Open Access Article
Open Access ArticleUse of exosomes as vectors to carry advanced therapies
María Sancho-Albero
 ab,
Ana Medel-Martínez
ab,
Ana Medel-Martínez
 ac and
Pilar Martín-Duque
ac and
Pilar Martín-Duque
 *bcd
*bcd
aDepartment of Chemical Engineering, Aragon Institute of Nanoscience (INA), Aragon Materials Science Institute (ICMA), University of Zaragoza, Campus Río Ebro-Edificio I+D, C/Poeta Mariano Esquillor s/n, 50018 Zaragoza, Spain
bNetworking Research Center on Bioengineering, Biomaterials and Nanomedicine (CIBERBBN), 28029 Madrid, Spain. E-mail: mpmartind.iacs@aragon.es
cAragon Health Sciences Institute(IACS)/IIS Aragón, Centro de Investigaciones Biomédicas de Aragón, Avda San Juan Bosco 13, 50009 Zaragoza, Spain
dAraid Fund., Av. de Ranillas 1-D, Planta 2a, Oficina B, 50018 Zaragoza, Spain
First published on 23rd June 2020
Abstract
Exosomes are microvesicles of nanometric size involved in the communication between cells and tissues. Inside their bilipidic membrane they carry nucleic acids such as cargos (DNA, miRNA, etc.). Some of the advantages that make exosomes very attractive therapeutic vehicles are (i) their tropism through different tissues, (ii) the ability to pass biological barriers and (iii) the protection of the encapsulated material from the immune system and degradation. Viruses are some of the most widely employed gene therapy vehicles; however, they are still facing many problems, such as inefficient tropism to damaged areas and their elimination by the immune system. One of the functions attributed to exosomes is the elimination of substances that could be harmful to the cell, including viruses. Recently it has been investigated whether complete viruses or part of them could be encapsulated in exosomes, for a new viral-exosome gene therapy approach. Moreover, nanotechnology is another type of advanced therapy (together with gene and cell therapies) that can be used, among other utilities, to transfer genetic material. Recently the field of encapsulation of nanomaterials in exosomes, with or without gene transfer, is increasing. In this review we will summarize all of those studies.
 María Sancho-Albero | Maria Sancho Albero: BSc in Biotechnology and MSc in Nanostructured Materials at the University of Zaragoza. PhD student in the Nanoscience Institute of Aragon (INA) in the NFP group. She works on the combinations of molecular biology and biochemistry with physicochemical, microfluidics and nanotechnolgical techniques in the field of extracellular vesicles for cancer treatment and diagnosis. She made two international research stays in the ETH of Zurich and in the Polytechnic University of Milan. She has published articles in high impact journals such as: M. Sancho-Albero, et al., Nat Catal., 2019, 2(10), 864–872, DOI: 10.1038/s41929-019-0333-4; M. Sancho-Albero, et al., Nanoscale, 2019, 11(40), 18825–18836, DOI: 10.1039/C9NR06183E; M. Sancho-Albero, et al., J Nanobiotechnol, 2019, 17(1), 1–16, DOI: 10.1186/s12951-018-0437-z. |
Exosomes: general overview and their role as therapeutic carriers
Exosomes are one subset of extracellular vesicles (EVs) secreted by almost all cells present in the organism.1 Apart from exosomes, other EVs including apoptotic bodies, ectosomes and microvesicles are also released to the extracellular environment.2 Exosomes are the smallest EVs from endocytic nature with a diameter from 30 to 100 nm and they are formed by the maturation of the early endosomes to the late endosomes, followed by the invagination of the endosomal membrane and the formation of the multivesicular bodies (MVBs).3 Afterwards, MVBs are fused with the plasmatic membrane and exosomes are finally released to the extracellular space.4 In fact, they could be found in almost all body fluids such as blood, serum, saliva, nasal secretions, urine, breast milk, pericardial fluid, etc.5Those “nanospheres” have a density between 1.08 and 1.22 g mL−1 and they are surrounded by a lipid bilayer membrane similar to the plasmatic one, but enriched in certain lipids such as sphingomyelin, phosphatidylserine, phosphatidylcholine, phosphatidylethanolamine, ceramide and cholesterol.6,7 Apart from lipids, exosome membrane also contains proteins (surface receptors and ligands) that might be different depending of the cell source. Among them, transporter proteins (CD13), heat shock proteins (Hsp70), fusion proteins (flotilin or annexin), proteins related to the biogenesis process and tetraspanins (CD63, CD81 or CD9) are some of the common ones.8 Those proteins were mostly found in exosomes, rather than in other EVs, allowing their distinction from other vesicles. In the cytosolic compartment, exosomes carry a wide variety of proteins identified as components of the Endosomal Sorting Complex Required for Transport (ESCRT) machinery, such as ALIX or VPS and RAB proteins. Moreover, metabolic enzymes (ATPase or aldehyde reductase), growth factors and cytokines (such as TNF) are also found inside exosomes.9 Apart from proteins, exosomes also contain different patterns of nucleic acids such as mRNAs and miRNAs (mi-214, miR-29a or miR-126)10,11 or even, double strand DNA.12 Although those biomolecules are contained in all types of exosomes, those vesicles also have an specific cargo that is closely associated with the parental cell, which determines exosome function and targets.13,14
Despite their homogeneity, exosomes mediate a wide range of functions and thus, and it has been demonstrated that those vesicles have different content on composition depending on the parental cell.3,15 In fact, Palma et al., showed that tumor and healthy cells release distinct populations of exosomes.16
On the past, the main role attributed to exosomes was the release of unnecessary or toxic materials for the cell (such as unfolded proteins).17 On the contrary, today those vesicles are considered as natural signaling vectors and they are thought to play an essential role for the intercellular local and distant communication.18 In fact, one of the most useful characteristic of exosomes is their ability to cross biological barriers including the blood brain barrier (BBB) or the cytoplasmic membrane.19 That fact allows us to transport selectively specific therapies to certain cell types or to inaccessible sites such as the central nervous system and in particular to the brain, which remains a major challenge. Apart from their role in cell–cell communication, they participate in the transport of small molecules amongst cells regulating a wide range of functions often immunity related (they have been proposed to be crucial in the development of organs or in some pathologies,20 such as neurodegenerative diseases of the brain21 and in cardiovascular diseases22). Due to their central role in the intercellular communication to recipient cells, exosomes are also essential for the interaction between tumor cells and their environment, and thus they are involved in cancer progression pathways, such as evasion of the immune system, generation of pro-tumoral niches, promotion of angiogenesis and tumor cell proliferation.23 For instance, recently Wang et al. demonstrated that healthy adipocytes incorporated hepatocarcinoma-derived exosomes. Then, those adipocytes changed their transcriptome and cytokine secretion patterns, and released exosomes creating a favorable microenvironment for tumor progression, enhancing angiogenesis and recruiting more macrophages.24
Nowadays, other synthetic delivery systems are employed, such as liposomes and polymeric nanoparticles, which are the most widely used25,26 However, both systems have enormous challenges: to evade the immune system, to increase the blood circulation time and to elevate their stability without causing secondary cytotoxic effects.27 Thus, exosomes are emerging as ideal candidates to deliver therapeutic drugs, proteins, nucleic acids or nanoparticles, providing several advantages compared with the liposomes or the polymer-based strategies. Firstly, they are naturally present in body fluids; therefore, they are stable in physiological conditions such as pH or temperature. Furthermore, they are less toxic and immunogenic compared to those synthetic vectors. Finally, they can deliver cargo to specific recipient cells due to their membrane proteins and lipids that are able to provide them with a fingerprint for the recognition and binding to specific receptors in the target cells. In particular, this characteristic is crucial to transport therapies to certain areas where polymeric or liposomes are unable to reach, such as the brain, via BBB. In fact only the 2% of the central nervous system drugs are capable of crossing the BBB with the conventional platforms, which determines their low efficacy and their limitations in the transference to clinical trials.28 Although further evidences are needed, some authors establish that exosomes may cross the BBB mainly following active endocytosis mechanisms.29,30 Furthermore, peripheral EVs can interact with the BBB leading to changes in the barrier's properties. A recent study, reports that in zebrafish neurons can remotely regulate blood–brain barrier integrity by delivering miR-132 through secretion of exosomes.31
For the development of exosome-based vehicles, the stability and the abundance of exosomes in body fluids would be very important points to consider.32 In two different studies, Sokolova et al. and Kalra et al., observed whether extracellular vesicles were stable at least for 3 months at 37 °C, 4 °C, −20 °C and −80 °C.33,34 Nevertheless, extracellular vesicles stored at −80 °C were highly stable compared with the other storage temperatures tested.33 To enable the development of novel exosome-based diagnostic and therapeutic tools, it is necessary to understand how modifications on the morphology and functionality of those vesicles might be made. Biological techniques (such as western blot, flow cytometry, polymerase chain reaction (PCR) or fluorescence microscopy) and physicochemical techniques (including dynamic light scattering (DLS), nanoparticle tracking analysis (NTA) or electronic microscopy) have to be employed to evaluate exosome content (proteins, lipids and nucleic acids) and also exosome morphology before and after customization.35 This characterization provides useful information on how exosome modification could affect to their trafficking internalization or targeting abilities in vivo (key factors involved in the efficacy of the encapsulated therapy within them).
In summary, exosomes are highly stable nanovesicles involved in a wide range of normal and pathological processes and can be isolated from almost every cell and are stable and able to remain for a long time in the majority of body fluids.32 Furthermore, it is known that exosomes secrete specific molecules to receptor cells and that they can reach different target tissues according to the characteristics of the cells from which they are originated, endowing them with exciting possibilities regarding selective targeting.36
However, there are major challenges to solve for the employment of exosomes as drug delivery agents in clinic. Firstly, a great difficulty for their translation to the clinic would be the low yield obtained with almost all the isolation methods, and also the presence of some impurities such as serum proteins or other extracellular vesicles.37,38 Secondly, when designing an exosome-based strategy, it is necessary to understand the implication of the exosomes in biological and pathological conditions in order to predict both short and long term safety issues and to increase the therapeutic effect of the exosomes cargo. Therefore, in vivo trafficking and behavior of exosomes as well as their interactions with the different organs need to be investigated more thoroughly before the translation of exosome-based strategies therapies which are currently performed all over the world.19 Nonetheless, 84 clinical trials involving exosomes for both therapy and diagnosis are currently performed around the globe.39
Overall, the important characteristics that makes exosomes suitable and feasible engineered vectors for the transport of theranostic tools are: (i) their content is protected by the lipid membrane. (ii) Their cargo and their membrane features are characteristic from the parental cell. (iii) They play an essential role in intercellular communication. (iv) They are highly stable in body fluids and (v) they can be stored for extended periods. For the reasons mentioned above, exosomes are ideal biological nanovectors for target therapeutic advanced therapies to specific pathological areas of the organism both for therapy and for diagnostic. In this review the recent combinations of genetic materials, viruses and nanotechnological tools together with exosomes are evaluated.
Exosomes to carry biological agents
Exosomes and transfer of proteins or nucleic acids
Introduction of therapeutic biomolecules within exosomes was not an easy task and several attempts were made. Haney et al. employed a passive incubating approach (incubation at room temperature without stirring or external forces) and a variety of active loading strategies (saponin, freeze–thaw cycles, sonication and extrusion) for the encapsulation of catalase (large protein of 240 kDa with antioxidant properties) in macrophage-derived exosomes. The encapsulation preserved its enzymatic activity, prolonged blood circulation time, and reduced immunogenicity, improving therapeutic efficacy.40 They observed that when using active approaches, they obtained higher efficiencies. When applying sonication pulses, the ultrasounds compromises the integrity of the exosomal membrane allowing the enzyme to diffuse into the exosomes without disturbing exosomal membrane proteins or lipids. In the case of freeze–thaw cycles, exosomes are rapidly frozen and then they are thawed at room temperature and even sometimes, it is heated upon 37 °C. Regarding to extrusion strategies, exosomes and enzymes are put in contact and the mixture is extruded in a 10–400 nm filter under controlled conditions. Finally, incubating exosomes with membrane permeabilizers allows the formation of transitory pores increasing membrane permeabilization.41Rather than proteins, nucleotides molecules are widely encapsulated in exosomes for the treatment of tumor or neurological diseases.42 For instance, Alvarez-Erviti et al. used electroporation to create small pores in exosome membranes in order to introduce siRNAs in dendritic cells derived exosomes for the treatment of Alzheimer disease. The loaded exosomes delivered a siRNA specifically to neurons, microglia, oligodendrocytes in the brain, knocking down BACE1 that is a therapeutic target in Alzheimer's disease.43 In other study, parental cells were engineered to express the transmembrane domain of epidermal growth factor receptor (EGFR). Then, as cancerous cells overexpress the epidermal growth factor (EGF), those exosomes were used therapeutically to target in vivo an specific antitumoral miRNA in tumor tissues.44 Recently, Han et al. improved the therapeutic effect of miR-675, which is downregulated in senescent cells, aging muscles and ischemic legs, by incorporating it into exosomes that were retained in a silk fibroin hydrogel. They observed a significantly improvement of the therapeutic effect of this mi-675 as antiaging factor and also preventing ischemic vascular dysfunction when it was encapsulating in exosomes.45
Exosomes to transfer viral material
Viruses are currently a leader way to direct gene therapy strategies to the target areas, especially in vivo. However, some limitations such as their high immunogenicity and some organ-specific barriers have led to the search for new virus-based vectors.46 Some strategies have tried to help by using liposomes or polymer-based vectors to offer a large hydrophilic lumen for the packaging of the virus.47 Although it was an interesting option, its high cytotoxicity and its difficulty on a large-scale production, have made of it an option to dismiss.For those reasons, the unique properties of EVs have encouraged to think about the novelty and advantages of the combination of viral vector systems together with exosomes, to solve the viral gene transfer issues.48–50 Interestingly, viruses can enter in the exosomes biogenesis pathway and incorporate its viral RNA genome, proteins, mRNAs and miRNAs.51 After the encapsulation, those exosomes would be able to transport the biologically active viral components from infected cells to the distant uninfected cells52–56 as it has been described how exosomes serve as protectors for virus miRNAs degradation.57
Regarding the type of virus to be transferred in exosomes, there is a subset of previous studies describing interactions of EVs with viruses such as picornavirus, retrovirus, flavivirus, parvovirus, or reovirus.58 It was demonstrated that dendritic cells infected with the human immunodeficiency virus (HIV) could secrete the virus out of the cells in association with exosomes.59–61 Also, in vitro exosomal transference of Epstein–Barr virus (EBV), human T-lymphotropic virus type 1 (HTLV-1), herpes simplex virus 1 (HSV-1) or Rift Valley fever virus (RVFV) mRNAs from the infected cells to the receptor cells was reported.62–66 But not only the mRNAs content was found in exosomes or EVs, as some authors have found the full-length viral RNA in some infections such as porcine reproductive and respiratory syndrome virus (PRRSV), foot-and-mouth disease virus (FMDV) and hepatitis A virus, as shown in Table 1.67,68
| Exosome origin (infected parental cell) | Virus | Viral genetic material | Function of viral material transfer | Reference |
|---|---|---|---|---|
| Hepatoma cells | HAV | Genomic RNA, viral particles | Viral spread | Longatti et al., 2015 (ref. 52) |
| Hepatoma cells | HBV | mRNA | Viral spread | Kapoor et al., 2017 (ref. 53); Yang et al., 2017 (ref. 54) |
| Hepatoma cells | HCV | Full-length genomic RNA | Viral spread | Ramakrishnaiah et al., 2013 (ref. 55); Dreux et al., 2012 (ref. 56) |
| Dendritic cells/lymphocytes | HIV-1 | Viral particles | Immune escape and viral spread | Wiley et al., 2006 (ref. 59); Arenaccio et al., 2015 (ref. 60); Sampey et al., 2016 (ref. 61) |
| Liver cancer cells | HSV-1 | Viral mRNA and microRNAs | Viral spread | Kalamvoki et al., 2014 (ref. 62) |
| T cells | HTLV-1 | mRNA and microRNAs | Viral spread | Jaworski et al., 2014 (ref. 63) |
| Nasopharyngeal carcinoma cells/lymphoblastoid B lymphocytes | EBV | mRNA and microRNAs | Immune escape and viral spread | Canitano et al., 2013 (ref. 64); Ahmed et al., 2014 (ref. 65); Pegtel et al., 2010 (ref. 57) |
| Kidney epithelial cells | RVFV | mRNA | Viral spread | Ashan et al., 2016 (ref. 66) |
| Kidney cells | FMDV | Genomic RNA | Inmune evasion | Zhang et al., 2019 (ref. 67) |
| Kidney cells | PRRSV | Genomic RNA | Viral spread an inmune evasion | Wang et al., 2018 (ref. 68) |
Those studies were the base for the use of EVs for transfer of viruses commonly used for gene therapy. Several groups documented a subpopulation of EV particles that could be employed as a carrier system to deliver oncolytic adenoviruses to human tumors.69,70 Moreover, several studies have tested the ability of a subpopulation of adenoassociated virus (AAV) vectors interacting with exosomes to deliver genes.71 Those exosomes loaded with adenoassociated virus (exo–AAV), contained AAV vectors encoding the gene of interest as well as vector encoded proteins. Those novel virus–exosome vectors open new approaches for gene therapy applications.
Therefore, new therapeutic approaches based on viral gene therapy mediated by exosomes were widely described, in particular, those studies focused on exosome production in virus-infected cells to exploit their increasing targeting efficacy. Koppers et al. were the first ones suggesting that viral factors incorporated into the appropriate delivery vesicles could increase virus-encoded RNA delivery and proposed to engineer exosomes to carry tissue specific virus-derived molecules.72 In another study, Maguire et al. employed stomatitis virus glycoprotein (VSVG) to enhance transgene delivery, pseudotyping extracellular vesicles with adeno-associated virus.73 Another virus glycoprotein, but from Rabies (RVG), was used for mirR-124 targeted delivery to the brain. In that work, the fusion of exosome protein LAMP2b, together with RVG, was used to modify exosomes, observing the knockdown expression of the target gene in a great area of the brain.74 Finally, VSVG9 was used to improve the ability of exosomes to transfer therapeutic proteins, showing decreased gene expression in U87 and 293T tumoral cells.75
Of all the viruses used on those approaches AAVs have taken most of the interest as they have been the preferential viruses used for gene therapy at preclinical or clinical trials, with many studies performed during decades that remarked their safety and efficacy. Indeed, its resistance neutralizing antibodies and the level of transgene expression was tested, and the primary results indicate that AAV vectors binding to exosomes increase transgene transfer levels as well as antibody resistance.72 The positive outcome obtained in those studies has led to subsequent studies to improve gene delivery of exo–AAV compared with non-enveloped AAV vectors in different tissues (brain, liver, heart, ear, and retina), which will be described in this review.
Most of the studies were performed applying systemic injection for the in vivo experiments, and they were able to transport transgenes to the distant uninfected organs and areas of disease. In the case of the brain, an in vivo model crossing the BBB using exo–AAV vectors was carried out. The in vivo intravenous delivery of exo–AAV vectors has been used to obtain successfully targeted transduction of interferon-beta cytokine in tumor stromal cells (astrocytes and tumor associated macrophages), contributing to the reduction of the tumor on mice.76 Along the same lines, other studies show a GFP detection-enhancement of exo–AAV8 and exo–AAV9 vs. their non-encapsulated controls in the brain after intravenous injection at low doses,77 representing a favourable approach for the genetic modification of the tumor microenvironment. Another organ in which exo–AAVs are actively used is the liver. In vivo efficiency of liver targeting of exo–AAV8 or exo–AAV5 vectors expressing human coagulation factor IX (hFIX) was checked on a mouse model of haemophilia B. An astonishing increase in hFIX transgene expression and hepatocyte distribution has been observed, especially at low vector doses, contributing to an exceptional correction of clotting time.78 When it comes to the heart, Liang et al. has detected a significant improved cardiac function in vivo when comparing AAV-containing exosomes vs. mice treated with regular AAV gene therapy, opening a window for cardiovascular diseases.79
However, other studies administered the exo-viral vector by local administration. Indeed cochlear vestibular hair cells have been transduced with exo–AAV1 and exo–AAV9 vectors both in vitro and in vivo,80 and a strengthen transduction of inner and outer cells in cochlear hair cells in culture has been found. Exo–AAV–LHFPL5 mediated expression has been studied in deaf mice, and surprisingly, after round window membrane (RWM) injection, a widespread expression of LHFPL5 throughout the cochlea and a restitution in sound and balance related abnormal movement are noticed. Thus, this study represents a novel strategy for rescue of inner-ear hearing. Apart from that, other studies have covered the intravitreal injection of exosomes in the retina of mice, and a powerful GFP expression has been detected around the optical disk, suggesting that intravitreal exosome-viral gene transfer could be a tool for basic research of the eye.81
Although the combination of exosomes with AAVs seems to be a promising strategy for high-efficient deliver gene therapy, the amount of the total exo–AAV vectors isolated from cell culture media is so low that still being a problem. In order to solve it, recent studies were focused on the design of approaches to increase exo–AAVs secretion, and they managed it by overexpressing CD9 in the parental cells.82 Exo–AAV–CD9 are more efficient in transduction of cells than standard exo–AAV, providing not only an excellent approach for the generation of exo–AAVs but also for stabilizing their interactions with AAVs. Moreover, the employment of AAV vectors has also other limitations including transgene's size limit or safety concerns.
However, although most of works focused on exosome production in virus-infected cells for targeting, the study of other applications of those virus–exosome complexes is an exciting new area of research. Applications as vaccines both for infective diseases83 or for cancer vaccines84 were studied. Currently, considering the evidences that antigens from some viruses from coronavirus family were found on plasma exosomes85 and taking into account the good results obtained with other coronavirus-exosomal vaccines86 there was a great interest in the development of vaccines for the new pandemic caused by SARS-CoV-2 virus. The speed of the investigations increased on this area and several groups are focused on testing exosomes for the development of an exosome-based vaccine for respiratory diseases with the first approaches, now just with empty vectors, in clinical trials.
In summary, preliminary results are promising, but some authors have claimed that preclinical studies with larger animal models are still necessary.57 Taking into account that several viruses mentioned above associate with extracellular vesicles and that many others export viral components such as mRNA, this field could be the subject of study of multiple gene therapy groups. In the future, the combination of viral vector systems together with exosomes could allow us to deliver a large amount of therapies into some more areas that are difficult to reach while supposing a resistant mechanism to inactivation through antibodies.
Exosomes and nanotechnology
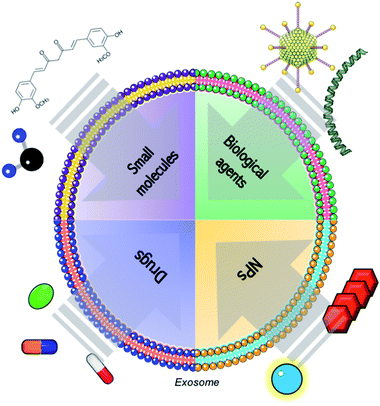
Even though most of the exosome-based tools consist in the combination of them with RNAs, proteins, viruses or chemotherapeutics, other types of advanced therapies can be loaded into those vesicles (Fig. 1).87Nanotechnology is considered the study, control and application of matter at nanoscale level and it has a large variety of applications, being nanomedicine one of the more important.88 This discipline applies the unique properties of the nanomaterials such as their reduced size and their high relation surface/volume ratio to develop novel tools for control, treatment and diagnosis of biological systems.89,90 In particular, nanotechnology provides artificial nanocarriers (such as PLGA NPs, liposomes, dendrimers, metallic and inorganic structures) to vehicle different types of therapies, including gene therapies, to pathological tissues.91 Although there are no doubts on the goals achieved in this field, some drawbacks still exist. There is a lack of knowledge about the toxicity and the in vivo interactions of those nanomaterials. This fact limits their further clinical translation due to their potential systemic toxicity and immunogenicity and further research is needed.92
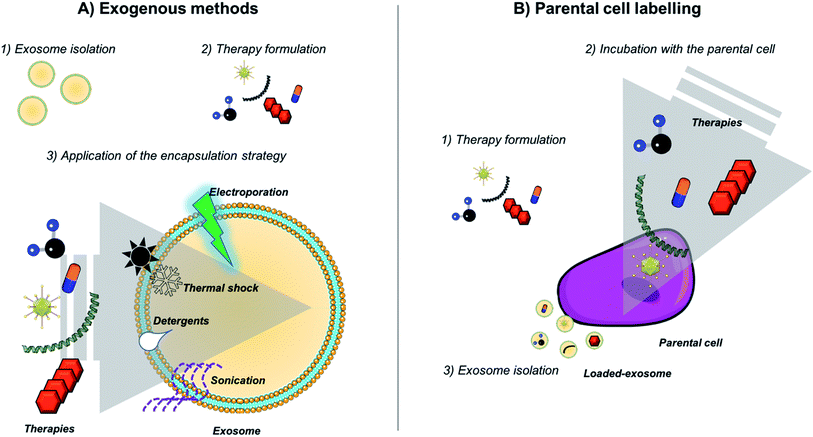
Extracellular vesicles and in particular exosomes, provides nanotechnology with robust and feasible biological nanovehicles. In particular, thanks to their long circulation time, small size, low immunogenicity and targeting capability, exosomes exhibit ideal characteristics as vectors to deliver therapeutic nanotechnological cargo to specific cells. Therefore, considering the exosomal characteristics previously mentioned, those vesicles constitute an emerging type of nanocarriers formed by a functional lipid membrane and a hydrophilic core. They provide a stable environment to transport theranostic nanomaterials avoiding the immune response and allowing them to fuse with the plasma membrane to enter directly in the target cells. Thus, in the last five years, several studies have been focused on the combination of nanotechnology with exosomes.93 Table 2 summarizes all the studies combing exosomes with nanotechnology. Fig. 2 shows a schematic representation of the different strategies widely employed for the loading of exosomes. Principally, two different approaches are being used to incorporate therapeutic agents into them (active or passive loading), resulting in different loading yields. The passive loading approach is the simplest way to incorporate nanoparticles and molecules within exosomes, and it consists in the incubation of the drug with the exosomes allowing them to enter following a concentration gradient. Although this method has very low loading efficiency (often less than 10%), a decade ago small lipophilic molecules were passively introduced in exosomes by incubating them at room temperature.94 Therefore, more complex molecules including anti-oxidants and anti-inflammatory compounds were loaded inside exosomes, even nanoparticles. For instance, Altanerova et al., showed that when labeling MSCs with carbohydrate-coated iron oxide nanoparticles, their released exosomes were loaded with those NPs. The magnetic nanoparticles packaged in MSCs-exosomes were successfully internalized by malignant cells and they served as magnetic hyperthermia antitumoral therapies killing cancerous cells in a dose-dependent manner.95
| Exosome origin (parental cell) | Type of advanced therapy | NPs size | Encapsulation strategy | Application | Reference |
|---|---|---|---|---|---|
| Mesenchymal stem cells | Venofer | 65 nm | Labelling parental cells | In vivo magnetic hyperthermia therapy | Altanerova et al., 2017 (ref. 95) |
| Adipose stem cells | USPIOs | 5 to 7 nm | Labelling parental cells | In vivo magnetic resonance imaging | Busato et al., 2016 (ref. 96) |
| Murine melanoma cells | SPIONs | 5 nm | Passive loading and electroporation | Labelling | Hood et al., 2014 (ref. 102) |
| Murine melanoma cell | SPIONs | 5 nm | Electroporation | In vivo magnetic resonance imaging | Hu et al., 2015 (ref. 103) |
| Glioma cells | SPIONs, curcumin and neuropilon-1 targeted peptide | 5 nm | Electroporation and click chemistry | In vivo targeted therapy and magnetic resonance imaging | Jia et al., 2018 (ref. 104) |
| Murine melanoma cells | PEG–AuNPs | 40–45 nm | Passive loading, electroporation, thermal shock, sonication, saponin-assisted loading and labelling parental cells | In vitro optical hyperthermia therapy and imaging by inherent reflection properties | Sancho-Albero et al., 2019 (ref. 107) |
| Placental stem cells | PEG–AuNPs | 40–45 nm | Labelling parental cells | Targeted in vitro optical hyperthermia therapy | Sancho-Albero et al., 2019 (ref. 14) |
| Mesenchymal stem cells | Glucose-coated AuNPs | From 5 nm to 20 nm | Active loading (internalized and/or attached to the external surface) | In vivo CT imaging | Betzer et al., 2017 (ref. 108) |
| PC-3 prostate cancer exosomes | AuNPs | 13 nm | Labelling parental cells | Nucleic acid delivery system (gene downregulating) | Alhasan et al., 2014 (ref. 109) |
| Breast adenocarcinoma cells | AuNPs with thiolted oligonucleotides | From 14 nm to 30 nm | Labelling parental cells | Nucleic acid delivery system (gene silencing) | Roma-Rodrigues et al., 2017 (ref. 110) |
| HeLa cells | MOFs | 250 nm | Fusion method | Smart and efficiency drug delivery | Illes et al., 2017 (ref. 112) |
| Lung cancer cells | Pd nanosheets | 2 nm | In situ generation inside exosomes | Targeted biorthogonal catalysis against cancer therapy | Sancho-Albero et al., 2019 (ref. 101) |
| Blood exosomes | Transferrin–SPIONs | 10 nm | Passive incubation | Cancer therapy | Qi et al., 2016 (ref. 105) |
| Genetically engineered mammalian cells (RVG peptide) | AuNPs | 48 nm | Extrusion | Enhancement of BBB penetration | Khongkow et al., 2018 (ref. 106) |
| Murine melanoma cells | Folic acid covered AuNPs–PEG | 12 nm | Labelling parental cells | Penetration of exosomes specifically in metastatic small tumors | Lara et al., 2020 (ref. 111) |
Adipose stem cell, primary neuronal cells or macrophages have been also incubated with iron nanoparticles (USPIOs or SPIONS) and then EVs loaded within them were purified in order to generate microvesicles for MRI imaging and photodynamic therapy in vivo.96–98 Although some authors suggest that the indirect labeling of source cells is not recommended for exosome loading,99–101 it is one of the most widely employed techniques to achieve the encapsulation of NPs within exosomes (together with electroporation strategies).
Active transport is also an alternative for the generation of nanoparticle–exosome hybrids. Hood et al. loaded 5 nm SPIONs nanoparticles by active transport and electroporation approaches for exosome loading. They observed that the application of a simple electroporation pulse in the presence of a sugar (trehalose) minimized B16-F10 derived exosomes aggregation and improved the loading of 5 nm diameter SPIONs within exosomes by electroporation.102 The same method was employed by Hu et al. observing the accumulation of the exosomes in lymph nodes in vivo and their suitable properties as MRI diagnostic tools.103
Moreover, the transfer of genetic material in exosome–nanoparticle vectors is still a possibility. Jia et al. created a multifunctional vector based on exosomes in which they encapsulated SPIONs, curcumin and neuropilon-1-targeted peptide for targeting and imaging specifically glioma cells in vivo. Firstly, they electroporated exosomes in the presence of SPIONs and curcumin and then they added the peptide by click chemistry following an EDC/NHS based method. The group successfully demonstrated the targeting capabilities of the modified exosomes toward glioma cells and their therapeutic effect in vivo.104
This year, Sancho-Albero et al., developed a breakthrough strategy to synthesize in situ the NPs directly inside the exosomes rather than encapsulating them. They incubated a Pd ionic precursor with the exosomes, introduced in the exosomes by passive diffusion toward their membrane. Afterwards, the Pd nanosheets were created successfully inside exosomes by the conversion of P2+ to P0 using CO as gaseous reducing agent. The mildness of the procedure yielded to catalytically active Pd nanoparticles loaded into exosomes. This new hybrid system mediated Pd-triggered dealkylation reactions inside cells and displays a preferential tropism and selectivity for their parental cells.101
Finally, instead of encapsulating nanoparticles into exosomes some studies reported the attachment of them by binding the NPs to the exosomal surface or by engineering the membrane of the vesicles to create new exosome-based nanoplatforms. As an example, Qi et al., created a dual-functional exosomes-based cluster by anchoring transferrin-conjugated SPIONs on exosome surface through interactions with the Tf–Tf receptor for cancer therapy.105 Rather than encapsulating AuNPs within exosomes and attaching them to their surface, some studies focused on the engineering of the exosomal membrane to decorate nanomaterials providing them with exosomal properties (stability and targeting). Khongkow et al. modified the AuNPs surface with brain-targeted exosomes following a serial extrusion approach. Thus, they equipped AuNPs with the functional lipidic membrane of exosomes demonstrating that the exosome modified AuNPs were internalized in vivo by brain cells thanks to the adhesion to the cell surface mediated by exosomes features.106
Concerning to the applications, mainly all the reported works focused on the combination of SPIONs with exosomes for diagnostic applications based on MRI or for therapy mediated by magnetic hyperthermia strategies, but they could have many other uses such as gene transfer added to them. Only few works reported the combination of exosomes with gold nanoparticles for both therapy and imaging applications. In one of them, authors encapsulated 40 nm of diameter hollow gold nanoparticles (capable to absorb in the NIR region) by different physicochemical methods including passive loading, electroporation, thermal shock, sonication, saponin-assisted loading and labelling parental cells. They reached relatively low encapsulations efficacies when using the physicochemical methods (probably attributed to the membrane damage) compared with the labeling of their parental cells.107 Specifically, encapsulations yields of 13.7%, 16.4%, 18.20%, 9.1%, 19.34% and 49.11% were obtained with passive and saponin assisted loading, two different thermal shock strategies, sonication and parental cell labeling, respectively. In other work, these authors encapsulate the same NPs within stem cells exosomes by incubating them with parental cells. They demonstrated that those exosomes maintain the tropism toward their source of cells, serving as specific and selective optical hyperthermia therapies.14 In the work reported by Betzer et al., exosomes were labeled directly with glucose coated gold nanoparticles (from 5 to 20 nm of diameter). AuNPs were incorporated on the exosomes assisted by a glucose-dependent active pathway. This new hybrid vector was then used for brain CT imaging in vivo.108 Alhasan et al., labelled parental cells (PC-3 prostate cancer cells) also with AuNPs conjugated to anti-miR21. Then, they isolated the exosomes from the supernatant and they transfected target cells, observing a selectively downregulation of miR21 (a miRNA related with the proliferation and progression of cancerous cells, which is significantly overexpressed in tumor tissue).109 Similarly, Roma-Rodrigues et al., also employed gold nanoparticles covered with oligonucleotides for selective silencing of RAB27A gene (crucial gene for the biogenesis and processing of exosomes) with a consequent decrease of exosomes production.110 This year, Lara et al., created a novel protocol for the incorporation of folic acid covered gold nanoparticles into exosomes derived from B16-F10 cells by incubating them with the parental cells. They observed a preferential tropism toward small metastatic tumors compared with other organs in vivo.111
Finally, not only iron and gold nanomaterials were incorporated into exosomes. Two years ago, Illes et al., combined the properties of metal–organic frameworks nanoparticles (MOFsNPs) for their structural, chemical diversity, high loading efficiency and intrinsic biodegradability, together with exosomes. In particular, they created a smart exosome-coated MOF with “onboard-trigger” combining the release mechanism of exosomes with the biodegradation of the nanocarrier.112
Nanoparticles have risen a great interest in several areas of the biomedical field. Many different approaches could be applied either for therapy (hyperthermia, gene therapy, drug delivery etc.) or for diagnostic (microfluidics, imaging etc.) offering a wide spectrum of options for the area of biomedicine. However, all these techniques have the same problems in common. Liver retention, lack of tropism to target areas and elimination during its circulation in the blood are factors that have prevented its transfer to the clinic. A large part of those problems could be solved by encapsulating those nanoparticles in vectors that bypassed those issues, such as exosomes.
Considering the attractiveness of combining the therapeutic and diagnostic applications that provide the nanotechnological tools and the ideal properties of exosomes as vehicles, it is not difficult to believe that in the following years exosome-based approaches could be translate into clinical settings. In fact, 84 clinical trials involving the use of exosomes were recorded (either active or completed). However, some drawbacks and limitations (such as more efficient purification strategies or the understanding of the interaction of exosomes with the living systems) have to be solved in order to successfully translate the findings obtained with animal models to the clinic.
Discussion
The use of exosomes as an ideal delivery system is attracting considerable attention on the last decade. Those natural carriers offer several advantages in terms of biocompatibility and specificity, an intrinsic long-term circulatory capability, low toxicity and ease of avoiding the immune system recognition and reducing clearance rates.81Furthermore, exosomes present a long-term accumulation in organs or tissues and specific tropism for some cell types.82 They exhibit an extraordinary ability to interact with and accumulate in target cells and are able to overcome various biological barriers such as the cytoplasmic membrane and the BB unlike other delivery systems, which makes them ideal as therapeutic delivery molecules.83
Taking all of this into account, exosomes are considered competitive targeted delivery vehicles for cellular or gene therapies with several advantages and a promising therapeutic value in the treatment of several disorders.113,114 However, the major drawbacks are the inefficiency on the extraction and isolation process, low encapsulation and loading efficiencies, low extraction yield, and potential delivery of unwanted cargo materials naturally present in the exosomes.84 Therefore, further research in this field is needed in order to standardize optimal methods for exosome isolation, purification and storage, avoiding lack of reproducibility. In fact, several methods for exosome extraction are labor intensive, complex and give low yield, issue that have been recently improved with the use of promising techniques such as magnetic adhesion and flow cytometry in addition to loading techniques such as sonication, electroporation and incubation. Nevertheless, offloading the content of exosome without distorting the structural integrity is very important in ensuring high loading and encapsulation efficiencies. This fact is particularly important in the case of using nanoparticles, because of their intrinsic size (2–50 nm) is too large to be encapsulated within exosomes. To solve this concern while achieving high encapsulations yields, some authors proposed the employment of passive methods in order to internalize an ionic precursor of the NPs followed by the in situ synthesis of the nanostructures directly inside the vesicles.101 Still early times for clinical trials but as the research in this field is growing very quickly, new information on preclinical studies would help to enhance the knowledge to be transfer to the clinic and to the market fairly soon.
Conflicts of interest
Authors declare no conflicts of interest.Acknowledgements
Authors would like to thank, Dr Jesús Santamaría and Dr Manuel Arruebo for their support and Jorge Alejo for his help in this review. We thank the Instituto de Salud Carlos III (PI19/01007 and COV20/00155) and ERC Advanced Grant (ERC-2016-ADG-742684-CADENCE) for the financial support. CIBER-BBN an initiative funded by the VI National R&D&i Plan 2008–2011 financed by the Instituto de Salud Carlos III with the assistance of the European Regional Development Fund. It was partially funded by the Aragon Government (T57_17R p) cofunded by Feder 2014–2020 “Building Europe from Aragon”. MS also acknowledge the Spanish Government for the receipt of a FPU predoctoral grant.Notes and references
- M. J. A. Wood, Nat. Rev. Drug Discovery, 2013, 12, 347–357 CrossRef PubMed.
- J. Meldolesi, Curr. Biol., 2018, 28, R435–R444 CrossRef CAS PubMed.
- M. Colombo, G. Raposo and C. Théry, Annu. Rev. Cell Dev. Biol., 2014, 30, 255–289 CrossRef CAS PubMed.
- B. J. Crenshaw, L. Gu, B. Sims and Q. L. Matthews, Open Virol. J., 2018, 12, 134–148 CrossRef CAS PubMed.
- S. Keller, J. Ridinger, A. K. Rupp, J. W. Janssen and P. Altevogt, J. Transl. Med., 2011, 9, 2–9 CrossRef PubMed.
- T. Skotland, K. Sandvig and A. Llorente, Prog. Lipid Res., 2017, 66, 30–41 CrossRef CAS PubMed.
- A. De Gassart, C. Ge, B. Fe and M. Vidal, Blood, 2015, 102, 4336–4345 CrossRef PubMed.
- G. Raposo and W. Stoorvogel, J. Cell Biol., 2013, 200, 373–383 CrossRef CAS PubMed.
- Y. Zhang, Y. Liu, H. Liu and W. H. Tang, Cell Biosci., 2019, 9, 1–18 CrossRef PubMed.
- X. Huang, T. Yuan, M. Tschannen, Z. Sun, H. Jaboc, M. Du, M. Liang, R. L. Dittmar, Y. Liu, M. Liand, M. Kohli, S. N. Thibodeau, L. Boardman and L. Wang, BMC Genomics, 2013, 14, 1–14 CrossRef PubMed.
- A. Waldenström and G. Ronquist, Circ. Res., 2014, 114, 315–324 CrossRef PubMed.
- B. K. Thakur, H. Zhang, A. Becker, I. Matei, Y. Huan, B. Costa-Silva, Y. Zheng, A. Hoshino, H. Brazier, J. Xiang, C. Williams, R. Rodriguez-Barrueco, J. M. Silva, W. Zhang, S. Hearn, O. Elemento, N. Paknejad, K. Manova-Todorova, K. Welte, J. Bromberg, H. Peinado and D. Lyden, Cell Res., 2014, 24, 766–769 CrossRef CAS PubMed.
- C. Rajagopal and K. B. Harikumar, Front. Oncol., 2018, 8, 1–13 CrossRef PubMed.
- M. Sancho-albero, N. Navascués, G. Mendoza, V. Sebastián, M. Arruebo, P. Martín-Duque and J. Santamaría, J. Nanobiotechnol., 2019, 17, 1–13 CrossRef PubMed.
- E. Willms, H. J. Johansson, I. Mäger, K. E. M. Blomberg, M. Sadik, A. Alaarg, C. I. E. Smith, J. Lehtiö, S. E. L. Andaloussi, M. J. A. Wood and P. Vader, Sci. Rep., 2016, 6, 1–12 CrossRef PubMed.
- J. Palma, S. C. Yaddanapudi, L. Pigati, M. A. Havens, S. Jeong, G. A. Winer, K. Mary, E. Weimer, B. Stern, M. L. Hastings, D. M. Duelli and S. Jeong, Nucleic Acids Res., 2012, 40, 9125–9138 CrossRef CAS PubMed.
- J. De Toro, L. Herschlik, C. Waldner and C. Mongini, Front. Immunol., 2015, 6, 1–12 CAS.
- F. Shahabipour, N. Barati, T. P. Johnston, G. Derosa, P. Maffioli and A. Sahebkar, J. Cell. Physiol., 2017, 232, 1660–1668 CrossRef CAS PubMed.
- D. Ha, N. Yang and V. Nadithe, Acta Pharm. Sin. B, 2016, 6, 287–296 CrossRef PubMed.
- G. Lachenal, K. Pernet-Gallay, M. Chivet, F. J. Hemming, A. Belly, G. Bodon, B. Blot, G. Haase, Y. Goldberg and R. Sadoul, Mol. Cell. Neurosci., 2011, 46, 409–418 CrossRef CAS PubMed.
- T. Xiao, W. Zhang, B. Jiao, C.-Z. Pan, X. Liu and L. Shen, Transl. Neurodegener., 2017, 6, 1–6 CrossRef PubMed.
- N. Amabile, P. E. Ratou, A. Tedgui and C. M. Boulanger, Semin. Thromb. Hemostasis, 2010, 36, 907–916 CrossRef CAS PubMed.
- C. Kahlert and R. Kalluri, J. Mol. Med., 2013, 91, 431–437 CrossRef CAS PubMed.
- S. Wang, M. Xu, X. Li, X. Su, X. Ciao, A. Keating and R. C. Zhao, J. Hematol. Oncol., 2018, 11, 1–14 CrossRef PubMed.
- L. Sercombe, T. Veerati, F. Moheimani, S. Y. Wu and S. Hua, Front. Pharmacol., 2015, 6, 1–13 Search PubMed.
- U. Agrawal, R. Sharma, M. Gupta and S. P. Vyas, Is nanotechnology a boon for oral drug delivery?, Drug Discovery Today, 2014, 19, 1530–1546 CrossRef CAS PubMed.
- K. Raemdonck, K. Braeckmans, J. Demeester and S. C. De Smedt, Chem. Soc. Rev., 2014, 43, 444–472 RSC.
- W. M. Pardridge, J. Cereb. Blood Flow Metab., 2012, 32, 1959–1972 CrossRef CAS PubMed.
- T. Druzhkova and A. A. Yakovlev, Neurochem. J., 2018, 12, 195–204 CrossRef CAS.
- C. Théry, A. I. Curie and U. Inserm, F1000 Biol. Rep., 2011, 8, 1–8 Search PubMed.
- Z. Zhao and B. Zlokovic, Cell Res., 2017, 27, 849–850 CrossRef CAS PubMed.
- S. Boukouris and S. Mathivanan, Proteomics: Clin. Appl., 2015, 9, 358–367 CAS.
- V. Sokolova, A. K. Ludwig, S. Jornung, O. Rotan, P. A. Horn, M. Epple and B. Giebel, Colloids Surf., B, 2011, 87, 146–150 CrossRef CAS PubMed.
- H. Kalra, C. G. Adda, L. Liem, C. S. Ang, A. Mechler, R. J. Simpson, M. D. Hulett and S. Mathivanan, Proteomics, 2013, 13, 3354–3364 CrossRef CAS PubMed.
- J. L. Hood, Nanomedicine, 2016, 11, 1745–1756 CrossRef CAS PubMed.
- S. Rana, S. Yue, D. Stadel and M. Zöller, Int. J. Biochem. Cell Biol., 2012, 44, 1574–1584 CrossRef CAS PubMed.
- Y. Toda, K. Takata, Y. Nakagawa, H. Kawakami, S. Fujioka, K. Kobayashi, Y. Hattori, Y. Kitamura, K. Akaji and E. Ashihara, Biochem. Biophys. Res. Commun., 2015, 456, 768–773 CrossRef CAS PubMed.
- K. E. Petersen, E. Manangon, J. L. Hood, S. A. Wickline, D. P. Fernandez, W. P. Johnson and B. K. Gale, Anal. Bioanal. Chem., 2014, 406, 7855–7866 CrossRef CAS PubMed.
- Clinical Trials, Pieejams, https://clinicaltrials.gov/ct2/results?term=Exosome%26cond=Cancer%26Search=Clear%26age_v=%26gndr=%26type=%26rslt=.
- M. J. Haney, N. L. Lyachko, Y. Zhao, R. Gupta, E. G. Plotnikova, Z. He, T. Patel, A. Piroyan, M. Sokolsky, A. V. Kabanov and E. V. Batrakova, J. Controlled Release, 2016, 21, 4062–4072 Search PubMed.
- I. Podolak, A. Galanty and D. Sobolewska, Phytochem. Rev., 2010, 9, 425–474 CrossRef CAS PubMed.
- M. Lu, H. Xing, Z. Xun, T. Yang, P. Ding, C. Cai, D. Wang and X. Zhao, Asian J. Pharm. Sci., 2018, 13, 1–11 CrossRef PubMed.
- L. Alvarez-Erviti, Y. Seow, H. Ying, C. Betts, S. Lakhal and M. J. A. Wood, Nat. Biotechnol., 2011, 29, 3–4 CrossRef PubMed.
- S. I. Ohno, M. Takanashi, K. Sudo, S. Ueda, A. Ishikawa, N. Matsuyama, K. Fujita, T. Mizutani, T. Ohgi, T. Ochiya, N. Gotoh and M. Kuroda, Mol. Ther., 2013, 21, 185–191 CrossRef CAS PubMed.
- C. Han, J. Zhou, B. Liu, C. Liang, X. Pan, Y. Shang, Y. Chang, Y. Wang, L. Shao, B. Shu, J. Wang, Q. Yin, X.-Y. Yu and Y. Li, Mater. Sci. Eng., C, 2019, 99, 322–332 CrossRef CAS PubMed.
- Y. Wang, Front. Immunol., 2018, 9, 1–8 CrossRef PubMed.
- Y. Wang, H. Huang, H. Zou, X. Tian, J. Hu, P. Qiu, H. Hu and G. Yan, Mol. Pharm., 2019, 16, 779–785 CrossRef CAS PubMed.
- S. G. van der Grein, K. A. Y. Defourny, E. F. J. Slot and E. N. M. N. Hoen, Semin. Immunopathol., 2018, 40, 491–504 CrossRef PubMed.
- Z. Yao, Y. Qiao, X. Li, J. Chen, J. Ding, L. Bai, F. Shen, B. Shi, J. Liu, L. Peng, J. Li and Z. Yuan, J. Virol., 2018, 92, 1–21 CrossRef PubMed.
- E. Nolte, T. Cremer, R. C. Gallo and L. B. Margolis, PNAS, 2016, 113, 9155–9161 CrossRef PubMed.
- M. Drahansky, Intech, 2016, 1, 13 Search PubMed.
- A. T. Longatti, Viruses, 2015, 7, 6707–6715 CrossRef CAS PubMed.
- N. Kapoor and R. Chadha, Virus Res., 2017, 14, 166–174 CrossRef PubMed.
- Y. Yang, Q. Han, Z. Hou, C. Zhang, Z. Tian and J. Zhang, Cell. Mol. Immunol., 2016, 13, 1–11 CrossRef PubMed.
- V. Ramakrishnaiah, C. Thumann, I. Fofana, F. Habersetzer, Q. Pan, P. E. de Ruiter, R. Willemsen, J. A. Demmers, V. S. Waj, G. Jenster, J. Kwekkeboom, H. W. Tilanus, B. L. Haagmans, T. F. Baumert and L. J. van der Laan, PNAS, 2013, 110, 13109–13113 CrossRef CAS PubMed.
- M. Dreux, U. Garaigorta, B. Boyd, E. Décembre, J. Chung, C. Whitten-bauer, S. Wieland and F. V. Chisari, Cell Host Microbe, 2013, 12, 558–570 CrossRef PubMed.
- D. M. Pegtel, K. Cosmopoulos, D. A. Thorley-lawson and M. A. J. Van Eijndhoven, PNAS, 2010, 107, 1–6 CrossRef PubMed.
- B. György and C. A. Maguire, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 10, 1–13 Search PubMed.
- R. D. Wiley and S. Gummuluru, PNAS, 2006, 2005, 1–6 Search PubMed.
- C. Arenaccio, S. Anticoli, F. Manfredi, C. Chiozzini, E. Olivetta and M. Federico, Retrovirology, 2015, 12, 1–17 CrossRef PubMed.
- G. C. Sampey, M. Saifuddin, A. Schwab, R. Barclay, S. Punya, M.-C. Chung, X. R. M. Hakami, M. A. Zadeh, B. Lepene, Z. A. Klase, N. El-hage, M. Young, S. Iordanskiy and F. Kashanchi, J. Biol. Chem., 2016, 291, 1251–1266 CrossRef CAS PubMed.
- M. Kalamvoki, T. Du and B. Roizman, PNAS, 2014, 111, 4991–4996 CrossRef PubMed.
- E. Jaworski, A. Narayanan, R. Van Duyne, S. Shabbeer- Meyering, S. Iordanskiy, M. Saifuddin, R. Das, P. V. Alfonso, G. C. Sampey, M. Chung, A. Popratiloff, B. Shrestha, M. Shegal, P. Jain, A. Vertes, R. Mahieux and F. Kashanchi, Biol. Chem., 2014, 289, 22284–22305 CrossRef CAS PubMed.
- A. Canitano, G. Venturi, M. Borghi, M. Grazia and S. Fais, Cancer Lett., 2013, 337, 193–199 CrossRef CAS PubMed.
- W. Ahmed, P. S. Philip, S. A. Tariq and G. Khan, PLoS One, 2014, 9, e99163 CrossRef PubMed.
- N. A. Ahsan, G. C. Sampey, B. Lepene, Y. Akpamagbo, R. A. Barclay, S. Iordanskiy, R. M. Hakami and F. Kashanchi, Front. Microbiol., 2016, 7, 139 CrossRef PubMed.
- K. Zhang, S. Xu, X. Shi, G. Xu, C. Shen, X. Liu and H. Zheng, Vet. Microbiol., 2019, 233, 164–173 CrossRef CAS PubMed.
- T. Wang, L. Fang, F. Zhao, D. Wang and S. Xiao, J. Virol., 2018, 92, e01734–17 CAS.
- L. Ran, X. Tan, Y. Li, H. Shang, R. Ma, T. Ji, W. Dong, T. Tong, Y. Liu, D. Chen, X. Yin, X. Liang, K. Tang, J. Ma, T. Y. Zhang, X. Cao, Z. Hu, X. Win and B. Huang, Biomaterials, 2016, 89, 56–66 CrossRef CAS PubMed.
- H. Saari, T. Turunen, A. Lõhmus, M. Turunen, M. Jalasvuori, S. J. Butcher, S. Ylä-herttuala, T. Viitala, V. Cerullo, R. Pia, M. Siljander and M. Yliperttula, J. Extracell. Vesicles, 2020, 9, 1–24 Search PubMed.
- Z. Fitzpatrick, M. H. W. Crommentuijn, D. Mu and C. A. Maguire, Biomaterials, 2014, 35, 7598–7609 CrossRef PubMed.
- D. Koppers-lalic, M. M. Hogenboom, J. M. Middeldorp and D. M. Pegtel, Adv. Drug Delivery Rev., 2013, 65, 348–356 CrossRef CAS PubMed.
- C. A. Maguire, L. Balaj, S. Sivaraman, M. H. Crommentujin, M. Ericsson, L. Mincheva-Nilsson, V. Baranoc, D. Gianni, B. A. Tannous, M. Sena-Esteves, X. O. Breakefield and J. Skog, Mol. Ther., 2012, 20, 960–971 CrossRef CAS PubMed.
- L. Alvarez-Erviti, Y. Seow, H. Yin, C. Betts, S. Lakhal and M. J. A. Wood, Nat. Biotechnol., 2011, 29, 341–345 CrossRef CAS PubMed.
- C. Meyer, J. Losacco, Z. Stickney, L. Li, G. Marriott and B. Lu, Int. J. Nanomed., 2017, 12, 3153–3170 CrossRef CAS PubMed.
- A. Volak, S. G. LeRoy, J. S. Natasan, D. J. Park, P. S. Cheah, A. Maus, Z. Fitzpatric, E. Hudry, K. Pinkham, S. Gandhi, B. T. Hyman, D. Mu, D. GuhaSarkar, A. O. Stemmer-Tachmimov, M. Sena-Esteves, C. E. Badr and C. A. Maguire, J. Neuro-Oncol., 2018, 139, 293–305 CrossRef CAS PubMed.
- E. Hudry, C. Martin, S. Gadhi, B. Gyögy, B. I. Sheffer, D. Mu, S. F. Merkel, F. Mingozzi, Z. Fitzbatrick, H. Dimant, M. Masek, T. Ragan, S. Tan, A. R. Brisson, S. H. Ramirez, B. T. Hyman and X. A. Maguire, Gene Ther., 2016, 23, 380–392 CrossRef CAS PubMed.
- A. Meliani, F. Boisgerault, Z. Fitzpatrick, S. Marmier, C. Leborgne, F. Collaud, M. S. Sola, S. Charles, G. Ronzitti, A. Vignaud, L. van Wittenbergcje, M. Marolleau, F. Jouen, S. Tan, O. Boyer, O. Christophe, A. R. Brisson, C. A. Maguire and F. Mingozzi, Blood Adv., 2017, 1, 2019–2031 CrossRef CAS PubMed.
- Y. Liang, P. Mathiyalagan, E. Kohlbrenner, E. Chepurko, D. Jeong, D. ceholski, N. Dubois, R. Hajjar and S. Sahoo, Circulation, 2017, 136, A15439 Search PubMed.
- B. György, C. Sage, A. A. Indshykulian, D. O. Sscheffer, A. R. Brisson, S. Tan, C. Wu, A. Volak, D. Mu, P. I. Tamvakologos, Y. Li, Z. Fitspatrick, M. Ericsson, X. O. Breakefield, D. P. Corey and C. A. Maguire, Mol. Ther., 2017, 25, 379–391 CrossRef PubMed.
- S. J. Wassmer, L. S. Carvalho, B. György, L. H. Vandenberghe and C. A. Maguire, Sci. Rep., 2017, 7, 1–10 CrossRef PubMed.
- L. T. Schiller, N. Lemus-Diaz, R. Rinaldi Ferreira, K. O. Böker and J. Gruber, Mol. Ther.--Methods Clin. Dev., 2018, 9, 278–287 CrossRef CAS PubMed.
- C. M. Bliss, A. J. Parsons, R. Nachbagauer, J. R. Hamilton, F. Cappuccini, M. Ulaszewska, J. P. Webber, A. Clayton, A. V. S. Hill and L. Coughlan, Mol. Ther.--Methods Clin. Dev., 2020, 16, 108–125 CrossRef CAS PubMed.
- Z. C. Hartman, J. Wei, O. K. Glass, H. Guo, G. Lei, X.-Y. Yang, T. Osada, A. Hobeika, A. Delcayre, J.-B. Le Pecq, M. A. Morse, T. M. Clay and H. K. Lyerly, Vaccine, 2011, 29, 9361–9367 CrossRef CAS PubMed.
- M. Gunasekaran, S. Bansal, R. Ravichandran, M. Sharma, S. Perincheri, F. Rodriguez, R. Hachem, C. E. Fisher, A. P. Limaye, A. Omar, M. A. Smith, R. M. Bremner and T. Mohanakumar, J. Heart Lung Transplant., 2020, 39, 379–388 CrossRef PubMed.
- S. Kuate, J. Cinatl, H. W. Doerr and K. Uberla, Virology, 2007, 362, 26–37 CrossRef CAS PubMed.
- K. B. Johnsen, J. M. Gudbergsson, M. N. Skov, K. Pilgaard, T. Moos and M. Doroux, Biochim. Biophys. Acta, Rev. Cancer, 2014, 1, 75–87 CrossRef PubMed.
- A. D. Maynard, Ann. Occup. Hyg., 2007, 51, 1–12 CAS.
- S. M. Moghimi, A. Christy Hunter and J. Clifford Murray, FASEB J., 2005, 19, 311–330 CrossRef CAS PubMed.
- O. V. Salata, J. Nanobiotechnology, 2004, 6, 1–6 Search PubMed.
- J. K. Patra, G. Das, L. F. Fraceto, E. V. R. Campos, M. P. Rodriguez-Torres, L. S. A.-T. R. Grillo, M. K. Swamy, S. Sharma, S. Habtemariam and H.-S. Shin, J. Nanobiotechnology, 2018, 16, 1–33 CrossRef PubMed.
- Y. Min, J. M. Caster, M. J. Eblan and A. Z. Wang, Chem. Rev., 2015, 115, 11147–11190 CrossRef CAS PubMed.
- B. Yang, Y. Chen and J. Shi, Adv. Mater., 2019, 31, 1–33 Search PubMed.
- A. Kalani, A. Tyagi and N. Tyagi, Mol. Neurobiol., 2009, 19, 389–399 Search PubMed.
- U. Altanerova, M. Babincova, P. Babinec, K. Benejova, J. Jakubechova, V. Altanerova, M. Zduriencikova, V. Rapiska and C. A. Taner, Int. J. Nanomed., 2017, 12, 7923–7936 CrossRef CAS PubMed.
- A. Busato, R. Bonafede, P. Bontempi, I. Scambi, L. Schiaffino, D. Benati, M. Malatesta, A. Sbarbati, P. Marzola and R. Mariotti, Curr. Protoc. Cell Biol., 2017, 75, 3–44 Search PubMed.
- J. Neubert and J. Glumm, Neural Regener. Res., 2016, 11, 61–63 CrossRef CAS PubMed.
- A. K. Silva, N. Luciani, F. Gazeaou, K. Aubertin, S. Bonneau, C. Chauvierre, D. Letourneur and C. Wihelm, Nanomedicine, 2015, 11, 645–655 CrossRef CAS PubMed.
- C. Liu and C. Su, Theranostics, 2019, 9, 1015–1028 CrossRef CAS PubMed.
- X. Luan, K. Sansanaphongpricha, I. Myers, H. Chen, H. Yuan and D. Sun, Acta Pharmacol. Sin., 2017, 38, 754–763 CrossRef CAS PubMed.
- M. Sancho-Albero, B. Rubio-Ruiz, A. M. Pérez-López, V. Sebastián, P. Martín-Duque, M. Arruebo, J. Santamaría and A. Unciti-Broceta, Nat. Catal., 2019, 2, 864–872 CrossRef CAS PubMed.
- J. L. Hood, M. J. Scott and S. A. Wickline, Anal. Biochem., 2014, 448, 41–49 CrossRef CAS PubMed.
- L. Hu, S. A. Wickline and J. L. Hood, Anal. Chem., 2015, 25, 368–379 Search PubMed.
- G. Jia, Y. Han, Y. An, Y. Ding, C. H. X. Wang and W. Tang, Biomaterials, 2018, 178, 302–316 CrossRef CAS PubMed.
- H. Qi, C. Liu, L. Long, Y. Ren, S. Zhang, X. Chang, X. Wian, H. Jia, J. Zhao, J. Sun, X. Hou, X. Yuang and C. Kang, ACS Nano, 2016, 10, 3323–3333 CrossRef CAS PubMed.
- M. Khongkow, T. Yata and S. Boonrungsiman, Sci. Rep., 2019, 9, 1–9 CrossRef CAS PubMed.
- M. Sancho-Albero, M. M. Encabo-Berzosa, M. Beltrán-Visiedo, L. Fernández-Messina, V. Sebastián, F. Sánchez-Madrid, M. Arruebo, J. Santamaría and P. Martín-Duque, Nanoscale, 2019, 11, 18825–18836 RSC.
- O. Betzer, N. Perets, A. Angel, M. Motiei, T. Sadan, G. Yadid, D. Offen and R. Popoctzer, ACS Nano, 2017, 11, 10883–10893 CrossRef CAS PubMed.
- A. H. Alhasan, P. C. Patel, C. H. J. Choi and C. A. Mirkin, Small, 2009, 6, 247–253 Search PubMed.
- C. Roma-Rodrigues, F. Pereira, A. P. Aves de Matos, M. Fernandes, P. V. Baptista and A. R. Fernandes, Nanomedicine, 2017, 13, 1389–1398 CrossRef CAS PubMed.
- P. Lara, S. P. Florez, E. S. Huenuleo, I. Polakovicova, S. Guerrero, L. L. Gonzalez, A. Campos, L. Muñoz, C. J. Cordero, M. V. Godoy, J. Cancino, E. Arias, J. Villegas, L. J. Cruz, F. Albericio, E. Araya, A. H. Corvalan, A. F. G. Quest and M. J. Kogan, J. Nanobiotechnology, 2020, 18, 1–17 CrossRef PubMed.
- B. Illes, P. Hirshle, S. Barnert, V. Cauda, S. Wuttke and H. Engelke, Chem. Mater., 2017, 29, 8042–8046 CrossRef CAS.
- W. J. Goh, S. Zou, W. Y. Ong, F. Torta, A. F. Alexandra, R. M. Schiffelers, G. Storm, J. W. Wang, B. Czarny and G. Pastorin, Sci. Rep., 2017, 7, 1–10 CrossRef CAS PubMed.
- P. Akuma, O. D. Okagu and C. C. Udenigwe, Front. Sustain. Food Syst., 2019, 3, 1–8 CrossRef.
| This journal is © The Royal Society of Chemistry 2020 |