Open Access Article
Open Access ArticleSensitive and enzyme-free fluorescence polarization detection for miRNA-21 based on decahedral sliver nanoparticles and strand displacement reaction†
Qingyue Zhu,
Hui Li and
Danke Xu *
*
State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, Jiangsu 210046, China. E-mail: xudanke@nju.edu.cn
First published on 30th April 2020
Abstract
A highly sensitive method for miRNA-21 analysis and detection has been developed, which relied on the enhanced effect of Ag10NPs, combined with the principle of strand displacement reaction to achieve asymmetric signal amplification. The detection probe was composed of Ag10NPs and two self-assembled complementary nucleic acid strands by thiol groups. The nucleic acid strand with a fluorescent group (named 2-FAM) was initially complementary to the chain in contact with Ag10NPs (named 1-SH), then it was replaced by a strand displacement reaction with miRNA-21, which exposed the toehold to react with the nucleic acid strand (named 3-fuel), and miRNA-21 was finally released into the next cycle of amplification, which eventually results in a significant increase in the fluorescence polarization signal change value. The linear range of this method was 100 pM to 16 nM while the detection limit was 93.8 pM, fast and sensitive detection could be achieved in 40 min. This method showed a certain anti-interference ability in the buffer and real biological samples, and achieved fast, accurate and sensitive detection.
Introduction
MiRNAs are endogenous non-coding and single-stranded RNA with a length of 18–23 nucleotides (nt). Mature miRNAs are combined with functional proteins to form an RNA-induced silencing complex (RISC), by identifying the corresponding mRNA, it can lead to degradation of the mRNA and decrease of the corresponding protein expression level.1,2 MiRNAs usually play an important role in many cell life activities, such as cell metabolism, apoptosis, and differentiation.3 What has attracted many researchers' interest is that the content of miRNAs will fluctuate during the occurrence and development of many cancers,4 some miRNAs are up-regulated such as miRNA-21, and some are down-regulated such as let-7a.1,5 Thus, miRNAs were regard as biomarkers for cancers' early diagnosis. As one of the earliest miRNAs found in human genome, miRNA-21 has high expression level in many kinds of tumor cells, including breast cancer, lung cancer and so on.6,7 Therefore, miRNA-21 is a recognized oncogene closely related to tumor, accurate and sensitive detection of miRNA-21 is very valuable for cancer diagnosis and treatment.The common detection methods of miRNA included northern blotting, microarrays, qRT-PCR and so on.8–10 Due to the distinguishing features such as short sequence, strong homology, and easy degradation of miRNAs, many existing detection methods still have certain limitations such as the dependence on complex steps, long time and thermal cycling,11 so in recent years, several detection methods were reported.12–14 Methods based on isothermal amplification like exponential amplification reaction (EXPAR), loop-mediated isothermal amplification (LAMP) and rolling circle amplification (RCA) can work without thermal cycling and amplify the targets to a significant quantity.15 Furthermore, through the construction of sophisticated biosensors, the signals for detecting various miRNAs can be presented by SERS,16 fluorescence,17 colorimetric assays18 and other methods in buffers, biological samples, cells and even living tissues.19,20 Among these methods, the fluorescence polarization (FP) assay is a reliable detection method for homogeneous systems by “mixing and reading” and sample separation was not required, it also has some advantages of high throughput, stability, sensitivity, fast and low-cost.21 Therefore, the fluorescence polarization method is suitable for different research such as exploring the interaction between molecules and the detection of contaminants in food and water.
The signal intensity of fluorescence polarization is related to the motion state of the fluorescent molecules, if the rate of molecular inversion or rotation is slow, the FP signal is high.22 Nanomaterials were usually used to improve sensitivity of fluorescence polarization because of the mass/volume, fluorophores are connected to the nanomaterials by means of nucleic acid chain assembly or adsorption, so that the rotation speed of the fluorophores significantly decreased, resulting in greatly increased fluorescence polarization.23,24 Silver nanoparticles have a unique optical effect due to the strong surface plasmon resonance (LSPR) phenomenon and metal enhanced fluorescence (MEF) effect, previous literature studies have shown that it can enhance the fluorescence intensity and the fluorescence polarization signal of fluorophores within a certain range.25 The decahedral silver nanoparticles (Ag10NPs) show the characteristics of NaCl tolerance, which can better combine with ssDNA through self-assembly of 5′ thiol group under the action of NaCl and keep stable,26 so the Ag10NPs can be used to fabricate fluorescent polarization detection probes. Several nucleic acid amplification techniques such as strand displacement reaction (SDR), hybridization chain reactions (HCR), and catalytic hairpin self-assembly reactions (CHA) have shown great effects and work well with nanomaterials.27–30 Strand displacement reaction (SDR) is an enzyme-free nucleic acid amplification techniques which does not need to be affected by enzyme activity and also refrains from the effect of enzymes and it is also an emerging hotspot.
In this report, an enzyme-free assay with decahedral silver nanoparticles (Ag10NPs) enhanced and strand displacement reaction (SDR) cyclically amplified fluorescence polarization detection has been developed. A detection probe was composed of Ag10NPs and two self-assembled complementary nucleic acid strands by thiol group, with the help of another fuel chain, a cycle of miRNA-21 was designed and a significant increase in the fluorescence polarization signal change value occurred. This assay achieved accurately and rapidly detect miRNA-21 in buffer and biological samples, and hope to play a certain role in the early diagnosis of cancer in the future.
Experimental
Materials and reagents
Trisodium citrate, sodium chloride (NaCl), Tween were got from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Ammonium persulfate (APS), tetramethylethylenediamine (TEMED), polyvinylpyrrolidone (PVP, MW = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), L-arginine, silver nitrate (AgNO3) and sodium borohydride (NaBH4) were purchased from Sigma-Aldrich (St. Louis, USA), 1× TBE (89 mM Tris, 89 mM boric acid, and 2 mM EDTA pH 8.2), 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4·12H2O, 2 mM KH2PO4, pH 7.4), 30% (w/v) acrylamide/methylene bisacrylamide solution, DNA molecular weight marker Marker A (25–500 bp) and diethypyrocarbonate (DEPC) water were purchased from Sangon Biotech Co., Ltd. (Shanghai, China), 2× SYBR® Premix Ex TaqTM was purchased from Takara Corporation (Japan), HEK-293 and MCF-7 cells were purchased from KeyGEN Biotech (Nanjing, China). The total RNA extraction kit was purchased from Omega BioTek (Norcross, USA). All the DNA used in the experiment were synthesized in Sangon Biotech Co., Ltd. (Shanghai, China), purified by high performance liquid chromatography (HPLC) and checked for tag concentration with the help of NanoDrop 2000. The main detection object was miRNA-21, and miRNA-7, miRNA-141, let-7a were used for control experiments, the nucleic acid sequences used are listed in Table 1. The above reagents were of analytical grade unless otherwise specified.
000), L-arginine, silver nitrate (AgNO3) and sodium borohydride (NaBH4) were purchased from Sigma-Aldrich (St. Louis, USA), 1× TBE (89 mM Tris, 89 mM boric acid, and 2 mM EDTA pH 8.2), 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4·12H2O, 2 mM KH2PO4, pH 7.4), 30% (w/v) acrylamide/methylene bisacrylamide solution, DNA molecular weight marker Marker A (25–500 bp) and diethypyrocarbonate (DEPC) water were purchased from Sangon Biotech Co., Ltd. (Shanghai, China), 2× SYBR® Premix Ex TaqTM was purchased from Takara Corporation (Japan), HEK-293 and MCF-7 cells were purchased from KeyGEN Biotech (Nanjing, China). The total RNA extraction kit was purchased from Omega BioTek (Norcross, USA). All the DNA used in the experiment were synthesized in Sangon Biotech Co., Ltd. (Shanghai, China), purified by high performance liquid chromatography (HPLC) and checked for tag concentration with the help of NanoDrop 2000. The main detection object was miRNA-21, and miRNA-7, miRNA-141, let-7a were used for control experiments, the nucleic acid sequences used are listed in Table 1. The above reagents were of analytical grade unless otherwise specified.
| Name | Sequence |
|---|---|
| 1-SH | 5′-SH-AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAATAGCTTATCAGAC-3′ |
| 2-FAM | 5′-FAM-TCAACATCAGTCTGATAAGCTATTTTT-3′ |
| 3-Fuel | 5′-AAAAATAGCTTATCAGACTGGAT-3′ |
| miRNA-21 | 5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| miRNA-7 | 5′-UGGAAGACUAGUGAUUUUGUUGU-3′ |
| miRNA-141 | 5′-UAACACUGUCUGGUAAAGAUGG-3′ |
| Let-7a | 5′-UGAGGUAGUAGGUUGUAUAGUU-3′ |
Apparatus
Four LED blue spotlights (7 W, Changzhou, China) were used for the synthesis of decahedral sliver nanoparticles, NanoDrop 2000 (BioTek) was used to measure the concentrations of nucleic acids, the BioTek spectrometer (Synergy H1, USA) was used to collect the fluorescence polarization signals, and the transmission electron microscope (TEM, JEM-200CX, Japan) was used to collect TEM images of the material. Bio-Rad Bole gel imaging system was used to image polyacrylamide gels.Synthesis and functional modification of Ag10NPs
Ag10NPs used in this work were synthesized and characterized by way of previously reported by our group31 and the characterization of Ag10NPs were shown in Fig. S1.† with 55.3 ± 4.0 nm diameters. The nucleic acid strands 1-SH (20 μM, 10.8 μl) and 2-FAM (20 μM, 10.8 μl) were mixed and heated at 95 °C for 5 min, placed on ice for 5 min, then 10 μl 10% Tween and Ag10NPs (500 μl, 0.56 nM) were added and vortexed well. After homogenization, 108 μl 2 M NaCl solution were added to promote the modification (final concentration was 0.3 M), then the mixture was placed in a 37 °C constant temperature shaker for 4 h and kept at room temperature for at least 24 h to obtain a functionally modified probe for the miRNA-21 detection. The probe needed to be centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and washed twice by phosphate buffer before use to ensure the best performance.
000 rpm and washed twice by phosphate buffer before use to ensure the best performance.
MiRNA-21 fluorescence polarization signals detection
The Ag10NPs probes were centrifuged, washed with 1× PBS and resuspend, then 15 μl probes, 100 μl miRNA solution and 5 μl 2 μM 3-fuel solution were added in a well of a 96-well plate and incubated at 37 °C in a constant temperature shaker for 40 min. The fluorescence polarization signals of the reaction solution were measured with a BioTek spectrometer (Synergy H1, USA) and the excitation wavelength was set to 480 nm and the emission wavelength was set to 520 nm.Gel electrophoresis analysis
The polyacrylamide gel concentration was 4–20%, the samples were run at 140 V for 30 min, the final concentration of each single strand was 125 nM, and the total volume was 12 μl for each lane. Before the PAGE experiment, the samples were placed in a constant temperature shaker at 37 °C and 220 rpm for 20 min. The buffer used in the gel electrophoresis experiments was 1× TBE (89 mM Tris, 89 mM boric acid, and 2 mM EDTA, pH 8.2).Preparation of total RNA extract
Human embryonic kidney cells HEK-293 and human breast cancer cells MCF-7 (KeyGEN Biotech, Nanjing, China) were grown in a clean incubator at 37 °C with a CO2 content of 5%. The medium was a DMEM high-sugar medium containing 100 U ml−1 penicillin, 100 μg ml−1 streptomycin (KeyGEN Biotech, Nanjing, China) and 10% fetal bovine serum (FBS, Gibco, Scotland, UK). Total RNA was extracted by the Total RNA Extraction Kit (Omega, Norcross, USA). Before the total RNA extraction, the medium in the culture dish was aspirated, and the TRK lysis buffer containing mercaptoethanol was directly added to the culture dish to obtain a cell lysate. In the immediate aftermath of the column of the Total RNA Extraction Kit was centrifuged and washed several times, the total RNA extracts of the two cells were obtained after appropriate dilution in DEPC water. NanoDrop 2000 was used to detect the concentration, and the extras were immediately put in a −80 °C refrigerator after indicating the label.Results and discussion
Detection principle
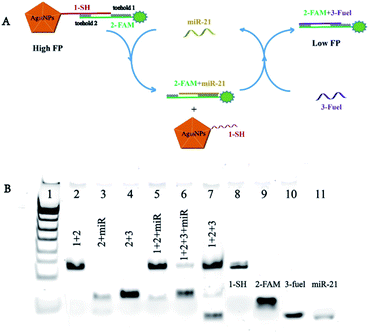
The assay for miRNA-21 detection was amplificated by Ag10NPs and cyclic strand displacement reaction (SDR), the detection principle was showed in Fig. 1A. In recent studies, a T7 exonuclease assisted dual-cycle signal amplification assay for sensitive detection of miRNA-141 by nanospheres-enhanced fluorescence polarization detection was proposed,32 comparing to enzyme-assisted assay, the enzyme-free amplification did not need to be affected by enzyme activity, so the application scenario could be further broadened.27 This enzyme-free process we designed mainly involved three nucleic acid chains: the chain named 1-SH connected to Ag10NPs through a 5′ thiol group, the chain named 2-FAM complementary paired with 1-SH and a 3-fuel chain were designed to initiate a toehold mediated strand displacement reaction (TMSD).33 The probe consisted of three parts: Ag10NPs, 1-SH and 2-FAM. At the beginning, the FAM fluorescent group and Ag10NPs maintained a certain distance to ensure that the metal enhanced fluorescence effect on fluorescence polarization can reach the best effectiveness. At the same time, Ag10NPs was connected to the fluorescent group to make its rotation slower, and achieve signal amplification through mass enhanced effect. In the absence of miRNA-21, the probe had a large fluorescence polarization value, when miRNA-21 in the solution reacted with the toehold 1 (5 nt) of 2-FAM, 2-FAM and the complementary part of 1-SH were loose to form a double-stranded structure of 2-FAM + miRNA-21, and the toehold 2 (9 nt) of the 2-FAM chain was then exposed; subsequently, the 3-fuel presented in the solution would trigger another toehold mediated strand displacement reaction by the toehold 2 with this double-stranded structure, hereby releasing miRNA-21 to enter the next chain substitution reaction process, and finally obtained 2-FAM + 3-fuel product. It was an asymmetric amplified strand displacement reaction and in theory, one target miRNA-21 chain could trigger multiple strand displacement reactions, causing effective amplification of the signal. After reacting with miRNA-21, the FAM fluorophores were far from the surface of Ag10NPs and were not affected by the metal enhanced fluorescence effect and mass enhanced effect, so the fluorescence polarization value were turned to lower. The difference between the fluorescence polarization value ΔFP of the initial probe and the final product had a linear relationship with the concentrations of miRNA-21, so it could be used to measure the concentration of miRNA-21 in solution.The results of PAGE (Fig. 1B) validated the principle of the above-mentioned strand displacement reaction. At first the 1-SH and 2-FAM chains were combined with each other to form a double-stranded structure, with a clear band in lane 1. When 1-SH + 2-FAM meet the target miRNA-21, a new band was generated in lane 5 because chain 2-FAM was replaced from the chain 1-SH by miRNA-21, forming a new 2-FAM + miRNA-21 structure and verifying the occurrence of the strand displacement reaction. Subsequently, 1-SH + 2-FAM was mixed with miRNA-21 and 3-fuel to simulate the cyclic reaction process in the experiment. It could be seen from the lane 6 that 2-FAM and 3-fuel mixed double-stranded products were finally obtained, and basically without the band of 2-FAM + miRNA-21, it proved that the designed strand displacement reaction could occur as planned. And the 1-SH + 2-FAM + 3-fuel were mixed to show the background of the sensors, 3-fuel chains were combined with part of the fallen 2-FAM chains to form a shallow band, but most of the 1-SH + 2-FAM double chain structure and 3-fuel were not wasted.
Feasibility study of miRNA-21 detection
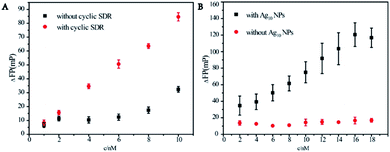
Mass enhanced effect and metal enhanced fluorescence effect of Ag10NPs and the designed cyclic amplification were important for the assay. The influence of the cyclic strand displacement reaction was verified by adding the 3-fuel. According to the above design, if 3-fuel was not present in the solution, a single strand miRNA-21 could only dissociate a pair of complementary 1-SH + 2-FAM strands, meanwhile the addition of 3-fuel made this process cycle several times to achieve the asymmetric amplification. Fig. 2A evidently illustrated the effect of the presence of cyclic strand displacement reaction (the reaction without cyclic amplification was showed in Fig. S2.†). Under the reaction conditions without cyclic strand displacement reaction, ΔFP signal was lower and less regular, and there was no obvious linear relationship between the fluorescence polarization values and the concentrations of miRNA-21. After adding a 3-fuel capable of initiating a strand displacement reaction cycle, the fluorescence polarization values were prominently increased, and the data showed a clear rule. The clear linear relationship between the fluorescence polarization change value ΔFP and the concentrations of miRNA-21 meant a good detection performance.The importance of Ag10NPs could be reflected by comparing the fluorescence polarization change data ΔFP with and without Ag10NPs (Fig. 2B). The detection effect was compared in the concentration range of 2–18 nM. After adding miRNA-21 at different concentrations, because of the larger mass and volume of Ag10NPs,26 the change of the fluorescence polarization were obviously effected and signals significantly increased, a linear relationship between ΔFP and the concentrations of miRNA-21 was significant. These experiments had proved that the cyclic strand displacement reactions and Ag10NPs were feasible to improve the sensitivity of miRNA-21 detection.
Optimization of reaction conditions
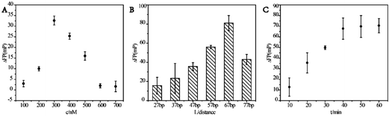
Some parameters were important for best detection results such as concentrations of nucleic acids that modified on the Ag10NPs, the distance between Ag10NPs and fluorophores and reaction time. In this work, Ag10NPs were vital and worked as amplifier. Firstly, we investigated the optical concentrations of nucleic acids that functionally modified on the surface of Ag10NPs. When the concentrations of nucleic acids modified on the surface of Ag10NPs was low, it was difficult to react with low concentrations of miRNA-21 in solution so the ΔFP signals were weak. As the concentration of nucleic acid on Ag10NPs surface increased, the ΔFP signal gradually increased, but too many nucleic acids on the surface of Ag10NPs caused mutual interference, making it difficult for miRNA-21 to squeeze chain 2-FAM by toehold 1 as expected. ΔFP reached the maximum value when the 300 nM nucleic acid chains 1-SH and 2-FAM were chosen, but ΔFP decreased because of higher nucleic acid concentration, which proved that high concentrations of nucleic acids were not conducive to the progress of the strand displacement reaction. When a suitable concentration of nucleic acid modification ratio was selected, a lower concentration target also returned a strong signal, so we chose 300 nM 1-SH and 2-FAM as the optical nucleic acid modification concentration.Secondly, in addition to the mass enhanced effect of Ag10NPs, the metal enhanced fluorescence effect could not only enhance the fluorescence signal, but also enhance the fluorescence polarization signal by reducing the overall rotation rate of the probe and reducing the fluorescence lifetime, which had been reported in previous research by our group.22 Because the metal enhanced fluorescence effect of Ag10NPs was affected by the distance between the fluorophores and Ag10NPs,25 it was mainly studied to achieve the best detection effect and changed by linking spacer arms of different lengths. As shown in Fig. 3B, the distance between the fluorophores and Ag10NPs was selected, at the beginning, the value of ΔFP gradually increased as the distance increased, but when the distance between the fluorophore and Ag10NPs increased from 67 bp to 77 bp, ΔFP decreases instead, so it could be determined that the optimal distance is 67 bases, which was about 22.3 nm between Ag10NPs and fluorophores.
In the meantime, the reaction time was a relatively important factor of this method, so we mixed the detection system with miRNA-21 for different times and measured at 10, 20, 30, 40, 50, 60 min to get corresponding ΔFP values. The ΔFP gradually stabilized and did not change significantly with time increasing (Fig. 3C), so we set 40 min was set as the optimal condition for the reaction time. At this time, it could ensure the smooth progress of the strand displacement reaction and obtained a strong and stable signal, which was conducive to improving the detection effect of the probe.
Analytical performance of miRNA-21 detection
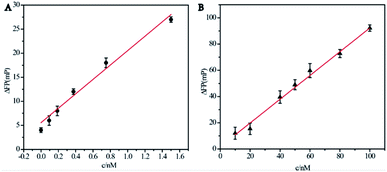
After several optimization experiments we finally chose the best detection conditions for the experiment. Due to the formation of the 2-FAM + 3-fuel double stranded structure, the nucleic acid chains attached to the FAM fluorophores left the surface of Ag10NPs and became free in solution. Therefore, the FP signals changed markedly and this method achieved a significant signal amplification effect.In order to further evaluate the detection sensitivity of the assay we designed, a series of concentrations of miRNA-21 were tested and recorded the fluorescence polarization value under optical conditions to calculate the corresponding ΔFP. As shown in Fig. 4A, we used the well-designed signal amplification detection system to detect miRNA-21 in the range of 0–1.6 nM in 1× PBS buffer. There was a clear linear relationship between the fluorescence polarization change value ΔFP and the target concentrations, showing a positive correlation. The fitted curve was y = 15.0x + 5.56, R2 = 0.948, and the LOD was 93.8 pM (S/N = 3). Although the concentration of miRNA-21 in the solution was very low at this time, it could still produce the target signal value that were able to be measured. Then we compared the effect of the system without cyclic strand displacement reaction showed in Fig. 4B, the fitting curve was y = 0.906x + 1.70, R2 = 0.994, and the LOD was 9.09 nM (S/N = 3). The results showed that although the mass enhanced effect and the metal enhanced fluorescence effect of Ag10NPs played a good signal amplification effect, the cyclic strand displacement reaction still further reduced the detection limit on this basis. The basis of reducing the detection limit of chain substitution reaction was that it was no longer a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 signal generation mode, but a single miRNA-21 chain could continuously trigger several strand displacement reactions, causing dramatic changes in ΔFP, achieving an asymmetric amplification effect. Whether from the simplicity of detection method or from the evaluation of detection effect, our detection method had good robustness and the feasibility, meanwhile, compared with other methods, it has certain advantages34–38 (Table 2).
1 signal generation mode, but a single miRNA-21 chain could continuously trigger several strand displacement reactions, causing dramatic changes in ΔFP, achieving an asymmetric amplification effect. Whether from the simplicity of detection method or from the evaluation of detection effect, our detection method had good robustness and the feasibility, meanwhile, compared with other methods, it has certain advantages34–38 (Table 2).
| Method | Dynamic range | Detection limit |
|---|---|---|
| Colorimetric microRNA detection strategy based on target-catalyzed toehold-mediated strand displacement reaction34 | 0–100 nM | 480 pM |
| Fluorescent biosensor based on MnO2 nanosheets and catalytic hairpin assembly amplification35 | 1–50 nM | 330 pM |
| Sensors based on phosphorescence resonance energy transfer and duplex-specific nuclease-assisted signal amplification36 | 0–40 nM | 160 pM |
| A biosensor based on competitive hybridization onto magnetic bead37 | — | 200 pM |
| An enzyme-free platform based on hybridization chain reaction engineered dsDNA for Cu metallization38 | 0.25–25 μM | 250 pM |
| This method | 0.16–16 nM | 93.8 pM |
Detection selectivity
In order to verify the specificity of this fluorescence polarization sensing method based on Ag10NPs, we compared three negative control samples: let-7a, miRNA-7 and miRNA-141 to evaluate it, each group was measured three times in parallel and calculated the mean and variance, the results are shown in Fig. 5. When the concentration of miRNA-21 was 2 nM, it could still produce strong ΔFP signals. By comparison, the same concentration of other three negative control miRNAs were tested and the signal value were far lower than miRNA-21. This experimental data showed that our method based on Ag10NPs and fluorescence polarization detection had good selectivity for miRNA-21, and it had the potential to play an ideal detection effect in complex system.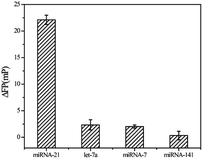 | ||
| Fig. 5 Specificity of the amplified fluorescence polarization assay to miRNA-21 by comparing it to some samples at the 2 nM: let-7a/miRNA-7/miRNA-141. | ||
Application of the amplified FP method to miRNA-21 detection in real samples
The total RNA extracts of HEK-293 and MCF-7 cells were used to evaluate the feasibility of the designed Ag10NPs probe to detect actual samples. In the experiment, the phosphate buffered saline 1× PBS was used as the blank control, and the total RNA extracts of HEK-293 and MCF-7 cells were measured according to the experimental methods described above. Total RNA extract was the total RNA solution extracted by total RNA kit, which was appropriately diluted by DEPC water and detected by NanoDrop 2000. It was depicted in Fig. S3.† that the signal value of total RNA extract of HEK-293 with low miRNA-21 expression level was significantly different from that of total RNA extract of MCF-7 with high miRNA-21 expression level,39 which further validated the established method based on Ag10NPs probe and fluorescence polarization detection by us appeared high specificity.Standard addition method was used to measure the miRNA-21 concentration in total RNA extracts to further verify the specificity and accuracy of this method.13 To the total RNA extracts of HEK-293 and MCF-7, miRNA-21 with different concentration gradients was added and detected (Fig. 6). For the HEK-293 total RNA extract, the fitting curve was y = 8.10x − 5.67, R2 = 0.966, and the intercept was negative, which proved that the probe was affected by the environment and some dissociation occurred thought a good linear relationship was maintained between the miRNA-21 concentrations and ΔFP signals. Furthermore, in the MCF-7 total RNA extract, the corresponding fitting curve was y = 0.0617x + 20.9 and R2 = 0.980. The miRNA-21 content was calculated to be about 1.02 amol ng RNA−1 (ESI†), which reached a good uniformity with the practical concentration reported in previous literature (1.13 amol ng RNA−1,39 1.17 amol ng RNA−1 (ref. 40)), so we believe that this method was promising to be used for the detection of miRNA-21 in complex biological samples.
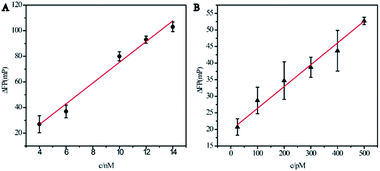 | ||
| Fig. 6 Linear relationships between the fluorescence polarization change (ΔFP) and the added concentrations of miRNA-21. (A) HEK-293 cells total RNA extract. (B) MCF-7 cells total RNA extract. | ||
Conclusions
We have developed a highly sensitive miRNA-21 analytical method based on the interaction of Ag10NPs enhanced fluorescence polarization detection and cyclic strand displacement reaction, which could detect miRNA quickly, accurately and sensitively in standard system and real biological samples. First of all, we used the mass enhanced effect and the metal enhanced fluorescence effect of Ag10NPs, and controlled the optimal concentration of nucleic acid modified on the surface of Ag10NPs and the appropriate distance of fluorophores to make Ag10NPs work well as amplifiers; secondly, we designed a strand displacement reaction system, using miRNA-21 as the initiator to achieve the asymmetric amplification, and the LOD was reduced by about two orders of magnitude after amplification. Again, this detection system was a homogeneous system without separation, which can be detected by mixing the detection probe with the solution and reading, the response time was short and the detection method was extremely simple. Finally, this detection method could show certain anti-interference ability in simple biological samples, which was expected to play a role in more complex biological samples such as blood and urine.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by National Natural Foundation of China (Grant No. 21775068 and 21974066).References
- J. Conde, E. R. Edelman and N. Artzi, Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: the jack-of-all-trades in cancer nanotheranostics, Adv. Drug Delivery Rev., 2015, 81, 169–183 CrossRef CAS PubMed.
- D. Kim, Y. M. Sung, J. Park, S. Kim, J. Kim and J. Park, et al., General rules for functional microRNA targeting, Nat. Genet., 2016, 48, 1517–1526 CrossRef CAS PubMed.
- D. P. Bartel, MicroRNAs: Target Recognition and Regulatory Functions, Cell, 2009, 136, 215–233 CrossRef CAS PubMed.
- S. Lin and R. I. Gregory, MicroRNA biogenesis pathways in cancer, Nat. Rev. Cancer, 2015, 15, 321–333 CrossRef CAS PubMed.
- J. Li, L. Huang, X. Xiao, Y. Chen, X. Wang and Z. Zhou, et al., Photoclickable MicroRNA for the Intracellular Target Identification of MicroRNAs, J. Am. Chem. Soc., 2016, 138, 15943–15949 CrossRef CAS PubMed.
- X. Xiao, X. Wang, Y. Wang, T. Yu, L. Huang and L. Chen, et al., Multi-Functional Peptide–MicroRNA Nanocomplex for Targeted MicroRNA Delivery and Function Imaging, Chem.–Eur. J., 2018, 24, 2277–2285 CrossRef CAS PubMed.
- B. Malla, D. M. Aebersold and A. Dal Pra, Protocol for serum exosomal miRNAs analysis in prostate cancer patients treated with radiotherapy, J. Transl. Med., 2018, 16, 223 CrossRef CAS PubMed.
- T. Kilic, A. Erdem, M. Ozsoz and S. Carrara, microRNA biosensors: opportunities and challenges among conventional and commercially available techniques, Biosens. Bioelectron., 2018, 99, 525–546 CrossRef CAS PubMed.
- C. Dell'aversana, C. Giorgio and L. Altucci, MicroRNA expression profiling using Agilent one-color microarray, Methods Mol. Biol., 2017, 1509, 169 CrossRef PubMed.
- W. Peng, Q. Zhao, M. Chen, J. Piao, W. Gao and X. Gong, et al., An innovative unlocked mechanism by a double key avenue for one-pot detection of microRNA-21 and microRNA-141, Theranostics, 2019, 9, 279–289 CrossRef CAS PubMed.
- J. Liu, M. Cui, H. Zhou and W. Yang, DNAzyme Based Nanomachine for in situ Detection of MicroRNA in Living Cells, ACS Sens., 2017, 2, 1847–1853 CrossRef CAS PubMed.
- K. Shi, B. Dou, C. Yang, Y. Chai, R. Yuan and Y. Xiang, DNA-Fueled Molecular Machine Enables Enzyme-Free Target Recycling Amplification for Electronic Detection of MicroRNA from Cancer Cells with Highly Minimized Background Noise, Anal. Chem., 2015, 87, 8578–8583 CrossRef CAS PubMed.
- S. Ye, X. Li, M. Wang and B. Tang, Fluorescence and SERS Imaging for the Simultaneous Absolute Quantification of Multiple miRNAs in Living Cells, Anal. Chem., 2017, 89, 5124–5131 CrossRef CAS PubMed.
- W. Li, W. Jiang, Y. Ding and L. Wang, Highly selective and sensitive detection of miRNA based on toehold-mediated strand displacement reaction and DNA tetrahedron substrate, Biosens. Bioelectron., 2015, 71, 401–406 CrossRef CAS PubMed.
- R. L. Wang, L. Lan, L. Liu and L. Cheng, Asymmetric polymerase chain reaction and loop-mediated isothermal amplification (AP-LAMP) for ultrasensitive detection of microRNAs, Chin. Chem. Lett., 2020, 31, 159–162 CrossRef CAS.
- M. Kaplan, T. Kilic, G. Guler, J. Mandli, A. Amine and M. Ozsoz, A novel method for sensitive microRNA detection: electropolymerization based doping, Biosens. Bioelectron., 2017, 92, 770–778 CrossRef CAS PubMed.
- N. Yan, X. Wang, L. Lin, T. Song, P. Sun and H. Tian, et al., Gold Nanorods Electrostatically Binding Nucleic Acid Probe for in vivo MicroRNA Amplified Detection and Photoacoustic Imaging-Guided Photothermal Therapy, Adv. Funct. Mater., 2018, 28, 1800490 CrossRef.
- X. M. Li, L. Gao, L. Feng, X. D. Hou and P. Wu, Universal and label-free photosensitization colorimetric assays enabled by target-induced termini transformation of dsDNA resistant to Exo III digestion, Chem. Commun., 2019, 55, 7211–7214 RSC.
- S. Bi, S. Yue and S. Zhang, Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- W. Dai, H. Dong, K. Guo and X. Zhang, Near-infrared triggered strand displacement amplification for MicroRNA quantitative detection in single living cells, Chem. Sci., 2018, 9, 1753–1759 RSC.
- G. Wang, S. Wang, C. Yan, G. Bai and Y. Liu, DNA-functionalized gold nanoparticle-based fluorescence polarization for the sensitive detection of silver ions, Colloids Surf., B, 2018, 167, 150–155 CrossRef CAS PubMed.
- Z. Chen, H. Li, W. Jia, X. Liu, Z. Li and F. Wen, et al., Bivalent Aptasensor Based on Silver-Enhanced Fluorescence Polarization for Rapid Detection of Lactoferrin in Milk, Anal. Chem., 2017, 89, 5900–5908 CrossRef CAS PubMed.
- H. Li, Y. Zhu, S. Dong, W. Qiang, L. Sun and D. Xu, Fast functionalization of silver decahedral nanoparticles with aptamers for colorimetric detection of human platelet-derived growth factor-BB, Anal. Chim. Acta, 2014, 829, 48–53 CrossRef CAS PubMed.
- S. J. Zhen, X. Xiao and C. H. Li, et al., An Enzyme-Free DNA Circuit-Assisted Graphene Oxide Enhanced Fluorescence Anisotropy Assay for MicroRNA Detection with Improved Sensitivity and Selectivity, Anal. Chem., 2017, 89, 8766–8771 CrossRef CAS PubMed.
- Y. Wang, Z. Li, H. Li, M. Vuki, D. Xu and H.-Y. Chen, A novel aptasensor based on silver nanoparticle enhanced fluorescence, Biosens. Bioelectron., 2012, 32, 76–81 CrossRef CAS PubMed.
- Y. Jiang, J. Tian, K. Hu, Y. Zhao and S. Zhao, Sensitive aptamer-based fluorescence polarization assay for mercury(II) ions and cysteine using silver nanoparticles as a signal amplifier, Microchim. Acta, 2014, 181, 1423–1430 CrossRef CAS.
- Y. H. Yuan, Y. D. Wu, B. Z. Chi, S. H. Wen, R. P. Liang and J.-D. Qiu, Simultaneously electrochemical detection of microRNAs based on multifunctional magnetic nanoparticles probe coupling with hybridization chain reaction, Biosens. Bioelectron., 2017, 97, 325–331 CrossRef CAS PubMed.
- Y. Li, C. Yu, C. Zhao, C. Ren and X. Zhang, Catalytic hairpin assembly induced dual signal enhancement for rapid detection of miRNA using fluorescence light-up silver nanocluster, Anal. Chem., 2019, 1084, 93–98 CAS.
- H. Zhang, S. Yang, K. De Ruyck, N. V. Beloglazova, S. A. Eremin and S. De Saeger, et al., Fluorescence polarization assays for chemical contaminants in food and environmental analyses, TrAC, Trends Anal. Chem., 2019, 114, 293–313 CrossRef CAS.
- Y.-C. He, B.-C. Yin, L. Jiang and B.-C. Ye, The rapid detection of microRNA based on p19-enhanced fluorescence polarization, Chem. Sci., 2014, 50, 6236–6239 CAS.
- H. Li, Y. Zhao, Z. Chen and D. Xu, Silver enhanced ratiometric nanosensor based on two adjustable Fluorescence Resonance Energy Transfer modes for quantitative protein sensing, Biosens. Bioelectron., 2017, 87, 428–432 CrossRef CAS PubMed.
- X. Li, N. Huang, L. L. Zhang, J. J. Zhao and S. L. Zhao, A T7 exonuclease assisted dual-cycle signal amplification assay of miRNA using nanospheres-enhanced fluorescence polarization, Talanta, 2019, 202, 297–302 CrossRef CAS PubMed.
- F. C. Simmel, B. Yurke and H. R. Singh, Principles and Applications of Nucleic Acid Strand Displacement Reactions, Chem. Rev., 2019, 119, 6326–6369 CAS.
- Y. Park, C. Y. Lee, S. Kang, H. Kim, K. S. Park and H. G. Park, Universal, colorimetric microRNA detection strategy based on target-catalyzed toehold-mediated strand displacement reaction, Nanotechnology, 2018, 29, 085501 CrossRef PubMed.
- W. Ouyang, Z. H. Liu, G. F. Zhang, Z. Chen, L. Guo, Z. Lin, B. Qiu and G. Chen, Enzyme free fluorescent biosensor for miRNA-21 detection based on MnO2 nanosheets and catalytic hairpin assembly amplification, Anal. Methods, 2016, 8, 8492–8497 RSC.
- J. J. Yang, Z. F. Zhang and G. Q. Yan, Facile detection of microRNA based on phosphorescence resonance energy transfer and duplex-specific nuclease-assisted signal amplification, Anal. Biochem., 2017, 539, 127–133 CrossRef CAS PubMed.
- E. Vargas, E. Povedano, V. R.-V. Montiel, R. M. Torrente-Rodríguez, M. Zouari and J. J. Montoya, et al., Single-Step Incubation Determination of miRNAs in Cancer Cells Using an Amperometric Biosensor Based on Competitive Hybridization onto Magnetic Bead, Sensors, 2018, 18, 863 CrossRef PubMed.
- Y. Zhang, Z. W. Chen, Y. Tao, Z. Z. Wang, J. S. Ren and X. G. Qu, Hybridization chain reaction engineered dsDNA for Cu metallization: an enzyme-free platform for amplified detection of cancer cells and microRNAs, Chem. Commun., 2015, 51, 11496–11499 RSC.
- X. Zhao, L. Xu and M. Sun, et al., RNA Detection: Gold-Quantum Dot Core-Satellite Assemblies for Lighting Up MicroRNA in vitro and in vivo, Small, 2016, 12, 4581 CrossRef CAS.
- W. Ma, P. Fu and M. Sun, et al., Dual Quantification of MicroRNAs and Telomerase in Living Cells, J. Am. Chem. Soc., 2017, 139, 11752–11759 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra01950j |
| This journal is © The Royal Society of Chemistry 2020 |