Open Access Article
Open Access ArticleExperimental and DFT studies of sulfadiazine ‘piano-stool’ Ru(II) and Rh(III) complexes†
Ahmed M. Mansour *a and
Krzysztof Radacki
*a and
Krzysztof Radacki b
b
aDepartment of Chemistry, Faculty of Science, Cairo University, Gamma Street, Giza, Cairo 12613, Egypt. E-mail: mansour@sci.cu.edu.eg; inorganic_am@yahoo.com
bInstitut für Anorganische Chemie, Julius-Maximilians-Universität Würzburg, Am Hubland, D-97074 Würzburg, Germany
First published on 12th March 2020
Abstract
While sulfadiazine (HLSZ) is extensively used to elaborate complexes of intriguing biological applications (e.g. topical antibiotic silvadene; silver sulfadiazine), the molecular structure modification of sulfadiazine or even other sulfa drugs by coordination to either η6-cymene Ru(II) or η5-Cp* Rh(III) motif has not been investigated. Here, half-sandwich organoruthenium(II) and organorhodium(III) compounds of the type [(η6-p-cymene)Ru(LSZ)2] (1) and [(η5-C5Me5)Rh(LSZ)2] (2) are synthesized, characterized and evaluated for their potential antimicrobial activity. Spectroscopic and single crystal X-ray analysis showed that LSZ is coordinated to Rh(III) via both the sulfonamide and pyrimidine nitrogen atoms forming “piano-stool” geometry. In 2, the NMR equivalence clearly pointed to participation of two LSZ molecules in a fluxional process in which the third bond of the base of the stool is oscillating between two equivalent sulfonamide nitrogen atoms. While 1 was biologically inactive, complex 2 was potent against Gram-positive bacteria, Candida albicans and Cryptococcus neoformans. Hen white egg lysozyme (HEWL), a model protein, reacted covalently with 2 via the loss of one LSZ molecule, while compound 1 decomposed during the interaction with that protein.
Introduction
Sulfonamide derivatives are extensively used as antibacterial agents because they are cheap, have low toxicity and exhibit excellent activity against several bacterial diseases.1 They slow down or stop bacterial growth in infected systems or in wounds without causing significant toxicity to the body tissues.2 In other words, Sulfonamides are bacteriostatic rather than bactericidal. Sulfonamides interfere with the conversion of p-amino benzoic acid into an essential nutrient necessary for the bacterial multiplication.3 In view of growing bacterial resistance towards Sulfonamides as well as the appearance of more effective modern antibiotics, the importance of Sulfonamides has been greatly reduced. Sulfadiazine(4-amino-N-2-pyrimidinylbenzene sulphonamide, HLSZ) was introduced in 1940 for curing of the bacterial infections in humans.4 It is used in combination therapy with pyrimethamine for treatment of chloroquine-resistant malaria parasite because of its long half-life time in the blood.5 From structural point of view, the presence of potential coordination sites in NaLSZ (–NH2, sulfonyl oxygen, sulfonamide (Nsfd) and pyrimidine nitrogen (Npym)) (Scheme 1) makes it versatile ligand in the field of coordination chemistry. Modified pharmacological and toxicological properties were observed upon coordination of HLSZ to metal ions and formation of metal complexes. Zinc(II) complex of HLSZ was used to inhibit the bacterial infection in burned animals,6 while silvadene(2-sulfanilamido-pyrimidine silver) was applied for treatment of topical burn.7 For HLSZ, several potential coordination modes were reported,6–13 in which HLSZ coordinated to Mn+ as mono-, bi- and tridentate ligand. Normally, HLSZ deprotonates during the interaction with Mn+ ions. The crystallographic data of silvadene showed a polymeric arrangement structure in which Ag(I) is coordinated to tridentate LSZ via Npym, Nsfd and OSO2.7 For Zn(II) sulfadiazine complex, Zn(II) is coordinated to three different molecules of LSZ in a tetrahedral polymeric arrangement. The ligands differently behaved towards Zn(II); the first LSZ molecule binds Zn(II) via Npym and sulfonyl oxygen, the second LSZ interacts with Zn(II) via Npym atom and the third ligand molecule coordinates to Zn(II) through Nsfd.6 Single-crystal X-ray analysis and spectroscopic characterization of other sulfadiazine complexes with Fe(III),8 Co(II),9 Ni(II),10 Cu(II),10 Cd(II),11 Pt(II)12 and Hg(II)13 ions were reported. For [Cu(NH3)(LSZ)2(OH2)],10a the crystal data showed that Cu(II) ion is surrounded by two LSZ in a distorted N4O1 square-pyramidal environment; the base of the pyramid is occupied by a given N,N-bidentate LSZ, OH2, and NH3, and its top is occupied with Npym of the adjacent LSZ molecule.In comparison with free HLSZ, the effect of complex formation on the antibacterial activity of HLSZ was evaluated.10,14 No negative effect on the antibacterial activity of HLSZ (against Staphylococcus aureus, Klebsiella pneumoniae and Escherichia coli) was detected by association to Cu(II) ions.10a Screening of Cu(II)-LSZ complexes against Staphylococcus aureus ATCC29213 and Escherichia coli ATCC 25922 indicated that the complexes were less effective than HLSZ.10d The –SO2NH group of HLSZ is essential for the antibacterial activity. Exchange of the ionizable proton with the metal ion may lead to diminishing of the toxicity. In other words, the truly active sulfonamide species is the ionic form; Sulfonamide drugs penetrate the bacterial cells in the neutral form and once they cross the cell wall, their toxicities may be attributed to the ionized form.14
In the present contribution, we disclose first examples of sulfa drugs based half-sandwich Ru(II) (1)15 and Rh(III) (2) organometallic compounds (Scheme 1) having either p-cymene or pentamethylcyclopentadienyl at the seat of piano-stool geometry. Although Sulfonamides constituted an important class of the antimicrobial agents in the world for a long time, it has not been used in synthesis of organometallic arene compounds.
The previous experimental evidences indicated that 98% of the overall administrated ruthenium, in the blood plasma, is in a protein bound form.16 The binding of cytotoxic Ru(II) complexes to the proteins may influence their toxicity, biodistribution as well as drug delivery. Glutathione S-transferase P1-1, transferrin and albumin were known as probable final targets for Ru(II) complexes.17 These arguments encouraged us to study and explore the interactions between the synthesized organometallic compounds and hen white egg lysozyme (HEWL) by means of ESI-MS. HEWL was widely studied in the context of metalation of proteins because of its small size and suitability to be studied by mass spectrometry. In addition, HEWL is a perfect protein to crystallize and thus suitable for X-ray crystallography analysis. Complexes (1, 2) and the metal precursors; [(η6-p-Cym)Ru(μ-Cl)Cl]2 and [{(η5-C5Me5)RhCl}2(μ-Cl)2] were evaluated for their inhibitory effect against the following microbes; Gram-positive bacterium (Staphylococcus aureus ATCC 43300), Gram-negative bacteria (Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa ATCC700603) and fungi (Candida albicans ATCC 90028 and Cryptococcus neoformans var. grubii H99; ATCC 208821). The complexes were assessed for their blood compatibility with the cell components and cell viability against non-malignant cells with rapid proliferation (noncancerous human embryonic kidney cells (HEK293)).
Results and discussion
Synthesis and characterization
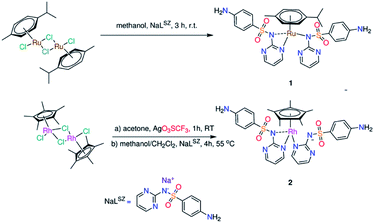
“Piano-stool” complex of type [(η6-p-Cym)Ru(LSZ)2] (1) (Scheme 1) was prepared from stirring of sodium sulfadiazine (NaLSZ) and [(η6-p-Cym)Ru(μ-Cl)Cl]2 (ref. 18) in methanol at room temperature for 3 h. The proposed structure of 1, isolated as air-stable yellow powder in 84% yield, was elucidated by elemental analysis, ESI-MS, IR and NMR (1H, 13C, {1H, 1H} COS90, and {1H, 13C} HSQC) (Fig. S1–S4†). In particular, the elemental composition, ESI-MS as well as the integral values of the 1H NMR resonances confirm isolation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (M
2 (M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p-Cym
p-Cym![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LSZ) complex. In the positive potential mode, the ESI-MS spectrum showed a unique peak at m/z = 735.1102, having a ruthenium isotope envelope, which may be allocated for the suggested mono-cationic monomeric complex of 1. The 1H NMR spectrum (in [D6] DMSO) of free NaLSZ shows a singlet signal at δ = 5.30 ppm (–NH2). The signals of the pyrimidine ring are shown as triplet and doublet at δ = 6.34 and 8.06 ppm, respectively. The two doublets at δ = 6.46 and 7.48 ppm are allocated to the phenyl ring.19 The 1H NMR signals of NaLSZ are shifted downfield upon coordination to (η6-p-Cym)Ru. The well-known 1H NMR pattern of p-Cym moiety is shown in the spectrum (Fig. S2†) of 1 as four doublets (at δ = 6.03, 5.89, 5.59 and 5.52 ppm) for the aryl-CH protons, a septet (at δ = 2.63 ppm) for CH(CH3)2, singlet methyl (at δ = 1.96 ppm) as well as two doublet methyl signals (centred at δ = 1.01 and 0.94 ppm).
LSZ) complex. In the positive potential mode, the ESI-MS spectrum showed a unique peak at m/z = 735.1102, having a ruthenium isotope envelope, which may be allocated for the suggested mono-cationic monomeric complex of 1. The 1H NMR spectrum (in [D6] DMSO) of free NaLSZ shows a singlet signal at δ = 5.30 ppm (–NH2). The signals of the pyrimidine ring are shown as triplet and doublet at δ = 6.34 and 8.06 ppm, respectively. The two doublets at δ = 6.46 and 7.48 ppm are allocated to the phenyl ring.19 The 1H NMR signals of NaLSZ are shifted downfield upon coordination to (η6-p-Cym)Ru. The well-known 1H NMR pattern of p-Cym moiety is shown in the spectrum (Fig. S2†) of 1 as four doublets (at δ = 6.03, 5.89, 5.59 and 5.52 ppm) for the aryl-CH protons, a septet (at δ = 2.63 ppm) for CH(CH3)2, singlet methyl (at δ = 1.96 ppm) as well as two doublet methyl signals (centred at δ = 1.01 and 0.94 ppm).
The 13C NMR spectrum of 1 displayed the expected resonances for the p-Cym moiety (Fig. S2†). As previously reported, HLSZ ligand exhibited different modes of coordination to Mn+ and might be interacted as mono-, bi- and tridentate ligands in either monomeric or polymeric arrangements.6–13 As the p-Cym occupies the top of the stool of 1 by three arms, the remaining three arms around Ru(II) ion should be occupied by two deprotonated LSZ molecules. Therefore, the tridentate mode of coordination for LSZ should be excluded. Alternatively, solution [1H–15N] heteronuclear multiple bond coherence (HMBC) NMR spectra of NaLSZ and 1 were recorded to evaluate if the primary amino group interacts with Mn+ ion or not. The Δ15Ncoord parameter shows the real influence of nitrogen coordination effect; the difference between the NMR resonances of the free nitrogen and metal–nitrogen bond. The position of the NH2 signal (δ = −313 ppm) in [1H–15N] NMR spectrum of NaLSZ doesn't change upon formation of 1, excluding the role of NH2 in the complex formation (Fig. S2 and S4†).
To get more insight into the coordination mode of HLSZ, the vibrational spectra of NaLSZ and 1 were compared. The IR spectrum of NaLSZ shows νas(NH2) at 3416 cm−1, while νas(NH2) is overlapped. The vibrations at 1630, 1292 and 1131 cm−1 may be assigned to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)/(C
N)/(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), νas(SO2) and νs(SO2), respectively.19 For 1, the observation of νas(NH2) and νs(SO2) at lower wavenumbers (3382 and 1259 cm−1) than those of NaLSZ may be attributed to presence of intermolecular H-bond of the type NH2⋯OSO2 (this is clearly shown in the crystal packing of 2, Fig. S5†). The shift of ν(C
C), νas(SO2) and νs(SO2), respectively.19 For 1, the observation of νas(NH2) and νs(SO2) at lower wavenumbers (3382 and 1259 cm−1) than those of NaLSZ may be attributed to presence of intermolecular H-bond of the type NH2⋯OSO2 (this is clearly shown in the crystal packing of 2, Fig. S5†). The shift of ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)/(C
N)/(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) of NaLSZ from 1630 to 1624 cm−1 in 1 may be taken as a sign of participation of pyrimidine nitrogen in complex formation. Observation of ν(S–N) mode at a higher wavenumber (972 cm−1) with respect to NaLSZ (977 cm−1) (ref. 20) reveals presence of some sort of interaction between Ru(II) and sulfonamide nitrogen. While the IR changes between NaLSZ and 1 are not so much as a result of change the sodium ion with Ru(II), the obtained IR data suggest coordination of LSZ to Ru(II) ion via Npym and Nsfd.
C) of NaLSZ from 1630 to 1624 cm−1 in 1 may be taken as a sign of participation of pyrimidine nitrogen in complex formation. Observation of ν(S–N) mode at a higher wavenumber (972 cm−1) with respect to NaLSZ (977 cm−1) (ref. 20) reveals presence of some sort of interaction between Ru(II) and sulfonamide nitrogen. While the IR changes between NaLSZ and 1 are not so much as a result of change the sodium ion with Ru(II), the obtained IR data suggest coordination of LSZ to Ru(II) ion via Npym and Nsfd.
To conclude, the coordination sphere around Ru(II) in 1 is formed from p-cymene ring and two N,N-bidentate LSZ molecules. As the seven-coordinates geometry is uncommon for Ru(II) complexes, the observed 1H NMR equivalence clearly pointed to presence of two LSZ coordinated ligands in a fluxional process in which the third bond of the base of the stool is oscillating between the two equivalent pyrimidine nitrogen atoms (Scheme 1).10a
Treatment of the acetone solution of [{(η5-C5Me5)RhCl}2(μ-Cl)2]21 with two equivalents of silver trifluoromethane sulfonate and the subsequent removal of silver chloride yielded [{(η5-C5Me5)RhCl(acetone)}2](CF3SO3). Reaction of the latter Rh(III) precursor with two equivalents of NaLSZ was undertaken in a mixture of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
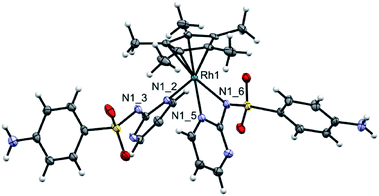
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) (CH2Cl2/CH3OH) at 55 °C for 4 h. Orange precipitate of the type [(η5-C5Me5)Rh(LSZ)2] (2) was obtained (Scheme 1). Analytical and spectroscopic tools (Fig. S3†) were used to get an idea about the structure of 2. However, a clear-cut proof of the structure of 2 was gained from X-ray crystallographic analysis (Fig. 1). A singlet signal assigned to C5Me5, with integration of fifteen equivalent protons, is shown in the 1H NMR spectrum (Fig. S3†) of 2 at δ = 1.69 ppm. The 13C NMR spectrum (Fig. S3†) of 2 shows a doublet signal at δ = 94.8 ppm (1JC–Rh = 7.5 Hz) due to coupling of η5-C5Me5 carbon atoms with Rh(III) metal centre having I = ½. Two characteristic fragments are observed in the ESI-MS(+) spectrum at m/z = 759.1007 {2 + Na}+, 487.0669 {2 − LSZ}+. Like 1, the 1H NMR equivalence indicated a fluxional behaviour between the two LSZ molecules towards Rh(III) ion. In comparison with NaLSZ, the pyrimidine-H3/H5 signals move downfield (δ = 8.47 ppm) in 2 due to coordination of LSZ with Rh(III). No change in the chemical shifts of the phenyl protons is detected, which rules out the role of either the primary amino group or SO2 group in the formation of 2.
1 (v/v) (CH2Cl2/CH3OH) at 55 °C for 4 h. Orange precipitate of the type [(η5-C5Me5)Rh(LSZ)2] (2) was obtained (Scheme 1). Analytical and spectroscopic tools (Fig. S3†) were used to get an idea about the structure of 2. However, a clear-cut proof of the structure of 2 was gained from X-ray crystallographic analysis (Fig. 1). A singlet signal assigned to C5Me5, with integration of fifteen equivalent protons, is shown in the 1H NMR spectrum (Fig. S3†) of 2 at δ = 1.69 ppm. The 13C NMR spectrum (Fig. S3†) of 2 shows a doublet signal at δ = 94.8 ppm (1JC–Rh = 7.5 Hz) due to coupling of η5-C5Me5 carbon atoms with Rh(III) metal centre having I = ½. Two characteristic fragments are observed in the ESI-MS(+) spectrum at m/z = 759.1007 {2 + Na}+, 487.0669 {2 − LSZ}+. Like 1, the 1H NMR equivalence indicated a fluxional behaviour between the two LSZ molecules towards Rh(III) ion. In comparison with NaLSZ, the pyrimidine-H3/H5 signals move downfield (δ = 8.47 ppm) in 2 due to coordination of LSZ with Rh(III). No change in the chemical shifts of the phenyl protons is detected, which rules out the role of either the primary amino group or SO2 group in the formation of 2.
Crystal structure
Diffusion of n-hexane into the dichloromethane solution of 2 afforded crystals suitable for single-crystal X-ray analysis. This compound crystallizes in the monoclinic space group P21/m. Applicable crystallographic parameters as well as the selected bond lengths (Å) and angles (°) are given in Fig. 1. The Rh(III) center is in a “three-legged piano-stool” coordination environment. While the seat of the stool is occupied by pentamethyl cyclopentadienyl ligand, the three legs are occupied with N,N-bidentate LSZ ligand [Rh–N1_5 = 2.137(2) Å, Rh–N1_6 = 2.172(2) Å] and pyrimidine N [Rh–N1_2 = 2.117(2) Å] from a second LSZ molecule. The Rh–C bond lengths are in the range of 2.132(2)–2.167(2) Å. The N1_3 is close to Rh by 3.120 Å and thus it cannot be considered as a true bond. The N1_3 is involved in two short contacts with the methyl groups with distances of 2.678 and 2.691 Å. The –SO2 group of a monodentate LSZ ligand is participated in two H-bonds of different strengths with –NH2 of the mono- and bidentate ligand as illustrated by the values of O⋯N = 2.887 and 3.027 Å, respectively.DFT and TD DFT calculations
Starting from the crystallographic data of 2, ground-state geometry optimization was carried out, without any symmetry restriction, by B3LYP/Genecp method (SDD for Rh and 6-31G(d) for the rest of the elements). The local minimum structure was checked as minimum on the potential energy surface by calculating the vibrational modes. No imaginary vibrations were found. Selected optimized bond lengths and angles of 2 are given in Table S1† in comparison to the crystal data. The root mean square error of the bond lengths is 0.037 Å. While the Rh–N bonds match well with the theoretical data, the Rh–C bonds deviate largely from the calculated values by about ∼0.05 Å. The optimized structure of 1 was obtained at B3LYP/Genecp (LANL2DZ for Ru and 6-31G(d) for the rest of the elementals) level of theory. Compound 1 adapts three-legged piano-stool geometry (Fig. 2). Cymene occupies the seat of the stool [Ru–C = 2.178–2.219 Å], while the two LSZ molecules coordinate to Ru through three arms; bidentate [Ru–N26 = 2.147 Å and Ru–N35 = 2.145 Å] and monodentate [Ru–N52 = 2.168 Å] modes of LSZ are observed. In contrast to the optimized and crystal structure of 2, the monodentate ligand LSZ coordinates to Ru(II) via sulfonamide N rather than pyrimidine N. The Ru–N bonds of the sulfonamide nitrogen are unequal; the bond to the bidentate LSZ is 0.23 Å longer than that bonded to monodentate LSZ. This indicates that there is not much space to accommodate two LSZ in a bidentate mode of coordination.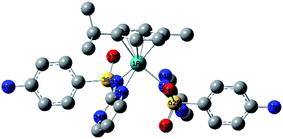 | ||
| Fig. 2 Local minimum structure of 1 calculated at B3LYP/Genecp level of theory (Genecp: LANL2DZ for Ru and 6-31G(d) for the rest of elements). | ||
Time dependent density functional calculations were carried out using hybrid exchange–correlation functional CAM-B3LYP, with a long-range correction term22 and LANL2DZ basis set to get an insight into the electronic structures of 1 and 2. The first 30 singlet excited states were taken into consideration. Table S2† including the wavelengths and energies of the calculated transitions and their assignments is given in the ESI.† In 190–360 nm region, the calculated electronic spectrum of 1 (Fig. S6†) shows two bands at 232 and 352 nm (broad) as well as a shoulder at 291 nm corresponding to HOMO−3 → LUMO, HOMO−7/HOMO−4 → LUMO+1 and HOMO → LUMO/LUMO+2, respectively. Besides, two transitions assigned to HOMO → LUMO+2 and HOMO−4 → LUMO+2 (broad band) are observed at 485 and 456 nm. The reversed order of the transitions is owing to some states experience smaller solvent effect than the others. HOMO is mainly composed of d(Ru) character (Fig. 3) and thus the transitions at 485 and 291 nm have partially a nature of MLCT. The other bands may be accounted for the intra ligand transitions.
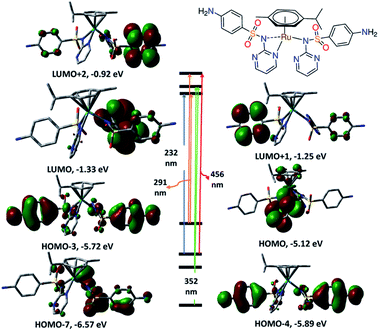 | ||
| Fig. 3 Selected frontiers molecular orbitals and electronic transitions of 1 calculated at CAM-B3LYP/LANL2DZ level of theory. | ||
The TD DFT spectrum of 2 displays four bands at 231, 284, 383 and 429 nm with oscillator strengths of 0.1196, 0.1290, 0.0159 and 0.0145 as a result of HOMO−3 → LUMO+3, HOMO/HOMO−2 → LUMO+3, HOMO−3 → LUMO+1 and HOMO−4 → LUMO in that order. HOMO−4 to HOMO−2 show d(Rh)/π ligand (LSZ) system character (Fig. 4). While LUMO and LUMO+1 are a mixture of d(Rh) and π(Cp*) orbitals, LUMO+3 has π(LSZ) character. HOMO–LUMO gap of 1 is 0.59 eV wider than 2. The transitions at 429 and 383 nm have ground-state composed of d(Rh)/π(LSZ)/π(Cp*) and excited state of d(Rh)/π(Cp*) forming d–d/MLCT.
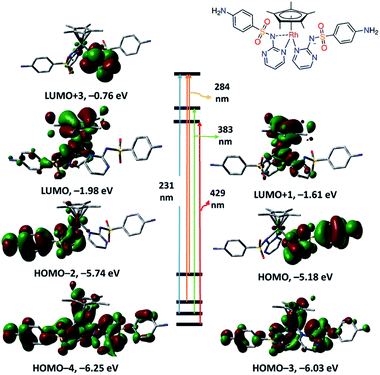 | ||
| Fig. 4 Selected frontiers molecular orbitals and electronic transitions of 2 calculated at CAM-B3LYP/LANL2DZ level of theory. | ||
Antimicrobial and cell viability
The Ru(II) and Rh(III) precursors; [RuCl(μ-Cl)(η6-p-Cym)]2, [{(η5-C5Me5)RhCl}2(μ-Cl)2] and their compounds (1 and 2) were screened for their potential antibacterial activity against Staphylococcus aureus ATCC 43300 (Gram-positive bacterium), Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, and Pseudomonas aeruginosa ATCC 27853 (Gram-negative bacteria) using the standard broth microdilution assays.23 The minimum inhibitory concentration (MIC, μg mL−1) values are given in Table 1. While most of ruthenium based complexes displayed significant biological activity,24 [RuCl(μ-Cl)(η6-p-Cym)]2 and its complex 1 exhibited no inhibitory activity against the tested bacteria at a concentration of 32 μg mL−1. Coordination of sulfadiazine to [RuCl(μ-Cl)(η6-p-Cym)]2 gave rise to biologically inactive compound (1). The inactivity may be accounted for formation of covalent bond between Ru(II) and sulfonamide nitrogen, which should be free for sulfa drugs to be active.14| E. coli | K. pneumoniae | A. baumannii | P. aeruginosa | |
|---|---|---|---|---|
| a Full names: Staphylococcus aureus ATCC 43300, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa ATCC, Candida albicans ATCC 90028 and Cryptococcus neoformans var. grubii H99; ATCC 208821.b [RuCl(μ-Cl)(η6-p-Cym)]2.c [{(η5-C5Me5)RhCl}2(μ-Cl)2]. | ||||
| Rub | >32 | >32 | >32 | >32 |
| Rhc | >32 | >32 | >32 | >32 |
| 1 | >32 | >32 | >32 | >32 |
| 2 | 16 | 16 | 32 | 16 |
While [{(η5-C5Me5)RhCl}2(μ-Cl)2] was inactive, compound 2 was potent against only the Gram-negative bacteria. Acinetobacter baumannii is two times more resistant to 2 (MIC 44 nM, equivalent to 32 μg mL−1) than the other tested Gram-negative bacteria. The inhibitory activity of 2 is associated to the cell wall structure of the bacteria, which is vital for the surviving of the bacteria. Inhibition of the synthesis of peptidoglycan may be the job of the antibiotics. Gram-positive bacteria possess a thick cell wall comprising of several layers of peptidoglycan and teichoic acids that would be easily destroyed by drugs. Gram-negative bacteria have a comparatively thin cell wall containing a few layers of peptidoglycan surrounded by lipopolysaccharides and lipoproteins (second lipid membrane). The differences in the structure of the cell wall can afford alterations in the toxicity against the two types of the bacteria.
Based on the results of the antibacterial activity of 1 and 2, we concluded that the antimicrobial activity cannot simply related to type and oxidation state of the metal ions, as well as type of arene moiety, other factors including lipophilicity, stability of the compounds, size of the receptor sites and electronic effect should be considered.
The antifungal activity of the synthesized complexes and their precursor metal salts was assessed against Candida albicans ATCC 90028 and Cryptococcus neoformans var. grubii H99; ATCC 208821. For fungi, the MIC values were determined as the lowest concentration at which the growth was completely reserved, well-defined by an inhibition of 80% and 70% for C. albicans and C. neoformans var. grubii H99, respectively.
While [RuCl(μ-Cl)(η6-p-Cym)]2 displayed antifungal activity against Candida albicans, its sulfadiazine complex is inactive. The antifungal activity of [{(η5-C5Me5)RhCl}2(μ-Cl)2], against Candida albicans (MIC 5.4 nM, equivalent to 4 μg mL−1), does not change by reaction with LSZ to form 2. For Cryptococcus neoformans, the antifungal activity of [{(η5-C5Me5)RhCl}2(μ-Cl)2] is two times higher than 2 (MIC 5.4 nM, equivalent to 4 μg mL−1). Generally, complex 2 has the same antifungal activity against the tested fungal species.
It is believable that many organic and inorganic compounds simply have general toxicity that is not related to microbes, but also affects human cells. To discourse this issue, toxicity of the title organometallic complexes was evaluated by measuring cell viability against noncancerous human embryonic kidney cells (HEK293) and blood compatibility with the cell components. Blood compatibility was assessed by calculating the HC10 (the concentration at which 10% haemolysis occurs; was calculated by curve fitting the inhibition values versus log(concentration)). Compound 2 was slightly potent to non-malignant HEK293 (CC50 = 30.48 μg mL−1) and moderate haemoglobin release (HC10 = 15.46 μg mL−1). Interestingly, the well-known Rh(III) precursor ([{(η5-C5Me5)RhCl}2(μ-Cl)2]) was safe to HEK293 and induced negligible haemoglobin release (HC10 > 32 μg mL−1). Based on the biological assays, greatest potential for further biological studies of [{(η5-C5Me5)RhCl}2(μ-Cl)2] as antifungal agent is highly recommended.
Interaction with hen white egg lysozyme
Following the administration of the drugs, there is a highly chance to bind covalently or non-covalently25,26 to the surface-accessible histidyl of proteins. Such interactions may affect the toxicity, biodistribution and pharmacokinetics. While it is established that the target of Pd(II) and Pt(II) based drugs is DNA, recent studies present conversant and reasonable evidences that the binding of Au(III) compounds to protein is the origin for their cytotoxicity.27 On the other hand, some model proteins could be used to carry the biologically active compounds into the living cells.28 Besides, studying the stability of the biologically active molecules in presence of coordinating protein is essential. Therefore, HEWL was chosen for the propose of studying the possibility of bioconjugation and to investigate the stability of our compounds in presence of protein. The interactions between [RuCl(μ-Cl)(η6-p-Cym)]2 and some model proteins were studied with the combination of X-ray crystallography, spectroscopic and spectrometric measurements.29–31 Refinement of [RuCl2(η6-p-Cym)(HEWL)] by 1.6 Å X-ray disclosed that the metalation process occurred at the His15 side-chain.29 Destabilization of the adducts was observed upon mixing lysozyme with [RuCl(μ-Cl)(η6-p-Cym)]2.31 The structure of the apo-ferritin nanocage adduct with [RuCl(μ-Cl)(η6-p-Cym)]2 showed presence of three Ru sites per ferritin monomer.30 Here, the HEWL binding affinity of 1 and 2 was investigated by positive mode electrospray ionization mass spectrometry. The reactivity of the complexes towards HEWL was investigated at the room temperature using 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (complex
1 (complex![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
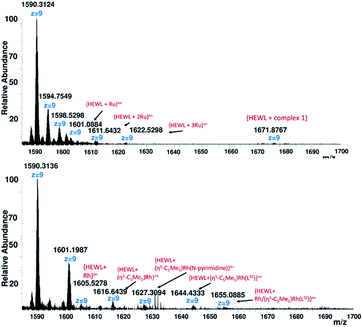
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HEWL). The mass spectrometry spectra were immediately recorded after addition of the organometallic compound to the protein. Soaking lysozyme with ten equivalents of 1 produces mainly three Run+ adduct peaks at m/z 1601.0884 {HEWL + Ru}n+, 1611.6432 {HEWL + (2 × Ru)}n+ and 1622.5298 {HEWL + (3 × Ru)}n+ as well as a weak noncovalent adduct peak25,26 at m/z 1671.8767 corresponding to bind of one molecule of 1 to lysozyme (Fig. 5). Destabilization of the HEWL adduct of 1 and presence of mainly three Ru binding sites per HEWL match the previously reported data of [RuCl(μ-Cl)(η6-p-Cym)]2.29–31
HEWL). The mass spectrometry spectra were immediately recorded after addition of the organometallic compound to the protein. Soaking lysozyme with ten equivalents of 1 produces mainly three Run+ adduct peaks at m/z 1601.0884 {HEWL + Ru}n+, 1611.6432 {HEWL + (2 × Ru)}n+ and 1622.5298 {HEWL + (3 × Ru)}n+ as well as a weak noncovalent adduct peak25,26 at m/z 1671.8767 corresponding to bind of one molecule of 1 to lysozyme (Fig. 5). Destabilization of the HEWL adduct of 1 and presence of mainly three Ru binding sites per HEWL match the previously reported data of [RuCl(μ-Cl)(η6-p-Cym)]2.29–31
Particularly, the ESI-MS of adduct 2 (Fig. 5), shows several adduct peaks at m/z 1605.5278 {HEWL + Rh}n+, 1616.6437 {HEWL + (η5-C5Me5)Rh}n+, 1616.6437 {HEWL + (η5-C5Me5)Rh(N-pyrimidine)}n+, 1644.4333 {HEWL + (η5-C5Me5)Rh(LSZ)}n+ and 1655.3140 {HEWL + Rh/(η5-C5Me5)Rh(LSZ)}n+. The obtained results indicated that metalation of HEWL by Rh(III) fragments occurs via the elimination of one or two sulfadiazine molecules. The fragment at m/z 1655.3140 showed presence of two metalation sites per Rh(III) HEWL adduct. In the absence of X-ray crystallography data of Ru(II) and Rh(III) adducts with lysozyme, it was difficult to assign the side-chain(s) binding sites.
Conclusion
Sulfa drugs, a well-known class of antibiotics, were extensively used to elaborate metal complexes of intriguing biological applications. However, they have not been used for synthesis of organometallic arene compounds. In the present contribution, first two examples of pentamethylcyclopentadienyl Rh(III) and p-cymene Ru(II) complexes bearing sulfadiazine drug (HLSZ) were reported. The influence of the organometallic formation on the antibacterial activity of sulfadiazine was investigated. Single crystal X-ray analysis and spectroscopic studies indicated formation of [(η6-p-Cym)Ru(LSZ)2] and [(η5-C5Me5)Rh(LSZ)2]. The NMR equivalence clearly pointed to involvement of two LSZ molecules in a fluxional process in which the third bond of the base of the “piano stool” is oscillating between two equivalent sulfonamide nitrogen atoms. The electronic structures of the complexes were investigated with the aid of quantum chemical calculations. While the coordination of sulfadiazine drug to cymene Ru(II) gave rise to antimicrobial inactive compound, Rh(III) organometallic compound showed interesting toxicity against Gram-positive bacteria as well as two fungal species; Candida albicans and Cryptococcus neoformans var. grubii. However, the Rh(III) compound showed slightly toxicity to non-malignant HEK293 (CC50 = 30.48 μg mL−1) and moderate haemoglobin release (HC10 = 15.46 μg mL−1). The precursor, [{(η5-C5Me5)RhCl}2(μ-Cl)2], was strongly potent against Candida albicans and Cryptococcus neoformans var. grubii, was safe to normal cells and was compatible with blood components. The interactions between the biologically active compound and protein(s) may affect the toxicity, resistance, biodistribution as well as pharmacokinetics. Recent studies present conversant evidences that binding of some metal complexes to protein is the origin of their cytotoxicity. While [(η6-p-Cym)Ru(LSZ)2] decomposed during the reaction with hen white egg lysozyme (HEWL), a model protein, giving several adducts peaks with ruthenium ion(s), Rh(III) analogue was covalently bio-conjugated to HEWL via the elimination of one sulfadiazine drug. Based on the biological assays, greatest potential for further biological studies of [{(η5-C5Me5)RhCl}2(μ-Cl)2] as antifungal agent is highly recommended.Experimental section
Materials and instruments
The solvents were degassed and purified according to the standard published methods. Sodium(4-aminophenyl)sulfonyl-pyrimidin-2-ylazanide (NaLSZ) was obtained from Sigma. [RuCl(μ-Cl)(η6-p-Cym)]2 (ref. 18) and [{(η5-C5Me5)RhCl}2(μ-Cl)2]21 were synthesized by following the published procedures. Elemental compositions of the organometallic compounds were determined with Elementar Vario MICRO cube CHN analyzer nor an EA 3000 elemental analyzer from HEKtech. Electrospray mass spectra were run with a Thermo-Fisher Exactive Plus instrument with an Orbitrap mass analyzer at a resolution of R = 70.000 and a solvent flow rate of 5 μL min. IR spectra were recorded in the solid state on a Nicolet 380 FT-IR spectrometer equipped with a smart iFTR accessory. Electronic spectra were recorded on an Agilent 8453 diode array spectrophotometer. NMR spectra were recorded with Bruker-Avance 500 (1H: 500.13 MHz and 13C{1H}: 125.77 MHz) and Bruker-Avance 400 (1H: 400.40 MHz, 13C{1H}: 100.70 MHz) spectrometers. Assignments of NMR chemical shifts were done were done with the aid of {1H, 1H} COS90 and {1H, 13C} HSQC.Synthesis
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/char_e0e1.gif) = 3382 (m, NH2), 3335 (m, NH2), 3231 (s), 1624 (w, CC/CN), 1592 (s), 1448 (vs), 1259 (s), 1131 (s), 1081 (s), 828 (m), 787 (s). 1H NMR ([D6]DMSO, 500.13 MHz): δ = 8.64 (br, 2H, pyrimidine-H3 or H5), 8.21 (br, 2H, pyrimidine-H3 or H5), 7.57 (d, 3JH,H = 6.1 Hz, 4H, phenyl-H2/6), 6.68 (br, 2H, pyrimidine-H4), 6.52 (d, 3JH,H = 8.5 Hz, 4H, phenyl-H3/5), 6.03 (d, 3JH,H = 6.0 Hz, 1H, p-Cym-H2 or H6), 5.89 (d, 3JH,H = 6.6 Hz, 1H, p-Cym-H6 or H2), 5.59 (d, 3JH,H = 4.1 Hz, 1H, p-Cym-H3 or H5), 5.67 (br, 4H, NH2), 5.53 (d, 3JH,H = 6.9 Hz, 1H, p-Cym-H5 or H3), 2.63 (sep, 3JH,H = 6.9 Hz, 1H, CH(CH3)2), 1.96 (s, 3H, CH3), 1.01 (d, 3JH,H = 7.3 Hz, 3H, CH(CH3)2), 0.94 (d, 3JH,H = 7.2 Hz, 3H, CH(CH3)2) ppm. 13C NMR ([D6]DMSO, 125.75 MHz): δ = 162.5 (pyrimidine-C1), 159.1 (pyrimidine-C3/C5), 151.5 (phenyl-C4), 129.50 (phenyl-C1), 128.7 (phenyl-C2/6), 112.3 (pyrimidine-C4), 112.0 (phenyl-C3/5), 102.6 (p-Cym-C4), 100.0 (p-Cym-C1), 83.3 (p-Cym-C2 or C6), 83.2 (p-Cym-C6 or C2), 82.4 (p-Cym-C3 or C5), 80.3 (p-Cym-C5 or C3), 30.63 (CH(CH3)2), 22.07 (CH(CH3)2), 21.7 (CH(CH3)2), 17.8 (CH3) ppm. ESI-MS (positive mode, acetone): m/z = 735.1102 {[(η6-p-Cym)Ru(LSZ)2] + H+}+, 485.0577 {[(η6-p-Cym)Ru(LSZ)2] + H+–LSZ}+. C30H32N8O4RuS2: C 49.10, H 4.40, N 15.27, S 8.74, found, C 48.79, H 4.45, N 15.02, S 8.97.
= 3382 (m, NH2), 3335 (m, NH2), 3231 (s), 1624 (w, CC/CN), 1592 (s), 1448 (vs), 1259 (s), 1131 (s), 1081 (s), 828 (m), 787 (s). 1H NMR ([D6]DMSO, 500.13 MHz): δ = 8.64 (br, 2H, pyrimidine-H3 or H5), 8.21 (br, 2H, pyrimidine-H3 or H5), 7.57 (d, 3JH,H = 6.1 Hz, 4H, phenyl-H2/6), 6.68 (br, 2H, pyrimidine-H4), 6.52 (d, 3JH,H = 8.5 Hz, 4H, phenyl-H3/5), 6.03 (d, 3JH,H = 6.0 Hz, 1H, p-Cym-H2 or H6), 5.89 (d, 3JH,H = 6.6 Hz, 1H, p-Cym-H6 or H2), 5.59 (d, 3JH,H = 4.1 Hz, 1H, p-Cym-H3 or H5), 5.67 (br, 4H, NH2), 5.53 (d, 3JH,H = 6.9 Hz, 1H, p-Cym-H5 or H3), 2.63 (sep, 3JH,H = 6.9 Hz, 1H, CH(CH3)2), 1.96 (s, 3H, CH3), 1.01 (d, 3JH,H = 7.3 Hz, 3H, CH(CH3)2), 0.94 (d, 3JH,H = 7.2 Hz, 3H, CH(CH3)2) ppm. 13C NMR ([D6]DMSO, 125.75 MHz): δ = 162.5 (pyrimidine-C1), 159.1 (pyrimidine-C3/C5), 151.5 (phenyl-C4), 129.50 (phenyl-C1), 128.7 (phenyl-C2/6), 112.3 (pyrimidine-C4), 112.0 (phenyl-C3/5), 102.6 (p-Cym-C4), 100.0 (p-Cym-C1), 83.3 (p-Cym-C2 or C6), 83.2 (p-Cym-C6 or C2), 82.4 (p-Cym-C3 or C5), 80.3 (p-Cym-C5 or C3), 30.63 (CH(CH3)2), 22.07 (CH(CH3)2), 21.7 (CH(CH3)2), 17.8 (CH3) ppm. ESI-MS (positive mode, acetone): m/z = 735.1102 {[(η6-p-Cym)Ru(LSZ)2] + H+}+, 485.0577 {[(η6-p-Cym)Ru(LSZ)2] + H+–LSZ}+. C30H32N8O4RuS2: C 49.10, H 4.40, N 15.27, S 8.74, found, C 48.79, H 4.45, N 15.02, S 8.97.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol/CH2Cl2 mixture and then the reaction mixture was stirred at 55 °C for 4 h. Filtration was done to remove water-soluble white precipitate (excess LSZ and sodium chloride). The volume of the solution was reduced to 7 mL and diethyl ether was added. The solution was left overnight. Yellow product was filtered off, washed with diethyl ether and dried under vacuum. Yield (based on metal precursor): 44% (129 mg, 0.18 mmol). IR (ATR, diamond):
1 methanol/CH2Cl2 mixture and then the reaction mixture was stirred at 55 °C for 4 h. Filtration was done to remove water-soluble white precipitate (excess LSZ and sodium chloride). The volume of the solution was reduced to 7 mL and diethyl ether was added. The solution was left overnight. Yellow product was filtered off, washed with diethyl ether and dried under vacuum. Yield (based on metal precursor): 44% (129 mg, 0.18 mmol). IR (ATR, diamond): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/char_e0e1.gif) = 3425 (w, NH2), 3389 (w, NH2), 3355 (m, NH2), 3242 (m, NH2), 1647 (m, CC/CN), 1592, 1459, 1372, 1281, 1128, 1087, 1013, 985, 824, 786. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 8.47 (br, 4H, pyrimidine-H3/H5), 7.50 (d, 3JH,H = 8.7 Hz, 4H, phenyl-H2/6), 6.77 (t, 3JH,H = 4.9 Hz, 2H, pyrimidine-H4), 6.48 (d, 3JH,H = 9.1 Hz, 4H, phenyl-H3/5), 5.62 (s, 4H, NH2), 1.69 (s, 15H, Cp*-CH3) ppm. 13C NMR ([D6]DMSO, 125.75 MHz): δ = 162.3 (pyrimidine-C1), 159.1 (pyrimidine-C3/C5), 151.2 (phenyl-C4), 130.1 (phenyl-C1), 129.0 (phenyl-C2/6), 111.9 (pyrimidine-C4), 111.0 (phenyl-C3/5), 94.8 (d, 1JC–Rh = 7.5 Hz, Cp*-CCH3), 9.0 (Cp*-CCH3). ESI-MS (positive mode, methanol): m/z = 1495.2134 {2 × [(η5-C5Me5)Rh(LSZ)2] + Na+}+, 759.1007 {[(η5-C5Me5)Rh(LSZ)2] + Na+}+, 487.0669 {[(η5-C5Me5)Rh(LSZ)2]-LSZ}+. C30H33N8O4RhS2·0.5CH2Cl2: C 47.02, H 4.55, N 14.38, S 8.23, found C 47.05, H 4.47, N 14.69, S 8.47.
= 3425 (w, NH2), 3389 (w, NH2), 3355 (m, NH2), 3242 (m, NH2), 1647 (m, CC/CN), 1592, 1459, 1372, 1281, 1128, 1087, 1013, 985, 824, 786. 1H NMR ([D6]DMSO, 500.13 MHz): δ = 8.47 (br, 4H, pyrimidine-H3/H5), 7.50 (d, 3JH,H = 8.7 Hz, 4H, phenyl-H2/6), 6.77 (t, 3JH,H = 4.9 Hz, 2H, pyrimidine-H4), 6.48 (d, 3JH,H = 9.1 Hz, 4H, phenyl-H3/5), 5.62 (s, 4H, NH2), 1.69 (s, 15H, Cp*-CH3) ppm. 13C NMR ([D6]DMSO, 125.75 MHz): δ = 162.3 (pyrimidine-C1), 159.1 (pyrimidine-C3/C5), 151.2 (phenyl-C4), 130.1 (phenyl-C1), 129.0 (phenyl-C2/6), 111.9 (pyrimidine-C4), 111.0 (phenyl-C3/5), 94.8 (d, 1JC–Rh = 7.5 Hz, Cp*-CCH3), 9.0 (Cp*-CCH3). ESI-MS (positive mode, methanol): m/z = 1495.2134 {2 × [(η5-C5Me5)Rh(LSZ)2] + Na+}+, 759.1007 {[(η5-C5Me5)Rh(LSZ)2] + Na+}+, 487.0669 {[(η5-C5Me5)Rh(LSZ)2]-LSZ}+. C30H33N8O4RhS2·0.5CH2Cl2: C 47.02, H 4.55, N 14.38, S 8.23, found C 47.05, H 4.47, N 14.69, S 8.47.X-ray diffraction analysis
Single crystals appropriate for X-ray crystallography analysis of 2 were isolated by slow diffusion of n-hexane into CH2Cl2 of the complex. Selected crystals of 2 were immersed in a film of perfluoropolyether oil, mounted on MiTeGen sample holders and transferred to a stream of cold nitrogen of the diffractometer. The crystal data were collected on a Bruker D8 QUEST diffractometer with a CMOS area detector and multi-layer mirror or graphite monochromated Mo-Kα radiation (λ = 0.71073 Å) at 100 K. All the diffracted intensities were collected using an APEX2-CCD detector and corrected for Lorentz-polarization and absorption using the Bruker AXS software. The structures were solved with the SHELXT program using intrinsic phasing method32 and refined with a full-matrix least-squares procedure using the SHELXL program.33 All the non-hydrogen atoms were refined anisotropically refined. Hydrogen atoms were included in the structure factors calculations. Crystallographic data have been deposited with the Cambridge Crystallographic Data Canter as ESI publication no. CCDC 1940322 for compound 2.DFT calculations
Ground-state geometry optimization of 1 and 2 were carried at B3LYP/Genecp (LANL2DZ for Ru, SDD for Rh, and 6-31G(d) for the rest of the elements) level of theory. Time-dependent density functional theory calculations were performed by CAM-B3LYP/LANL2DZ method using PCM model to introduce the solvent effect. All the calculations were done using Gaussian 03 package34 and the obtained data were visualized using Gaussview.35Interaction with hen white egg lysozyme
The reactivity of the synthesized organometallic compounds, dissolved in pure DMSO towards HEWL (2 mg mL−1, H2O) was studied by orbitrap high resolution mass spectrometer (ThermoFisher Exactive plus orbitrap) equipped with the conventional electrospray ionization source. The reaction mixtures were immediately measured after mixing. The working conditions were as follows: spray voltage 3.80 kV, capillary voltage 45 V, and capillary temperature 320 °C. For acquisition, Thermo Xcalibur qual was used.Biological activity testing
[RuCl(μ-Cl)(η6-p-Cym)]2, [{(η5-C5Me5)RhCl}2(μ-Cl)2] and compounds (1 and 2) were initially screened at a fixed concentration (usually 32 μg mL−1 or 20 μM), with a maximum of 0.5% DMSO, final in assay concentration, against a series of bacterial and fungal pathogens; Staphylococcus aureus ATCC 43300, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa ATCC 27853, Candida albicans ATCC 90028 and Cryptococcus neoformans var. grubii H99; ATCC 208821. For active compound, a follow-up hit confirmation is triggered, where the activity is measured by means of a dose–response assay against the same strains.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Antimicrobial screening was performed by CO-ADD (The Community for Antimicrobial Drug Discovery), funded by the Wellcome Trust (UK) and the University of Queensland (Australia).References
- J. H. Maren, Annu. Rev. Pharmacol., 1976, 16, 309 CrossRef PubMed.
- Burger's Medicinal Chemistry and Drug Discovery, ed. M. E. Wolff, Wiley, Laguna Beach, 5th edn, 1996, vol. 2, pp. 528–576 Search PubMed.
- S. Roland, R. Ferone, R. J. Harvey, V. L. Styles and R. W. Morrison, J. Biol. Chem., 1979, 254, 10337 CAS.
- A. Bult, Met. Ions Biol. Syst., 1982, 16, 261 Search PubMed.
- W. H. Wernsdorfer, Acta Trop., 1994, 56, 143 CrossRef CAS PubMed.
- (a) R. Yuan, R. Xiong, Z. Chen, P. Zhang, H. Ju, Z. Dai, Z. Guo, H. Fun and X. You, J. Chem. Soc., Dalton Trans., 2001, 774 RSC; (b) N. C. Baenziger, S. L. Modak and C. L. Fox, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1983, 39, 1620 CrossRef; (c) C. J. Brown, D. S. Cook and L. Sengier, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1985, 41, 718 CrossRef.
- (a) N. C. Baenziger and A. W. Struss, Inorg. Chem., 1976, 15, 1807 CrossRef CAS; (b) D. S. Cook and M. F. Turner, J. Chem. Soc., Perkin Trans. 2, 1975, 1021 RSC.
- M. K. Hassan, R. M. Hassan and M. A. Abd-Alla, Monatsh. Chem., 1991, 122, 829 CrossRef CAS.
- P. A. Ajibade, G. A. Kolawole, P. O'Brien, M. Helliwell and J. Raftery, Inorg. Chim. Acta, 2006, 359, 3111 CrossRef CAS.
- (a) J. Tommasino, F. N. R. Renaud, D. Luneau and G. Pilet, Polyhedron, 2011, 30, 1663 CrossRef CAS; (b) K. K. Narang and J. K. Gupta, Transition Met. Chem., 1977, 2, 83 CrossRef CAS; (c) A. Bult, J. D. Uitterdijk and H. B. Klasen, Transition Met. Chem., 1979, 4, 285 CrossRef CAS; (d) E. Kremer, G. Facchin, E. Estévez, P. Alborés and E. J. Baran, J. Inorg. Biochem., 2006, 100, 1167 CrossRef CAS PubMed.
- (a) L. Menabue and M. Saladini, J. Inorg. Biochem., 1993, 49, 201 CrossRef CAS; (b) K. K. Narang and J. K. Gupta, Transition Met. Chem., 1977, 2, 181 CrossRef CAS.
- A. Pasini and E. Bersanetti, Inorg. Chim. Acta, 1983, 80, 99 CrossRef CAS.
- A. García-Raso, J. J. Fiol, G. Martorell, A. López-Zafra and M. Quirós, Polyhedron, 1997, 16(4), 613 CrossRef.
- W. O. Foye, T. L. Lemke, and D. A. Williams, Principles of Medicinal Chemistry, Williams & Williams, 4th edn, 1995, pp. 709–713 Search PubMed.
- (a) J. Li, M. Tian, Z. Tian, S. Zhang, C. Yan, C. Shao and Z. Liu, Inorg. Chem., 2018, 57, 1705 CrossRef CAS PubMed; (b) W. Ma, L. Guo, Z. Tian, S. Zhang, X. He, J. Li, Y. Yang and Z. Liu, Dalton Trans., 2019, 48, 478 RSC.
- J. M. Radmaker, D. van den Bongard, D. Pluim, J. H. Beijnen and J. H. Schellens, Clin. Cancer Res., 2004, 10, 3717 CrossRef CAS PubMed; D. A. Wolters, M. Stefanopoulou, P. J. Dyson and M. Groessel, Metallomics, 2012, 4, 1185 RSC.
- G. Sava, S. Zorzet, C. Turrin, F. Vita, M. Soranzo, G. Zabucchi, M. Cocchietto, A. Bergamo, S. Di Giovine, G. Pezzoni, L. Sartor and S. Garbisa, Clin. Cancer Res., 2003, 9, 1898 Search PubMed; L. Messori, P. Orioli, D. Vullo, E. Alessio and E. Iengo, Eur. J. Biochem., 2000, 267, 1206 CrossRef CAS PubMed; F. Frausin, V. Scarcia, M. Cocchietto, A. Furlani, B. Serli, E. Alessio and G. Sava, J. Pharmacol. Exp. Ther., 2005, 313, 227 CrossRef PubMed.
- S. B. Jensen, S. J. Rodger and M. D. Spicer, J. Organomet. Chem., 1998, 556, 151 CrossRef CAS.
- A. M. Mansour, F. A. Soliman, O. R. Shehab and N. T. Abdel-Ghani, RSC Adv., 2017, 7, 39989 RSC; O. R. Shehab, F. A. Soliman, N. T. Abdel-Ghani and A. M. Mansour, Spectrochim. Acta, Part A, 2020, 28, 117821 CrossRef CAS PubMed.
- (a) G. M. Golzar-Hossain, A. J. Amoroso, A. Banu and K. M. A. Malik, Polyhedron, 2007, 26, 967 CrossRef; (b) A. M. Mansour, Inorg. Chim. Acta, 2013, 394, 436 CrossRef CAS; (c) A. M. Mansour, J. Coord. Chem., 2013, 66(7), 1118 CrossRef CAS.
- C. White, A. Yates, P. M. Maitlis and D. M. Heinekey, Inorg. Synth., 1992, 29, 228 CAS.
- T. Yanai, D. P. Tew and N. C. Handy, Chem. Phys. Lett., 2004, 393, 51 CrossRef CAS.
- M. A. Blaskovich, J. Zuegg, A. G. Elliott and M. A. Cooper, ACS Infect. Dis., 2015, 1, 285 CrossRef CAS PubMed.
- F. Li, F. J. G. Collins and R. Keene, Chem. Soc. Rev., 2015, 44, 2529 RSC.
- (a) A. M. Mansour and O. R. Shehab, Dalton Trans., 2018, 47, 3459 RSC; (b) A. M. Mansour, O. R. Shehab and K. Radacki, Eur. J. Inorg. Chem., 2020, 299 CrossRef CAS; (c) A. M. Mansour and K. Radacki, Polyhedron, 2020, 175, 114175 CrossRef CAS.
- G. Ferraro, A. M. Mansour and A. Merlino, Dalton Trans., 2018, 47, 10130 RSC.
- L. Engman, M. McNaughton, M. Gajewska, S. Kumar, A. Birmingham and G. Powis, Anticancer Drugs, 2006, 17, 539 CrossRef CAS PubMed.
- (a) M. Chaves-Ferreira, I. S. Albuquerque, D. Matak-Vinkovic, A. C. Coelho, S. M. Carvalho, L. M. Saraiva, C. C. Romão and G. J. L. Bernardes, Angew. Chem., Int. Ed., 2015, 54, 1172 CrossRef CAS PubMed; (b) H. Tabe, K. Fujia, S. Abe, M. Tsujimoto, T. Kuchimaru, S. Kizaka-Kondoh, M. Takano, S. Kitahawa and T. Ueno, Inorg. Chem., 2015, 54, 215 CrossRef CAS PubMed.
- I. W. McNae, K. Fishburne, A. Habtemariam, T. M. Hunter, M. Melchart, F. Wang, M. D. Walkinshaw and P. J. Sadler, Chem. Commun., 2004, 1786 RSC.
- Y. Takezawa, P. Bockmann, N. Sugi, Z. Wang, S. Abe, T. Murakami, T. Hikage, G. Erker, Y. Watanabe, S. Kitagawa and T. Ueno, Dalton Trans., 2011, 40, 2190 RSC.
- M. P. Sullivan, M. Groessel, S. M. Meier, R. L. Kingstone, D. C. Goldstone and C. G. Hartinger, Chem. Commun., 2017, 53, 4246 RSC.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, V. G. Zakrzewski, J. A. Montgomery, R. E. Stratmann, J. C. Burant, S. Dapprich, J. M. Millam, A. D. Daniels, K. N. Kudin, M. C. Strain, O. Farkas, J. Tomasi, V. Barone, M. Cossi, R. Cammi, B. Mennucci, C. Pomelli, C. Adamo, S. Clifford, J. Ochterski, G. A. Petersson, P. Y. Ayala, Q. Cui, K. Morokuma, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. Cioslowski, J. V. Ortiz, A. G. Baboul, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. Gomperts, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, C. Gonzalez, M. Challacombe, P. M. W. Gill, B. G. Johnson, W. Chen, M. W. Wong, J. L. Andres, M. Head-Gordon, E. S. Replogle and J. A. Pople, GAUSSIAN 03 (Revision A.9), Gaussian, Inc., Pittsburgh, 2003 Search PubMed.
- A. Frisch, A. B. Nielson and A. J. Holder, Gaussview User Manual, Gaussian, Inc, Pittsburgh, PA, 2000 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1940322. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0ra01085e |
| This journal is © The Royal Society of Chemistry 2020 |