which generates a good reproducible signal. In addition, the adjacent nanoparticles create hotspots, providing an enhanced SERS signal.13–16Gold nanoparticles were widely investigated for several biological and chemical sensing applications, due to their unique chemical and physical properties including ease of synthesis, biocompatibility, chemical stability, and plasmonic tunability.17–21 Moreover, surface-modified gold nanoparticles can conjugate with various biomolecules, such as antibodies, enzymes, oligonucleotides, which provide an opportunity to develop a new biosensor platform with high specificity and selectivity to detect biological and chemical molecules.22–24 On the other hand, label free SERS sensing technique has great potential for direct bio-detection without using any tag molecules.25 There have been many efforts in analysis of biomolecules including low molecular weight substances (amino acids, vitamins, nucleotides etc.), nucleic acid and proteins in biofluids, based on label free SERS sensing strategy.25–28 Among them, as an example, to better understand processes in the brain, determining the level of dopamine, an key neurotransmitter, is of great importance. The low concentrations (0.01–1 μM) dopamine in the extracellular fluid and the presence of interfering molecules are challenging factors in detection of dopamine.29
Nanoparticles with anisotropic shapes, such as nanorods; nanotriangles; and nanostars with tips, edges, and corners, have intrinsic hotspots, which generate highly enhanced EM field.9,30,31 Synthetic methods for controlling the shape and size of gold nanoparticles include using surfactant and polymers for synthesis.32–34 These chemicals allow for shape orientation and growth prevention in specific directions. However, these nanoparticles, may exhibit poor SERS performance, because the surfactant layer can obstruct the interaction between the target analyte and nanoparticles.17 Another technique, which does not employ any surfactant or polymer, involves adding silver ions into gold seed solution to synthesize gold nanostars for biomedical applications such as cell imaging and biosensing.35–37 The branch growth mechanism is similar to that of nanorods formation. Silver ions are effective for the formation and differentiation of branches.36,38
Nanostars have attracted much attention, due to their unique plasmonic properties.39,40 Although many studies have already been conducted on nanostars, their effectiveness in SERS-based sensor is not fully understood. Comparative information about SERS signal enhancement using different sizes and shapes, such as spherical and star, of gold nanoparticles was presented in this study. By using the surfactant free method, branch formation and growth were controlled by changing the silver ion concentration in aqueous solution. The surfactant free synthesis method developed by Vo Dinh's et al. was followed in our preparation for nanoparticles.35 However, the recommended silver concentration (at least 5 μM) of their method was further reduced in our work. In our case the nanoparticles were synthesized at a concentration of 2 μM silver in order to obtain nanoparticles with fewer branches and smaller protrusions. These nanoparticles showed a more enhanced SERS signal than longer and more branched nanostars obtained in concentrations of 5 μM and above. SERS measurements were performed for the first time on the white epoxy ink-frosted label side of the microscope glass slide. This substrate with hydrophobic morphology contributed to the uniform distribution of aggregated nanoparticles and the formation of hotspots, which result in enhanced and reproducible SERS signal. To demonstrate the applicability of our sensing platform, dopamine spiked in FBS solution was used.
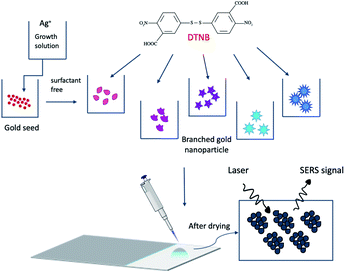
Materials and methods
Hydrogen tetrachloroaurate(III) trihydrate (HAuCl4·3H2O), silver nitrate (AgNO3), trisodium citrate (Na3C6H5O7), ascorbic acid (AA), hydrochloric acid (HCl), 5,5-dithio-bis-(2-nitrobenzoic acid) (DTNB), dopamine hydrochloride ((HO)2C6H3CH2CH2NH2·HCl), purchased from Sigma-Aldrich. All reagents were of analytical grade and used as received. Deionized water (DI) was used in all synthesis. All glassware was precleaned by aqua regia and then rinsed thoroughly with DI water. The morphology of nanoparticles was examined by Transmission Electron Microscope (TEM, Hitachi, H7650, Japan) at a voltage of 100 kV. In order to prevent aggregation of nanoparticles on the copper grid in the drying process, samples were prepared in 1 mg ml−1 BSA solution for TEM measurements.41 The mean size of nanoparticles was determined using Image J software (open source) and TEM images. UV-visible spectroscopy (Varian Cary 50 Bio, Agilent Technologies, USA) was used to obtain absorbance spectrum of the prepared colloidal nanoparticles. Samples were numbered S1–S8. Scanning Electron Microscope (SEM, Hitachi S 4700) was used to determine of aggregated nanoparticles on the substrate. The surface charge of nanoparticles was determined with Zetasizer (Malvern, Nanoseries Nano-ZS). Schematic of the preparation process of the sensing platform is shown in Fig. 1.Synthesis of citrate-stabilized gold nanoparticles
Gold nanoseed solution (S1) was synthesized following Turkevich method.42 Aqueous solution of HAuCl4 (100 ml) with a concentration of 1 mM was prepared and stirred while being heated until boiling in reflux condition. Subsequently, a 1% trisodium citrate solution (15 ml) was added to the mixture that was then stirred vigorously for 30 minutes. The same protocol was applied by reducing the citrate ion concentration in the solution to 600 μl to prepare bigger nanoparticles (S2).Preparation of branched gold nanoparticles
Branched gold nanoparticles were prepared without using any surfactant or solvent. The protocol defined by Vo Dinh's et al. was followed in the preparation.35 However, the limits defined in the protocol were expanded by using silver ions at a concentration of less than 5 μM in final concentration. HCl (10 μl) at a concentration of 1 M and the seed solution (100 μl) were added to 0.25 mM HAuCl4 aqueous solution (10 ml) and the mixture was then stirred at 400 rpm. After stirring for 5 minutes, AgNO3 of different concentrations (S3–S8; 0–20 μM, respectively), i.e., the final concentration in 10 ml solution, as well as 50 μl of 100 mM AA were simultaneously added to the main solution. Once the color of the solution changed, the solution was centrifuged at 4000 rpm for 20 min. Next the precipitated nanoparticles were dispersed in DI water.Preparation of self-assembled monolayer (SAM) DTNB on gold nanoparticles
DTNB with a concentration of 1 mM was prepared in 0.1 M PBS solution. DTNB was slowly added to the colloidal gold solutions in a 1.75 ml Eppendorf tube and was left under stirring overnight. The solution was centrifuged at 8000 rpm for 20 min to remove unbonded molecules, and the precipitate was then dispersed in DI water.Preparation of gold nanoparticles–dopamine biosensor
Dopamine of different concentrations (0.1 mM to 1 pM) were added in the colloidal gold solution at pH 8.5. The pH value was adjusted by using 0.1 M NaOH. The solutions were left overnight for incubation.Pretreatment of biological samples
To demonstrate the applicability of the proposed biosensor, dopamine detection in serum samples was performed with various dopamine concentrations spiked into FBS solution. In order to minimize the complex matrix effect, the methanol extraction method was employed on the serum samples. Briefly, 100 μl of sample was mixed with 900 μl HPLC grade methanol followed by centrifugation at 5000 rpm for 10 min. The resulting clear supernatant was removed to use in the SERS measurement.SERS measurement
Gold Seal Rite-On glass slide was purchased from Thermo-Fisher Scientific. The white epoxy ink-frosted part of the glass slide was used as a substrate. 3 μl of analyte/gold nanoparticle solution was placed on the substrate. After drying samples, the SERS signal was collected from 15 different locations in each sample to determine average data and standard deviation. SERS measurements were performed using a Raman spectrometer (Thermo Scientific DXR-USA) with 5 mW of 780 nm wavelength laser. The scattered light was collected by a 10× objective lens (NA = 0.25) into a charge-coupled device detector. A grating of 400 lines per mm was used to disperse the scattered light. The spot size of the laser was estimated to be 3.1 μm. The Raman shift was calibrated using the signal of 1001 cm−1 from a polystyrene. All DTNB spectra reported here were the results of signal accumulation for 2 s within a range of 50–1850 cm−1 with total data points of 1868.Results and discussion
Size and shape of gold nanoparticles
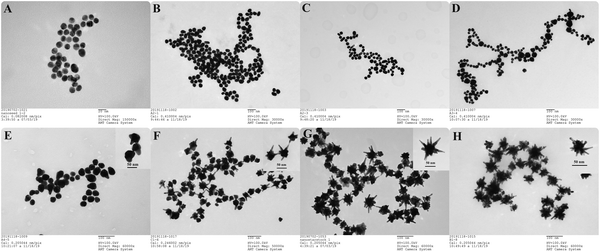
The size and shape of gold nanoparticles were analyzed by TEM. Fig. 2A–H illustrate the results of S1–S8, respectively. The average dimensions of the particles, such as core diameter (dcore), branch length (Lbranch), number of branches (Nbranch) were calculated using TEM images and Image J software. The change in size and shape of nanoparticles is clearly reflected in Fig. 2 and Table 1. While the nanoparticles (S3, S4, S5, S6, S7, S8) were grown by using (S1) gold seeds with 15 ± 1 nm and AA as reducing agent, S2 solution was prepared directly by citrate reduction without seeds. Fig. 2B shows that the size distribution in the S2 solution was homogenous with spherical shaped nanoparticles of 52 ± 5 nm diameter. Seed-prepared nanoparticles (S3) formed without using AgNO3 were mainly composed of spherical shaped particles with a diameter of 38 ± 6 nm, but some rod-shaped nanoparticles were also observed. Fig. 2D shows that adding AgNO3 (1 μM) into seed solution did not cause any apparent branching in the particles. S4 particles had an anisotropic shape with a diameter of 54 ± 17 nm based on their angular structures. Upon increasing the silver concentration to 2 μM, one to three branches were clearly observed, and the size distribution became more homogeneous (38 ± 4 nm) compared to the previous structure (Fig. 2D and E). On the other hand, increasing the silver concentration to 5 μM resulted in the formation of particles with five to eight branches and an increase in both the number of branches and their length compared to the previous structure. In addition, the structure of nanoparticle (S6) changed into nanostar (Fig. 2F). By further increasing the silver ion concentration to 10 μM, the number of branches and their length as well as the particle's core diameter increased, which was also the case when the silver concentration was further increased to 20 μM. However, side protrusions started to appear on the branches of nanostars in S8. Small changes in silver concentration considerably affected the morphology of gold nanoparticles. Therefore, the number of branches and their length increased with the silver concentration. However, there was no correlation between silver concentration and core diameter. Similar to the previous study, nanostars were observed at a silver concentration of 5 μM and above.35 It was reported that underpotential deposition of metallic silver ions on different crystal surfaces of gold leads to symmetry breaking and formation of gold nanorods.43–45Fig. 2E shows that the formation of protrusions started in one direction upon the addition of a small amount of silver ions to the solution. Thus, the formation of branched nanoparticles shows a similarity to the growth of nanorods.| Sample nu. | Silver concentration | d core | L branch | N branch |
|---|---|---|---|---|
| S1 | — | 15 ± 1 nm | — | — |
| S2 | — | 52 ± 5 nm | — | — |
| S3 | — | 38 ± 6 nm | — | — |
| S4 | 1 μM | 54 ± 17 nm | — | — |
| S5 | 2 μM | 38 ± 4 nm | 10 ± 3 nm | 1–3 |
| S6 | 5 μM | 33 ± 3 nm | 16 ± 5 nm | 5–8 |
| S7 | 10 μM | 45 ± 3 nm | 23 ± 5 nm | 10–12 |
| S8 | 20 μM | 48 ± 4 nm | 33 ± 7 nm | ≥12 |
Absorption measurements of nanoparticles
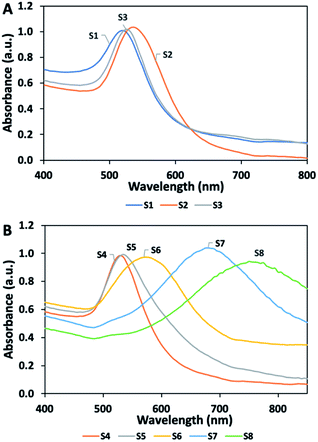
Fig. 3 presents the UV-vis absorbance spectra of the nanoparticles, in which optical properties offer information about their size and shape. Fig. 3A shows two intense absorption peaks for S1 and S2 at 520 and 535 nm, respectively. As the gold nanoparticle size grows, the maximum extinction of Localized Surface Plasmon Resonance shifted to the red region in the EM spectrum. As expected, the size of the gold nanoparticle (S2) increased with decreasing the citrate ion concentration relative to nanoseed (S1). In case of nanoparticles (S3) prepared in seed solution with no AgNO3, the ascorbic anions transfer electrons to the seed particles so that particles grow with reduction of gold ions. S3 shows maximum absorbance at 525 nm. Fig. 3B shows the UV-vis absorption spectra obtained by increasing the silver ion concentration between 1 and 20 μM in the presence of seed particles, AA, and gold ions.Increasing the silver concentration causes an increase in the maximum absorbance wavelength and a shift toward the NIR region, due to an increase in the size of particles. A single absorption peak was observed on nanoparticles (S1–S5). In contrast, the multi-branched nanostars in S6, S7, and S8 resulted in two absorption peaks, of which one was a weak peak originating from the core body at around 520 nm. In addition, these nanostars exhibited broad absorption peak, due to their complexity.
SAM DTNB on nanoparticles
DTNB was chosen as a probe molecule, because of its high affinity to gold nanoparticles. It was reported that DTNB exhibits poor stability in basic environment.46,47 Therefore, the solution was prepared in 0.1 M PBS to maintain neutral pH. The concentration of DTNB was optimized to form SAM on nanoparticles, while maintaining solution stability. Fig. 3 shows that the maximum absorbance intensity of all colloidal gold solutions was adjusted to approximately 1.0. The changing in stability of colloidal solutions was observed after adding DTNB by absorbance measurements. Different concentrations of DTNB solution (20–200 μM) was added to the colloidal gold solutions. S2 solution showed poor stability and formed aggregates after adding a solution with a concentration of 20 μM. The maximum absorbance intensity of S2 significantly decreased because of aggregation (Fig. S1†). Similarly, SERS probe molecules were previously reported to change the electronic double layer formed by stabilizers surrounding the nanoparticle, resulting in particle aggregation.48–51 On the other hand, branched nanoparticles (i.e., S5, S6, and S7) without surfactant displayed better stability compared to S2, even though a more concentrated DTNB solution was added in the preparation. The maximum absorbance wavelength for the branched nanoparticles (Fig. S2 and S3†) shifted to the blue region, due to the increase in the particle size with DTNB molecules. The maximum concentration of DTNB for these particles reached 100 μM. As the critical concentration of the solution was further increased, the stability of the solution started to significantly deteriorate and visible aggregates started to form.SERS measurements
The Raman spectrum of the DTNB molecule shows a strong peak at 1342 cm−1 resulting from the symmetric stretching of the nitro group as well as a peak at 1556 cm−1 originated from the mode of the aromatic ring in the molecule. However, SERS spectrum of TNB exhibits some peak shifts (1342–1333 cm−1) compared to the Raman spectrum of DTNB.51 The interval of 1342–1333 cm−1 and the band at 1556 cm−1 were considered the fingerprint region of the molecule in all measurements.Effect of surface properties of the glass slide on SERS signal
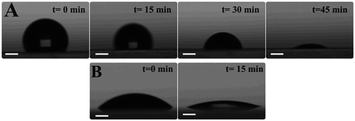
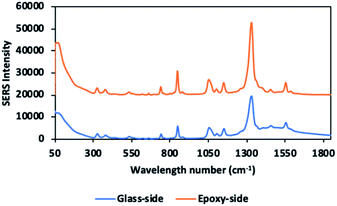
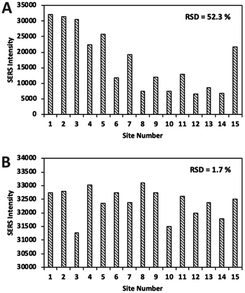
Comparing the white epoxy ink-coated side of the glass to bare glass, the droplet covered a smaller area in the epoxy-coated side, due to its hydrophobic structure (Fig. 4). The drying process ensured a uniform distribution of the aggregated nanoparticles in the confined area and the formation of a higher number of hotspots. During evaporation, the color of the sample point changed from pink to gray, due to the nanoparticles gathering in the limited area (Fig. S4†). As seen in the SEM image (Fig. 5), the nanoparticles were positioned closer to each other, forming hotspots and enhancing the SERS signal. In contrast, the particles on the bare glass randomly dispersed in a wider area.Fig. 6 indicates that the SERS signal was approximately three times stronger on the epoxy part compared to that on the bare glass. In addition, the RSD values of the glass and epoxy side were found to be 52.3% and 1.7%, respectively (Fig. 7). This means that almost the same signal value was obtained from each point on the epoxy side. Therefore, both enhanced and low signal variations can be achieved with this substrate. Thus, a reproducible signal was obtained for SERS measurements.
Effect of shape and size of nanoparticles on SERS signal
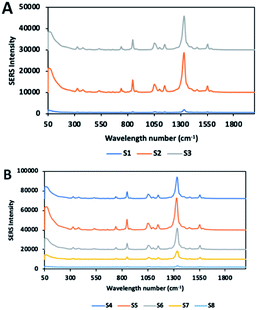
DTNB solutions (100 μM) were added to all colloidal solutions (S1–S8), and a 3 μl volume of each solution was placed onto the epoxy side of glass. Fig. 8A and B illustrate the SERS spectra of all solutions. Previous studies showed that the particle size of gold nanoparticles affects the SERS signal enhancement.52–55 Some studies achieved maximum SERS enhancement with spherical nanoparticles of a 50 nm diameter.54–56 Similar to these results, the highest average SERS signal intensity among the spherical nanoparticles (i.e., S1, S2, S3) was achieved using nanoparticles with a diameter of about 52 nm (S2), even though the signal variation was quite high. The effective conductivity and light scattering properties required for SERS enhancement are known to decrease with very small particles such as S1.55–57 S3 displayed more SERS enhancement compared to S1, due to the increasing size. This observation supports that the theory in which EM field enhancement depends on the number of excited electrons and the volume of nanostructures.55,56 Nanoparticles with sharp corners or high curvature, such as tips, affect the SERS enhancement, due to their hotspots, which generate highly enhanced EM field.25 However, it is noteworthy that the position of the nanostars relative to each other is also effective on the EM enhancement. In the case of aggregation, nanostars with long branch exhibited less EM enhancement than short branched particles (Fig. 8 and 9). This is in good agreement with previous studies, due to its complex geometry and orientation, nanostars probably collapse into each other, breaking the hot spots on the branches.34,58,59 As the number of branches further increased, the signal significantly decreased. When the laser strikes the centre of a long branched nanostar, such as S7 and S8, energy is dissipated by the branches, and it becomes difficult to contain it in a certain area. In addition, the branches create fewer junctions, which act as hotspots, because the two individual particles are prevented from getting close enough to each other. In case of long branched nanoparticles, the majority of the analyte is attached to the core body and the arms of the tips. As the concentration of the molecule retained on the tips is very small compared to the entire surface area and since the unusual SERS enhancement effect occurs on the tips, this effect is considered negligible compared to the signals collected from the entire surface.34 Thus, low signal intensity in long branched nanostars may be attributed to these factors. Whereas, S5 with small protrusions shows the strongest EM enhancement. This enhancement may be attributed to the strong electrical field near-surface in short-branched nanoparticles synthesized by different methods in previous study.31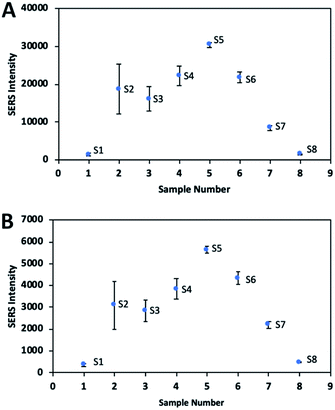 | ||
| Fig. 9 The SERS signal intensity including signal variation of DTNB for all nanoparticle solutions (S1–S8) in specific band at (A) 1337 cm−1 and (B) 1556 cm−1. | ||
Comparison of the SERS signal intensities of the nanoparticles for DTNB
Fig. 9 shows the signal intensities of the band at 1337 cm−1 and the band at 1556 cm−1 resulting from symmetric stretching of the nitro group and aromatic ring stretching mode respectively while Tables S1 and S2† show the RSD values. S2 showed the poorest reproducible SERS signal, due to some aggregation in the solution, which caused a heterogeneous distribution on the substrate and consequently increased the variation of the signal. In order to increase the reproducibility and accuracy of the signal, it is necessary to ensure a homogeneous distribution of the nanoparticles on the substrate. Although S3 solution can maintain the stability with no aggregation, SERS signal variation of the particles is relatively high. Because S3 solution has particles of different shapes, such as rods and spheres, signal variation may occur. On the other hand, branched nanoparticles exhibited good reproducible SERS signals with low RSD value, due to their good stability and uniform distribution on the substrate.SERS EF calculation
The SERS EF of DTNB on nanoparticles was determined for S5 solution which showed the highest signal intensity. EF was calculated for the characteristic bands at 1337 cm−1 as well as at 1556 cm−1. The following equation was used to obtain the EF value:| EF = [ISERS]/[IRaman] × [NRaman]/[NSERS] |
The number of analyte molecules (Nbulk) in 1 mM DTNB solution was calculated, while taking into account the laser spot size (d = 3.1 μm) and depth of field (h = 29.3 μm), to be 2 × 108. The shape of the S5 nanoparticles was assumed to be spherical, even though they have small branches or protrusions, because S5 comprised mainly spherical structures (Fig. 2E). Accordingly, the concentration of spherical nanoparticles was determine by using an equation, which employed the UV-vis absorbance intensity at 450 nm.60 The concentration of nanoparticles in S5 solution was determined to be 7.68 × 1010 molecules per ml. Assuming that the coverage of the analyte monolayer on the nanoparticle and TNB molecular footprint was 0.26 nm2 per molecule, NSERS was estimated to be 7.6 × 103. Ibulk and ISERS were obtained from the spectra of DTNB illustrated in Fig. S5.† Using the equation given above, EF was calculated to be 3.8 × 105 and 4.2 × 105 at 1337 cm−1 and 1556 cm−1 respectively.
Label free SERS detection of dopamine
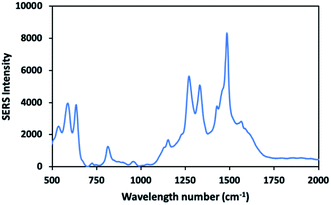
Molecules containing functional group such as thiols, phosphines and amines have high affinity towards the surface of gold nanoparticles, based on the hard and soft acids and bases (HSAB) theory. Positively charged amine group of dopamine tend to bind to the negatively charged surface of gold nanoparticles.61,62 In previous studies, gold nanoparticles were reported to show better electrostatic interaction with more deprotonated dopamine at alkaline environment.63,64 Similarly, low branched nanoparticles with a zeta potential of (−38.3 mV) exhibited good interaction with dopamine molecules at pH 8.5 in our study. Fig. 10 shows the SERS signal of the 10−4 M of dopamine molecules adsorbed onto the low branched gold nanoparticles. The characteristic bands of dopamine molecules were mainly observed at 1270 cm−1, 1333 cm−1 and 1480 cm−1. The spectra of dopamine exhibited a good similarity with that of previous works.29,65 The intense bands at 1270 cm−1, 1333 cm−1 and 1480 cm−1 were assigned to the catechol carbon–oxygen stretching, the C–O stretching and the catechol ring breathing respectively.29,66 It is important to note that there is no interference peak arising from the gold nanoparticle/substrate without adding any analyte. The SERS measurements of 10−6 M to 10−10 M dopamine were performed and the peak intensity for the marker band at 1480 cm−1 was followed in Fig. 11. As the dopamine concentration increased, the density of the specific peak in 1480 cm−1 increased. Dopamine was detected at picomolar concentration level by using sensing platform consisting of low-branched nanoparticles aggregated on the label side of the glass. The detected concentration of dopamine is one of the lowest levels achieved by label free SERS detection. In previous studies, dopamine was detected at the picomolar level by fabricating special substrates such as graphene-Au nanopyramids and Au/Ag nanocluster electrodeposited on ITO glass.67,68 In contrast, the similar detection level can be achieved by using a simple method and low-cost substrate in this study.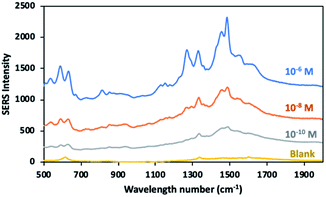 | ||
| Fig. 11 SERS spectra of dopamine molecules adsorbed on low branched gold nanoparticles in three different concentration (10−6 to 10−10 M) at pH 8.5. | ||
Detection of dopamine in spiked serum samples
FBS is a complex medium including various molecules such as proteins, growth factors, vitamins, trace elements, hormones.69 To minimize interference effect of matrix, the methanol extraction method was applied to remove proteins.15,61,70Fig. 12 shows the SERS spectra of the dopamine with different concentration in the FBS samples and as well as the spectrum of control sample (FBS) without dopamine. Specific peaks of dopamine were clearly observed at 1270 cm−1, 1333 cm−1 and 1480 cm−1 from dopamine at the nanomolar level was detected in the FBS. Although no dominant peak was observed, the presence of weak peaks at 733 cm−1 and in the range of 1100 cm−1 to 1700 cm−1 indicate a small amount of molecule residues in the control sample. The fact that the detection limit in FBS remains at nanomolar (10 nM) levels can be attributed to the interference effect of molecule residues in FBS despite the use of methanol extraction method.67,71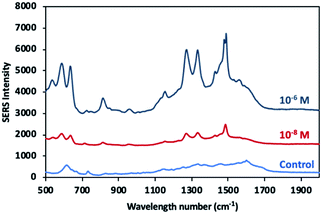 | ||
| Fig. 12 SERS spectra of dopamine molecules in two different concentrations (10−6 and 10−8 M) in FBS and control sample. | ||
Conclusion
In this study, new gold nanoparticles with one to three small protrusions were produced by using surfactant-free synthesis. The number of branches and their length in the gold nanoparticle increased with the silver ion concentration. The gold nanoparticles with small protrusions exhibited the highest SERS signal intensity for DTNB molecule among the spherical and multi-branched nanoparticles. In contrast, multi-branched nanoparticles with longer spikes showed lower SERS signal intensities. In addition, the white epoxy ink-frosted labeling side of the glass slide was proven to be a new efficient solid SERS substrate that contributes to the formation of hotspots and enables uniform distribution of aggregated nanoparticles, due to its hydrophobic morphology. Thus, highly reproducible and enhanced SERS signal was obtained on the solid substrate. The developed method has been successfully applied for sensitive detection of dopamine spiked into FBS as the real application. In summary, the combined use of the new gold nanoparticles with small protrusions and the new solid SERS substrate holds great potential for high sensitivity SERS-based sensing of traces of biomolecules.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Professor Shu-Yi Lin (Department of Nanomedicine of National Health Research Institutes (NHRI)) for providing equipment. This work was supported by the fellowship from NHRI in Taiwan.Notes and references
- B. Sharma, R. R. Frontiera, A.-I. Henry, E. Ringe and R. P. Van Duyne, Mater. Today, 2012, 15, 16–25 CrossRef CAS.
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis, J. Dorney, K. Esmonde-White, N. J. Fullwood, B. Gardner, P. L. Martin-Hirsch and others, Nat. Protoc., 2016, 11, 664 CrossRef CAS PubMed.
- I. Bruzas, W. Lum, Z. Gorunmez and L. Sagle, Analyst, 2018, 143, 3990–4008 RSC.
- S.-Y. Ding, E.-M. You, Z.-Q. Tian and M. Moskovits, Chem. Soc. Rev., 2017, 46, 4042–4076 RSC.
- E. C. Le Ru, E. Blackie, M. Meyer and P. G. Etchegoin, J. Phys. Chem. C, 2007, 111, 13794–13803 CrossRef CAS.
- L. Jensen, C. M. Aikens and G. C. Schatz, Chem. Soc. Rev., 2008, 37, 1061–1073 RSC.
- N. Valley, N. Greeneltch, R. P. Van Duyne and G. C. Schatz, J. Phys. Chem. Lett., 2013, 4, 2599–2604 CrossRef CAS.
- T. Vo-Dinh, Y. Liu, A. M. Fales, H. Ngo, H.-N. Wang, J. K. Register, H. Yuan, S. J. Norton and G. D. Griffin, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 17–33 CAS.
- A. M. Fales, H. Yuan and T. Vo-Dinh, J. Phys. Chem. C, 2014, 118, 3708–3715 CrossRef CAS PubMed.
- W. J. Cho, Y. Kim and J. K. Kim, ACS Nano, 2012, 6, 249–255 CrossRef CAS PubMed.
- W. Q. Li, G. Wang, X. N. Zhang, H. P. Geng, J. L. Shen, L. S. Wang, J. Zhao, L. F. Xu, L. J. Zhang, Y. Q. Wu, R. Z. Tai and G. Chen, Nanoscale, 2015, 7, 15487–15494 RSC.
- T. Y. Chan, T. Y. Liu, K. S. Wang, K. T. Tsai, Z. X. Chen, Y. C. Chang, Y. Q. Tseng, C. H. Wang, J. K. Wang and Y. L. Wang, Nanoscale Res. Lett., 2017, 12, 344 CrossRef PubMed.
- F. De Angelis, F. Gentile, F. Mecarini, G. Das, M. Moretti, P. Candeloro, M. L. Coluccio, G. Cojoc, A. Accardo, C. Liberale, R. P. Zaccaria, G. Perozziello, L. Tirinato, A. Toma, G. Cuda, R. Cingolani and E. Di Fabrizio, Nat. Photonics, 2011, 5, 682–687 CrossRef CAS.
- F. Xu, Y. Zhang, Y. Sun, Y. Shi, Z. Wen and Z. Li, J. Phys. Chem. C, 2011, 115, 9977–9983 CrossRef CAS.
- H. Wu, Y. Luo, Y. Huang, Q. Dong, C. Hou, D. Huo, J. Zhao and Y. Lei, Front. Chem., 2018, 6, 482 CrossRef CAS PubMed.
- C. Zhang, T. You, N. Yang, Y. Gao, L. Jiang and P. Yin, Food Chem., 2019, 30, 363–368 CrossRef PubMed.
- M. A. Wall, S. Harmsen, S. Pal, L. Zhang, G. Arianna, J. R. Lombardi, C. M. Drain and M. F. Kircher, Adv. Mater., 2017, 29, 1605622 CrossRef PubMed.
- P. D. Howes, R. Chandrawati and M. M. Stevens, Science, 2014, 346, 1247390 CrossRef PubMed.
- C. Burda, X. Chen, R. Narayanan and M. A. El-Sayed, Chem. Rev., 2005, 105, 1025–1102 CrossRef CAS PubMed.
- H. Jans and Q. Huo, Chem. Soc. Rev., 2012, 41, 2849–2866 RSC.
- C. Kohout, C. Santi and L. Polito, Int. J. Mol. Sci., 2018, 19, 3385 CrossRef PubMed.
- L. Dykman and N. Khlebtsov, Chem. Soc. Rev., 2012, 41, 2256–2282 RSC.
- J.-H. Lee, H.-Y. Cho, H. K. Choi, J.-Y. Lee and J.-W. Choi, Int. J. Mol. Sci., 2018, 19, 2021 CrossRef CAS PubMed.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- X. S. Zheng, I. J. Jahn, K. Weber, D. Cialla-May and J. Popp, Spectrochim. Acta, Part A, 2018, 197, 56–77 CrossRef CAS PubMed.
- J. Chao, W. Cao, S. Su, L. Weng, S. Song, C. Fan and L. Wang, J. Mater. Chem. B, 2016, 4, 1757–1769 RSC.
- S. Yüksel, L. Schwenkbier, S. Pollok, K. Weber, D. Cialla-May and J. Popp, Analyst, 2015, 140, 7254–7262 RSC.
- X. Pan, L. Li, H. Lin, J. Tan, H. Wang, M. Liao, C. Chen, B. Shan, Y. Chen and M. Li, Biosens. Bioelectron., 2019, 145, 111713 CrossRef CAS PubMed.
- M. Kaya and M. Volkan, Anal. Chem., 2012, 84, 7729–7735 CrossRef CAS PubMed.
- J. Reguera, J. Langer, D. J. de Aberasturi and L. M. Liz-Marzán, Chem. Soc. Rev., 2017, 46, 3866–3885 RSC.
- W. Lv, C. Gu, S. Zeng, J. Han, T. Jiang and J. Zhou, Biosensors, 2018, 8, 113 CrossRef CAS PubMed.
- P. S. Kumar, I. Pastoriza-Santos, B. Rodriguez-Gonzalez, F. J. G. De Abajo and L. M. Liz-Marzán, Nanotechnology, 2007, 19, 15606 CrossRef PubMed.
- S. Atta, M. Beetz and L. Fabris, Nanoscale, 2019, 11, 2946–2958 RSC.
- S. Barbosa, A. Agrawal, L. Rodríguez-Lorenzo, I. Pastoriza-Santos, R. A. Alvarez-Puebla, A. Kornowski, H. Weller and L. M. Liz-Marzán, Langmuir, 2010, 26, 14943–14950 CrossRef CAS PubMed.
- H. Yuan, C. G. Khoury, H. Hwang, C. M. Wilson, G. A. Grant and T. Vo-Dinh, Nanotechnology, 2012, 23, 75102 CrossRef PubMed.
- A. D'Hollander, E. Mathieu, H. Jans, G. Vande Velde, T. Stakenborg, P. Van Dorpe, U. Himmelreich and L. Lagae, Int. J. Nanomed., 2016, 11, 3703 CrossRef PubMed.
- A. S. D. S. Indrasekara, S. Meyers, S. Shubeita, L. C. Feldman, T. Gustafsson and L. Fabris, Nanoscale, 2014, 6, 8891–8899 RSC.
- Y. Xia, Y. Xiong, B. Lim and S. E. Skrabalak, Angew. Chem., Int. Ed., 2009, 48, 60–103 CrossRef CAS PubMed.
- H. Yuan, C. G. Khoury, C. M. Wilson, G. A. Grant, A. J. Bennett and T. Vo-Dinh, Nanomedicine, 2012, 8, 1355–1363 CrossRef CAS PubMed.
- W. Niu, Y. A. A. Chua, W. Zhang, H. Huang and X. Lu, J. Am. Chem. Soc., 2015, 137, 10460–10463 CrossRef CAS PubMed.
- B. Michen, C. Geers, D. Vanhecke, C. Endes, B. Rothen-Rutishauser, S. Balog and A. Petri-Fink, Sci. Rep., 2015, 5, 9793 CrossRef CAS PubMed.
- J. Kimling, M. Maier, B. Okenve, V. Kotaidis, H. Ballot and A. Plech, J. Phys. Chem. B, 2006, 110, 15700–15707 CrossRef CAS PubMed.
- M. Liu and P. Guyot-Sionnest, J. Phys. Chem. B, 2005, 109, 22192–22200 CrossRef CAS PubMed.
- G. Kawamura, Y. Yang, K. Fukuda and M. Nogami, Mater. Chem. Phys., 2009, 115, 229–234 CrossRef CAS.
- M. Grzelczak, J. Pérez-Juste, P. Mulvaney and L. M. Liz-Marzán, Chem. Soc. Rev., 2008, 37, 1783–1791 RSC.
- P. W. Riddles, R. L. Blakeley and B. Zerner, Anal. Biochem., 1979, 94, 75–81 CrossRef CAS PubMed.
- V. J. Elia and J. P. Danehy, J. Org. Chem., 1971, 36, 1394–1398 CrossRef.
- J. Ni, R. J. Lipert, G. B. Dawson and M. D. Porter, Anal. Chem., 1999, 71, 4903–4908 CrossRef CAS PubMed.
- D. S. Grubisha, R. J. Lipert, H. Y. Park, J. Driskell and M. D. Porter, Anal. Chem., 2003, 75, 5936–5943 CrossRef CAS PubMed.
- S. Xu, X. Ji, W. Xu, B. Zhao, X. Dou, Y. Bai and Y. Ozaki, J. Biomed. Opt., 2005, 10, 031112 CrossRef PubMed.
- C. C. Lin, Y. M. Yang, Y. F. Chen, T. S. Yang and H. C. Chang, Biosens. Bioelectron., 2008, 24, 178–183 CrossRef CAS PubMed.
- P. N. Njoki, I. I. S. Lim, D. Mott, H. Y. Park, B. Khan, S. Mishra, R. Sujakumar, J. Luo and C. J. Zhong, J. Phys. Chem. C, 2007, 111, 14664–14669 CrossRef CAS.
- V. Joseph, A. Matschulat, J. Polte, S. Rolf, F. Emmerling and J. Kneipp, J. Raman Spectrosc., 2011, 42, 1736–1742 CrossRef CAS.
- S. Hong and X. Li, J. Nanomater., 2013, 790323 Search PubMed.
- N. D. Israelsen, C. Hanson and E. Vargis, Sci. World J., 2015, 124582 Search PubMed.
- K. G. Stamplecoskie, J. C. Scaiano, V. S. Tiwari and H. Anis, J. Phys. Chem. C, 2011, 115, 1403–1409 CrossRef CAS.
- M. Moskovits, J. Raman Spectrosc., 2005, 36, 485–496 CrossRef CAS.
- S. Abalde-Cela, P. Aldeanueva-Potel, C. Mateo-Mateo, L. Rodríguez-Lorenzo, R. A. Alvarez-Puebla and L. M. Liz-Marzán, J. R. Soc., Interface, 2010, 7, S435–S450 CrossRef CAS PubMed.
- H. Yuan, A. M. Fales, C. G. Khoury, J. Liu and T. Vo-Dinh, J. Raman Spectrosc., 2013, 44, 234–239 CrossRef CAS PubMed.
- W. Haiss, N. T. K. Thanh, J. Aveyard and D. G. Fernig, Anal. Chem., 2007, 79, 4215–4221 CrossRef CAS PubMed.
- N. Mohseni and M. Bahram, Spectrochim. Acta, Part A, 2018, 193, 451–457 CrossRef CAS PubMed.
- M. C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293–346 CrossRef CAS PubMed.
- Y. Bu and S. W. Lee, Int. J. Nanomed., 2015, 47–54 CAS.
- Y. J. Yuan, Z. Xu and Y. Chen, AIP Adv., 2019, 9, 035028 CrossRef.
- L. Qin, X. Li, S. Z. Kang and J. Mu, Colloids Surf., B, 2015, 126, 210–216 CrossRef CAS PubMed.
- M. L. B. Figueiredo, C. S. Martin, L. N. Furini, R. J. G. Rubira, A. Batagin-Neto, P. Alessio and C. J. L. Constantino, Appl. Surf. Sci., 2020, 522, 146466 CrossRef CAS.
- P. Wang, M. Xia, O. Liang, K. Sun, A. F. Cipriano, T. Schroeder, H. Liu and Y. H. Xie, Anal. Chem., 2015, 87, 10255–10261 CrossRef CAS PubMed.
- V. D. Phung, W. S. Jung, T. A. Nguyen, J. H. Kim and S. W. Lee, Nanoscale, 2018, 10, 22493–22503 RSC.
- J. van der Valk, K. Bieback, C. Buta, B. Cochrane, W. G. Dirks, J. Fu, J. J. Hickman, C. Hohensee, R. Kolar, M. Liebsch, F. Pistollato, M. Schulz, D. Thieme, T. Weber, J. Wiest, S. Winkler and G. Gstraunthaler, ALTEX, 2017, 35, 99–118 Search PubMed.
- T. M. Alshammari, A. A. Al-Hassan, T. B. Hadda and M. Aljofan, Saudi Pharm. J., 2015, 23, 689–697 CrossRef PubMed.
- A. H. Nguyen, E. A. Peters and Z. D. Schultz, Rev. Anal. Chem., 2017, 36, 1–29 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Top view of droplet on white epoxy ink side of glass slide during evaporation process. UV-vis absorbance graphs of DTNB modified nanoparticles. SERS and Raman spectra of DTNB molecule. RSD results of DTNB modified nanoparticles on the labelling part of glass slide. See DOI: 10.1039/d0ra02490b |
| This journal is © The Royal Society of Chemistry 2020 |