Open Access Article
Open Access ArticleUltra-low level detection of hepatocellular carcinoma global methylation using a AuNP modified carbon fiber microelectrode†
Bobo Huang‡
a,
Bin Zhang‡b,
Bo Liang *a,
Lu Fangc and
Xuesong Ye
*a,
Lu Fangc and
Xuesong Ye *a
*a
aBiosensor National Special Laboratory, Key Laboratory of Biomedical Engineering of Ministry of Education, College of Biomedical Engineering and Instrument Science, Innovation Center for Minimally Invasive Technique and Device, Zhejiang University, Hangzhou 310027, PR China. E-mail: boliang1986@zju.edu.cn; yexuesong@zju.edu.cn
bKey Laboratory of Laparoscopic Technology of Zhejiang Province, Department of General Surgery, Sir Run-Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou 310016, PR China
cCollege of Automation, Hangzhou Dianzi University, Hangzhou 310018, PR China
First published on 23rd April 2020
Abstract
Hepatocellular carcinoma (HCC) is one of the most common cancerous diseases, with a low 5 year survival rate. Global hypomethylation drives genomic instability, which is regarded as one biomarker for early diagnosis. Long interspersed nucleotide element-1 (LINE-1) makes up around 17% of the genome, and could be regarded as a surrogate marker for global DNA methylation. In this work, a gold nanoparticle (AuNP) modified carbon fiber microelectrode (CFME) with a diameter of 7 μm was applied for the first time to detect the methylation level of LINE-1, by distinguishing adsorption affinities between different DNA bases and AuNPs. Several parameters, including AuNP electrodeposition time, sample adsorption time, and DNA concentration have been analyzed and optimized. The detection limit of our assay was 0.1 nM with only 2 μL sample solution. And the CFME had an excellent sensitivity of 10% methylation change and had the capacity to distinguish only one methylated CpG site. The global DNA methylation level of real samples including cell lines and clinical tissues was tested. Higher signals of HCC cell lines and cancer tissues were observed respectively, compared with normal hepatic cell lines and normal tissues. This work provides a promising approach for HCC early diagnosis and prognosis.
Introduction
Hepatocellular carcinoma (HCC) has become a major health problem worldwide with a high mortality rate.1 The leading causes of HCC are cirrhosis, hepatitis virus infection, alcohol abuse and metabolic syndrome.2 Patients at early stage can be treated with liver resection and radiofrequency ablation (RFA), with a 5 year survival rate of more than 70%, while patients at advanced stage have a poor prognosis.3–6 Thus, this indicates that early detection strategies are very important for the treatment of HCC.7,8 DNA methylation is an important nucleic acid modification that can regulate gene expression and silencing.10,11 Under the catalysis of DNA transmethylase, a methyl group is covalently bonded to the fifth carbon of the cytosine of a CpG dinucleotide. Previous studies have demonstrated that DNA methylation alteration is a cause of carcinogenesis.15–17 DNA methylation is regarded as one promising biomarker for early diagnosis of HCC.19–21Global hypomethylation could activate retrotransposition, leading to genomic instability.22 Regional hypermethylation is often observed in the promoter region of tumor suppressor genes, leading to gene silencing.23,24 Long interspersed nucleotide element-1 (LINE-1), which is widespread in the genome, makes up around 17% of the human genome.25 LINE-1 is one kind of retrotransposition, which can reverse transcript themselves and integrate into a genome. Studies had shown that hypomethylation of LINE-1 was linked to the development of various cancers, including colorectal cancer,26 breast cancer,27 gastric cancer,28 melanoma,29 and HCC.30 And researchers had confirmed that LINE-1 hypomethylation was associated with shorter overall survival and poor prognosis in HCC.31 Therefore, LINE-1 is a surrogate marker for estimating the level of global DNA methylation in HCC.
Traditional methods for global DNA methylation detection, including HPLC-UV, LUMA, ELISA-based, mass spectrometry-based etc.,32 are limited by expensive instruments, costly reagent, time consuming, and large clinical sample usage. More recently, several electrochemical approaches were used to analyse global DNA methylation. Affinity capturing by methylcytosine (5mC) antibody was a common approach to quickly detect global DNA methylation.33–36 These approaches were costly, due to the antibody and conjugation kit, and required long incubation time.
Carbon fiber is conductive, high mechanical strength, and inexpensive,37 which makes it a good substrate for electrode. The carbon fiber microelectrode (CFME) with diameter in several micrometers serves as a unique tool in microvolume and microlocation measurements. Since the fast response time, the CFME was placed into or nearby living cell to monitor chemical releasing.38–40 The high sensitivity and selectivity of the CFME facilitate detection of low concentrations of DNA.41,42 The surface modification can further increase the sensitivity and selectively of the CFME. Gold nanoparticles (AuNPs) are unique material for chemical sensors, since they are highly stable, high surface-to-volume ratio, easily tuning of size and shape.43 The electrostatic interaction between DNA and AuNPs depends on the base, following the adsorption trend as adenine > cytosine > guanine > thymine,44,45 which have been used for DNA methylation detection.18,46 But these methods still consumed a large amount of samples.
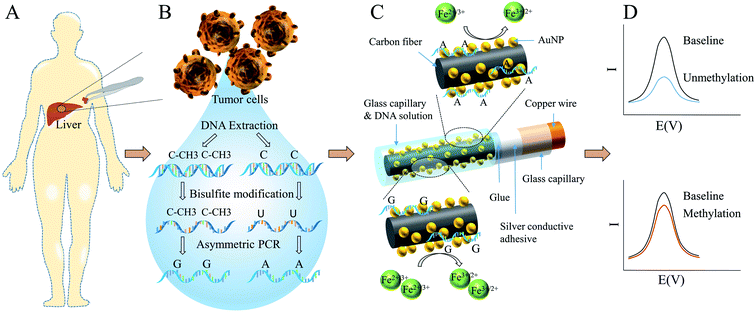
Herein, we demonstrated an AuNPs modified CFME for HCC global methylation detection in ultra-low level for the first time. As illustrated in Scheme 1, tissue samples were derived from patients during surgery. DNA was extracted and purified from tumor cells, followed by bisulfite treatment and asymmetric PCR. Without the protection of methyl groups, cytosine residues convert to uracil. Therefore, methylation and unmethylation sequences were substituted by guanine-enriched and adenine-enriched amplicons respectively after asymmetric PCR. Carbon fiber was bonded to a copper wire with silver conductive adhesive. And then the glass capillary and carbon fiber were sealed with glue. AuNPs were electrodeposited onto the CFME in HAuCl4 solution using the amperometry technique. In order to fix the volume of sample solution used in each experiment and improve the reproducibility, AuNPs-CFME was carefully insert into another larger glass capillary. The DNA solution was added into the other side of capillary and coated the CFME for adsorption. Compared with guanine-enriched sequences, adenine-enriched sequences had higher electrostatic interaction with AuNPs and displayed stronger electrostatic repulsion with bulk ferricyanide [Fe(CN)6]3− ions, leading to lower differential pulse voltammetry (DPV) signals.
Experimental
Chemicals and reagents
Chloroauric acid were purchased from Aladdin Bio-Chem Technology Co., Ltd. (Shanghai, China). DNase/RNase-free distilled water (Dalian Meilun Biotechnology Co., Ltd, China) was used throughout the experiments. The oligonucleotides and primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. Dulbecco's Modified Eagle's Medium (DMEM) and RPMI Medium 1640 basic were obtained from GIBCO, Thermo Fisher Scientific Co., Ltd (Rockford, USA). Fetal bovine serum was provided by CellMax. Carbon fiber was purchased from Jilin Carbon Co., Ltd. All other chemicals not mentioned here were of analytical reagent grade.Apparatus and measurements
The morphologies, structures, and elemental distribution of the AuNPs-CFME were characterized using a field emission scanning electron microscope (FESEM, Hitachi SU8010). AuNPs deposition and electrochemical measurements were carried out on an electrochemical workstation (IVIUM, CompactStat.h). A conventional three-electrode system was used for all electrochemical experiments, which consisted of a working electrode, a platinum wire counter electrode and an Ag/AgCl reference electrode. The electrolyte buffer consisted of 10 mM phosphate buffer solution (pH 7.4) containing 2.5 mM [Fe(CN)6]3−/[Fe(CN)6]4− (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and 0.1 M KCl. The cyclic voltammetry (CV) was carried out with a scan rate of 10 mV s−1. DPV signals were recorded from −0.1 V to 0.6 V with a pulse amplitude of 50 mV and a pulse width of 50 ms.
1) and 0.1 M KCl. The cyclic voltammetry (CV) was carried out with a scan rate of 10 mV s−1. DPV signals were recorded from −0.1 V to 0.6 V with a pulse amplitude of 50 mV and a pulse width of 50 ms.
Fabrication of CFME
A single carbon fiber (7 μm) was attached to one terminal of a copper wire (length: 7 cm, diameter: 80 μm), which was coated with silver conductive adhesive. After dried in the oven at 80 °C, it was carefully inserted into a glass capillary (length: 5 cm, inter diameter: 0.3 mm). Subsequently, the glass capillary was sealed with a drop of non-conductive glue and make sure the carbon fiber was in the middle of the capillary. The exposed carbon fiber was cut to 3 mm with a scissor. For the purpose of removing the attachments on the surface of the carbon fiber, it was cleaned by ultrasonic with ethanol and DNase/RNase-free distilled water separately and then dried in the oven at 90 °C.Preparation of AuNPs modified CFME
The prepared CFME was then immersed into 0.3 mM HAuCl4 solution containing 0.1 M KNO3 and treated by a constant potential at −0.24 V for 50 s to deposit AuNPs onto the CFME. Then the electrode was rinsed with DNase/RNase-free distilled water and gently dried with nitrogen.Preparation of genomic DNA and clinical samples
HCC cell lines including L-02, SK-Hep-1, Hep G2 and SUN-449 were kindly provided by Key Laboratory of Laparoscopic Technology of Zhejiang Province, Department of General Surgery, Sir Run-Run Shaw Hospital. HCC cell lines were cultured in DMEM medium replenished with 10% fetal bovine serum at 37 °C with 5% CO2 under a 95% humidified atmosphere. Clinical samples were treated followed protocols. Briefly, three patients who suffered from HCC and two non-cancer patients were investigated. The research project was carried out according to the principles of the Declaration of Helsinki and consented by the Institutional Review Board of the Sir Run-Run Shaw Hospital. All patients were informed and agreed to the study. During surgery, cancer and normal tissue samples were collected and stored at −80 °C. Genomic DNA was extracted and purified from the cultured cells and clinical samples using the Axygen® AxyPrep Multisource Genomic Miniprep DNA (Corning, USA), according to the manufacturer's instructions. First, cultured cells or clinical samples were suspended and lysed. After centrifuging, DNA was extracted in a spin column and purified DNA was eluted from the column. Finally, purified DNA was stored at −20 °C.Bisulfite treatment and asymmetric PCR
Bisulfite conversion was carried out as described previously.47 MSP was performed using AmpliTaq Gold® 360 Master Mix (Applied biosystems). MSP and asymmetric PCR primes for LINE-1 were listed in Table S1.† Asymmetric PCR was carried out according to previous study.48 The reward primer amount was 0.2 μM. And primer ratio (forward primer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) reward primer) was 1
reward primer) was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50. Up to 40 PCR cycles were used in this experiment.
50. Up to 40 PCR cycles were used in this experiment.
DNA adsorption onto AuNPs-CFME
The exposed carbon fiber was carefully inserted into another glass capillary, in order to fix the amount of samples for each assay, thereby improving the stability and repeatability of the experiments. Also, the glass capillary could protect the protruding carbon fiber from breaking. Due to the capillary action, 2 μL DNA solution was added into the capillary from the other end. The carbon fiber was immersed in the solution and kept static for 15 min at room temperature. Then the CFME was washed three time with 10 mM phosphate buffer. Lower DPV signals were observed on the electrodes which were adsorbed with adenine-enriched sequences, due to the stronger electrostatic repulsion with bulk ferricyanide [Fe(CN)6]3− ions. Relative signal change (%I), which was defined in eqn (1), was used to describe the absorption of the DNA, due to differences in surface area of different electrodes. Δ%I characterized the difference of %I between methylated and unmethylated DNA sequences, which was defined in eqn (2).| %I = (IBaseline − ISample)/IBaseline × 100% | (1) |
| Δ%I = %IUM − %IM | (2) |
Results and discussion
Characterization of AuNPs-CFME
SEM was used to examine the surface morphologies of carbon fiber with/without AuNPs modification. As shown in Fig. S1,† the carbon (red) distributed all over the electrode. The Au (green) almost covered all the electrode, expect the parts surrounded by the red dotted lines. And there was an Au peak after electrodeposition. It could be confirmed that the particles on the CFME were AuNPs. As seen in Fig. 1, the SEM imaging shows that the deposited gold particles were roughly 50 nm in diameters that tend to be randomly distributed on the electrode surface. High-magnification SEM images show that diameter and density of the AuNPs were increased with the rising deposition time, as shown in Fig. S2.†Electrochemical performance of AuNPs-CFME
As shown in Fig. 2A, the sigmoid-shaped voltammograms were acquired on each electrode, indicating that a nonlinear diffusion process was involved in the electrochemical process and revealing the changes of the carbon fiber surface. A low signal was observed on the bare CFME (red), for the small surface area and good sealing of the CFME. The steady-state limiting current of the AuNPs modified electrode increased obviously and the peak-to-peak separation decreased, compared with the bare CFME. As shown in Fig. 2B, the DPV signals of AuNPs modified CFMEs were much larger than the bare CFME (red). As the electrodeposition time increased, the amplitude of the signal changes decreased. These results had demonstrated that the larger surface area after the deposition of AuNPs, leading to more [Fe(CN)6]3−/4− ions undergo redox reactions on the electrode. With increasing deposition time, the increase of specific surface area was limited because the carbon fiber was all covered with AuNPs.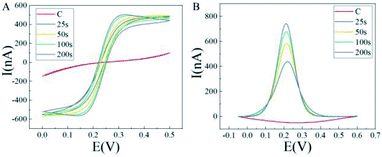 | ||
| Fig. 2 (A) CV signals of bare CFME and AuNPs-CFMEs. (B) DPV signals before and after AuNPs deposition. | ||
Optimization of experimental conditions
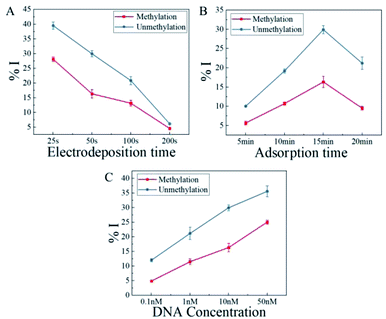
In order to optimize the relevant experimental variables, oligonucleotide sequences containing 7 CpG sites within the LINE-1 gene were synthesized, listed in Table S1.† The guanine-enriched and adenine-enriched sequences represented fully methylated and unmethylated sequences respectively, which underwent bisulfite treatment and asymmetrically PCR-amplification.In order to optimize the electrodeposition time, various periods of time ranging from 25 s to 200 s were applied to modify CFMEs. As seen in Fig. 3A, with the increase of electrodeposition time, both the %I of methylation and unmethylation reduced. %I described the changes of DPV signal after DNA adsorption. With the same quantity of DNA during the experiments, methylated or unmethylated sequences could cover more reaction sites of the modified CFME, while there was a small amount of AuNPs. The degree of coverage of DNA decreased with the increasing electrodeposition time, leading to a decline in %I. In order to distinguish methylated and unmethylated sequences, the difference of %I between both sequences should be the highest. The highest Δ%I was acquired with 50 s of electrodeposition. According to these results, 50 s was selected as the optimal electrodeposition time for a better discrimination between current responses of methylated and unmethylated DNA for this experiment.
In order to maximize the signal difference between methylated and unmethylated oligonucleotides, the AuNPs-CFMEs were immersed in 10 nM DNA solution for various time, ranging from 5 min to 20 min. As seen in Fig. 3B, an adsorption time of only 5 min is sufficient to distinguish these two sequences with a 4.37 Δ%I. 15 min adsorption time led to the maximum level of 13.60 Δ%I, which was decreased with the increasing adsorption time. Long immersion time (>15 min) would cause the nucleic acid to desorb and reduce the signal. At the same time, the two kinds of oligonucleotides on the electrode tend to be saturated, which resulted in a similar level of coulomb repulsion to the bulk [Fe(CN)6]3− ions, providing two DPV signals with almost identical magnitudes leading to a small Δ%I change. Therefor, 15 min was chosen as the optimal parameter for a better discrimination of methylated and unmethylated sequences for this work.
As shown in Fig. 3C, methylated and unmethylated oligonucleotides of various concentrations ranging from 0.1 nM to 50 nM were tested. 15 min and room temperature were applied in the experiment. Even at 0.1 nM, a 7.15 Δ%I was observed. High concentration of DNA would cause similar amount of adsorbed DNA on the surface, which led to identical level of coulomb repulsion between the bulk [Fe(CN)6]3− ions and the surface-bound methylated or unmethylated sequences. So, 10 nM was the optimized sample concentration.
Sensitivity of AuNPs-CFME
For real samples detection, the tissues obtained from patients are usually a mixture of cancer cells and normal cells. Even within the tumor tissue, the micro environments of cancer cells are significantly different, which results various DNA methylation patterns. Therefore, it is necessary to detect the degree of methylation in mixed samples. In order to evaluate the performance of AuNPs-CFME for analysis heterogeneous DNA methylation, mixed samples were prepared. %Methylation (Vmethylation/(Vmethylation + Vunmethylation) × 100%) mean the volume ratios of methylated and unmethylated oligonucleotides while keeping the total amount of nucleic acid in the mixed sample constant. Samples of various %methylation including 0%, 10%, 25% 50%, 75% and 100%, were used to add into the glass capillary under optimized parameters and followed by DPV measurements. As seen in Fig. 4A, %I was decreased with the rising volume ration of methylated oligonucleotides in the mixture samples. The linear regression equation was y = −0.15x + 29.49, with a correlation coefficient (r2) of 0.99. The result demonstrated that adenine-enriched sequences, which represent unmethylated copies, had stronger electrostatic adsorption capacity with the AuNPs, leading to a higher %I. A decreased of 1.82 %I was observed from 0% methylation to 10% methylation, showing as low as 10% methylation change could be distinguished by this assay. A high sensitivity had acquired with the AuNPs-CFME under the optimized conditions.The heterogeneity of cellular methylation is not only reflected in the proportion of methylated nucleic acids but also in the number of methylated CpG sites. As shown in Fig. 4B, synthetic oligonucleotides containing 0, 1, 4 and 7 CpG sites were investigated. As the number of methylated CpG sites increased, the signal of %I decreased, which supported the principle of guanine had weaker adsorption ability on the AuNPs, leading to a higher DPV signal after sample adsorption and lower %I. The linear regression equation was y = −1.98x + 29.52, with a correlation coefficient (r2) of 0.98. A decreased of 2.55 %I was observed from 0 CpG site to 1 CpG site, showing the assay was sensitive to single site methylation. The results indicated that AuNPs-CFME had high specificity and resolution.
Several DNA methylation sensors that based on DNA adsorption affinity with other materials had been summarized in Table 1 with respect to the electrode, sample volume, sample concentration, limit of detection, sensitivity, and specificity. The AuNPs-CFME could distinguish methylation and unmethylation samples at ultra-low level. The limit of detection of the AuNPs-CFME was 5 times lower than other microelectrode and 100 times lower than macroelectrode. As little as 2 μL samples was enough to cover the carbon fiber. Compared with screen print electrode, the CFME was dramatically smaller in size, which was more conducive to integration for the detection of multiple channels or multiple parameters. Microelectrodes normally exhibit improved signal-to-noise ratio.49,50 And the high surface-to-volume ratio of AuNPs further improved the performance.51 Moreover, the CFME had an excellent sensitivity of 10% methylation change and had the capacity to distinguish only one methylated CpG site. Briefly, the AuNPs-CFME could potentially be useful in cancer diagnosis and prognosis.
| Electrode | Electrode size | Sample volume | Lowest synthetic sample concentration | Lowest real sample concentration | Limit of detection | Sensitivity | Specificity | Ref. |
|---|---|---|---|---|---|---|---|---|
| Au-SPR | — | 250 μL | 200 nM | — | — | 25% | — | 9 |
| Au-SPE | 4 mm in diameter | 30 μL | 10 nM | 50 nM | 10 nM | 10% | — | 12 |
| Graphene-SPE | 4 mm in diameter | 8 μL | 100 nM | — | — | 5% | 1 CpG site | 13 |
| Au-microelectrode | 100 μm in diameter | 10 μL | — | 30.9 nM | 0.5 nM | 10% | — | 14 |
| AuNPs-SPE | 2.5 mm in diameter | 5 μL | 12.5 nM | 24 nM | 12.5 nM | 10% | 1 CpG site | 18 |
| AuNPs-CFME | 3 mm in length, 7 μm in diameter | 2 μL | 0.1 nM | 10 nM | 0.1 nM | 10% | 1 CpG site | This work |
Detection of cell lines and clinical samples
To test the application potential and analytical reproducibility of the AuNPs-CFME assay on real samples, LINE-1 sequences extracted from cell lines or clinical tissues were investigated. The concentration of extracted and purified DNA was estimated and then diluted to 10 nM before the assay. The assay was carried out for 15 min at room temperature.As seen in Fig. 5A, significant relative current changes were observed for three HCC cell lines (SK-Hep-1, Hep G2, SUN-449) and normal liver cell line (L-02), indicating the different methylation level of LINE-1. %I of HCC cell lines (red) were higher than normal cell lines (blue), indicating hypomethylation at LINE-1 sequences of HCC cell lines. The results indicated that the AuNPs-CFME could be used to detect abnormal methylation level of LINE-1 in cell derived samples.
We analyzed the global methylation level in five clinical tissue, which were derived from non-carcinoma patients and patients with HCC, in order to further demonstrated the capability of the AuNPs-CFME. As shown in Fig. 5B, the %I of HCC tissues (red) were significantly different with the normal tissue (blue), indicating the LINE-1 were at a higher methylation level in normal tissue compared with HCC tissue. Therefore, the AuNPs-CFME could be applied to discriminate DNA methylation of clinical samples with a little amount of DNA solution, which was very essential in HCC early detection.
Conclusions
In summary, we demonstrated an AuNPs modified CFME for HCC global methylation detection at ultra-low level for the first time. Several parameters including AuNPs electrodeposition time, sample adsorption time, and sample concentration had been analyzed and optimized. The results showed that our assay maintained high sensitivity at low concentration, and the detection limit of our AuNPs-CFME was 0.1 nM. Also, the assay had a good sensitivity of 10% methylation change and could distinguish only one methylated CpG site. Moreover, we had demonstrated the feasibility of this assay to analyze global methylation level of cell lines and clinical samples, indicating that the proposed approach might be a potential method for robust early stage diagnostic and prognostic of HCC.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Natural Science Foundation of Zhejiang Province (LQ20F010011, LY18H180006); Key Research and Development Program of Zhejiang Province (2019C03066); National Natural Science Foundation of China (61501400, 81501555).Notes and references
- L. A. Torre, F. Bray, R. L. Siegel, J. Ferlay, J. Lortet-Tieulent and A. Jemal, Ca-Cancer J. Clin., 2015, 65, 87–108 CrossRef PubMed.
- J. M. Llovet, J. Zucman-Rossi, E. Pikarsky, B. Sangro, M. Schwartz, M. Sherman and G. Gores, Nat. Rev. Dis. Primers, 2016, 2, 16018 CrossRef PubMed.
- J. H. Wang, C. C. Wang, C. H. Hung, C. L. Chen and S. N. Lu, J. Hepatol., 2012, 56, 412–418 CrossRef PubMed.
- A. Villanueva, B. Minguez, A. Forner, M. Reig and J. M. Llovet, Annu. Rev. Med., 2010, 61, 317–328 CrossRef CAS PubMed.
- M. H. Hyun, Y. S. Lee, J. H. Kim, C. U. Lee, Y. K. Jung, Y. S. Seo, H. J. Yim, J. E. Yeon and K. S. Byun, Hepatology, 2018, 68, 977–993 CrossRef CAS PubMed.
- E. G. Giannini, L. Bucci, F. Garuti, M. Brunacci, B. Lenzi, M. Valente, E. Caturelli, G. Cabibbo, F. Piscaglia, R. Virdone, M. Felder, F. Ciccarese, F. G. Foschi, R. Sacco, G. Svegliati Baroni, F. Farinati, G. L. Rapaccini, A. Olivani, A. Gasbarrini, M. Di Marco, F. Morisco, M. Zoli, A. Masotto, F. Borzio, L. Benvegnu, F. Marra, A. Colecchia, G. Nardone, M. Bernardi, F. Trevisani and Italian Liver Cancer group, Hepatology, 2018, 67, 1784–1796 CrossRef CAS.
- E. N. De Toni, A. Schlesinger-Raab, M. Fuchs, W. Schepp, U. Ehmer, F. Geisler, J. Ricke, P. Paprottka, H. Friess and J. Werner, Gut, 2020, 69, 168–176 CrossRef.
- A. G. Singal, A. Pillai and J. Tiro, PLoS Med., 2014, 11, e1001624 CrossRef.
- A. A. Sina, L. G. Carrascosa, R. Palanisamy, S. Rauf, M. J. Shiddiky and M. Trau, Anal. Chem., 2014, 86, 10179–10185 CrossRef CAS PubMed.
- M. Esteller, N. Engl. J. Med, 2008, 358, 1148–1159 CrossRef CAS.
- M. V. C. Greenberg and D. Bourc'his, Nat. Rev. Mol. Cell Biol, 2019, 20, 590–607 CrossRef CAS PubMed.
- K. M. Koo, A. A. Sina, L. G. Carrascosa, M. J. Shiddiky and M. Trau, Analyst, 2014, 139, 6178–6184 RSC.
- M. H. Haque, V. Gopalan, S. Yadav, M. N. Islam, E. Eftekhari, Q. Li, L. G. Carrascosa, N. T. Nguyen, A. K. Lam and M. J. A. Shiddiky, Biosens. Bioelectron., 2017, 87, 615–621 CrossRef CAS PubMed.
- A. A. Sina, M. T. Foster, D. Korbie, L. G. Carrascosa, M. J. A. Shiddiky, J. Gao, S. Dey and M. Trau, Analyst, 2017, 142, 3573–3578 RSC.
- Y. Delpu, P. Cordelier, W. C. Cho and J. Torrisani, Int. J. Mol. Sci., 2013, 14, 15029–15058 CrossRef PubMed.
- K. D. Robertson and A. P. Wolffe, Nat. Rev. Genet., 2000, 1, 11–19 CrossRef CAS PubMed.
- A. Koch, S. C. Joosten, Z. Feng, T. C. de Ruijter, M. X. Draht, V. Melotte, K. M. Smits, J. Veeck, J. G. Herman and L. Van Neste, Nat. Rev. Clin. Oncol., 2018, 15, 459 CrossRef CAS PubMed.
- B. Huang, L. Ji, B. Liang, Q. Cao, T. Tu and X. Ye, Analyst, 2019, 144, 3282–3288 RSC.
- W. C. Mah and C. G. Lee, Biomark. Res., 2014, 2, 5 CrossRef PubMed.
- R. H. Xu, W. Wei, M. Krawczyk, W. Wang, H. Luo, K. Flagg, S. Yi, W. Shi, Q. Quan, K. Li, L. Zheng, H. Zhang, B. A. Caughey, Q. Zhao, J. Hou, R. Zhang, Y. Xu, H. Cai, G. Li, R. Hou, Z. Zhong, D. Lin, X. Fu, J. Zhu, Y. Duan, M. Yu, B. Ying, W. Zhang, J. Wang, E. Zhang, C. Zhang, O. Li, R. Guo, H. Carter, J. K. Zhu, X. Hao and K. Zhang, Nat. Mater., 2017, 16, 1155–1161 CrossRef CAS PubMed.
- S. Udali, P. Guarini, S. Moruzzi, A. Ruzzenente, S. A. Tammen, A. Guglielmi, S. Conci, P. Pattini, O. Olivieri, R. Corrocher, S. W. Choi and S. Friso, Hepatology, 2015, 62, 496–504 CrossRef CAS PubMed.
- A. Eden, F. Gaudet, A. Waghmare and R. Jaenisch, Science, 2003, 300, 455 CrossRef CAS PubMed.
- B. Yang, M. Guo, J. G. Herman and D. P. Clark, Am. J. Pathol., 2003, 163, 1101–1107 CrossRef CAS PubMed.
- Y. H. Shim, G. S. Yoon, H. J. Choi, Y. H. Chung and E. Yu, Cancer Lett., 2003, 190, 213–219 CrossRef CAS PubMed.
- Y. Nagai, E. Sunami, Y. Yamamoto, K. Hata, S. Okada, K. Murono, K. Yasuda, K. Otani, T. Nishikawa, T. Tanaka, T. Kiyomatsu, K. Kawai, H. Nozawa, S. Ishihara, D. S. B. Hoon and T. Watanabe, Oncotarget, 2017, 8, 11906–11916 CrossRef PubMed.
- M. Antelo, F. Balaguer, J. R. Shia, Y. Shen, K. Hur, L. Moreira, M. Cuatrecasas, L. Bujanda, M. D. Giraldez, M. Takahashi, A. Cabanne, M. E. Barugel, M. Arnold, E. L. Roca, M. Andreu, S. Castellvi-Bel, X. Llor, R. Jover, A. Castells, C. R. Boland and A. Goel, PLoS One, 2012, 7, e45357 CrossRef CAS PubMed.
- A. Q. van Hoesel, C. J. van de Velde, P. J. Kuppen, G. J. Liefers, H. Putter, Y. Sato, D. A. Elashoff, R. R. Turner, J. M. Shamonki and E. M. de Kruijf, Breast Cancer Res. Treat., 2012, 134, 1103–1114 CrossRef PubMed.
- H. Shigaki, Y. Baba, M. Watanabe, A. Murata, S. Iwagami, K. Miyake, T. Ishimoto, M. Iwatsuki and H. Baba, Gastric Cancer, 2013, 16, 480–487 CrossRef CAS PubMed.
- S. Hoshimoto, C. T. Kuo, K. K. Chong, T. L. Takeshima, Y. Takei, M. W. Li, S. K. Huang, M. S. Sim, D. L. Morton and D. S. Hoon, J. Investig. Dermatol., 2012, 132, 1689–1697 CrossRef CAS PubMed.
- K. Harada, Y. Baba, T. Ishimoto, A. Chikamoto, K. Kosumi, H. Hayashi, H. Nitta, D. Hashimoto, T. Beppu and H. Baba, Ann. Surg. Oncol., 2015, 22, 1280–1287 CrossRef PubMed.
- C. C. Yeh, A. Goyal, J. Shen, H. C. Wu, J. A. Strauss, Q. Wang, I. Gurvich, R. A. Safyan, G. A. Manji, M. V. Gamble, A. B. Siegel and R. M. Santella, Ann. Surg. Oncol., 2017, 24, 3788–3795 CrossRef PubMed.
- S. Kurdyukov and M. Bullock, Biology, 2016, 5, 3 CrossRef PubMed.
- M. H. Haque, R. Bhattacharjee, M. N. Islam, V. Gopalan, N. T. Nguyen, A. K. Lam and M. J. A. Shiddiky, Analyst, 2017, 142, 1900–1908 RSC.
- R. Bhattacharjee, S. Moriam, N.-T. Nguyen and M. J. Shiddiky, Biosens. Bioelectron., 2019, 126, 102–107 CrossRef CAS PubMed.
- R. Bhattacharjee, S. Tanaka, S. Moriam, M. K. Masud, J. J. Lin, S. M. Alshehri, T. Ahamad, R. R. Salunkhe, N. T. Nguyen, Y. Yamauchi, M. S. A. Hossain and M. J. A. Shiddiky, J. Mater. Chem. B, 2018, 6, 4783–4791 RSC.
- E. Povedano, V. R. V. Montiel, A. Valverde, F. Navarro-Villoslada, P. Yanez-Sedeno, M. Pedrero, A. Montero-Calle, R. Barderas, A. Pelaez-Garcia, M. Mendiola, D. Hardisson, J. Feliu, J. Camps, E. Rodriguez-Tomas, J. Joven, M. Arenas, S. Campuzano and J. M. Pingarron, ACS Sens., 2019, 4, 227–234 CrossRef CAS PubMed.
- S. B. Hocevar, B. Ogorevc, K. Schachl and K. Kalcher, Electroanalysis, 2004, 16, 1711–1716 CrossRef CAS.
- F. Y. Du, W. H. Huang, Y. X. Shi, Z. L. Wang and J. K. Cheng, Biosens. Bioelectron., 2008, 24, 415–421 CrossRef CAS PubMed.
- Y. Chen, Q. W. Li, H. Jiang and X. M. Wang, J. Electroanal. Chem., 2016, 781, 233–237 CrossRef CAS.
- X. Li, S. Majdi, J. Dunevall, H. Fathali and A. G. Ewing, Angew. Chem., 2015, 54, 11978–11982 CrossRef CAS PubMed.
- X. Zhao, X. Lu, W. T. Tze and P. Wang, Biosens. Bioelectron., 2010, 25, 2343–2350 CrossRef CAS PubMed.
- E. Dogru, E. Erhan and O. A. Arikan, Electroanalysis, 2017, 29, 287–293 CrossRef CAS.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- J. J. Storhoff, R. Elghanian, C. A. Mirkin and R. L. Letsinger, Langmuir, 2002, 18, 6666–6670 CrossRef CAS.
- H. Kimura-Suda, D. Y. Petrovykh, M. J. Tarlov and L. J. Whitman, J. Am. Chem. Soc., 2003, 125, 9014–9015 CrossRef CAS PubMed.
- Y. Z. Lin and P. L. Chang, ACS Appl. Mater. Interfaces, 2013, 5, 12045–12051 CrossRef CAS PubMed.
- Q. Tao, L. J. Swinnen, J. Yang, G. Srivastava, K. D. Robertson and R. F. Ambinder, Am. J. Pathol., 1999, 155, 619–625 CrossRef CAS PubMed.
- S. K. Poddar, Mol. Cell. Probes, 2000, 14, 25–32 CrossRef CAS PubMed.
- J. Fang, Z. G. Xie, G. Wallace and X. G. Wang, Appl. Surf. Sci., 2017, 412, 131–137 CrossRef CAS.
- E. J. Wee, S. Rauf, M. J. Shiddiky, A. Dobrovic and M. Trau, Clin. Chem., 2015, 61, 163–171 CrossRef CAS PubMed.
- L. Soleymani, Z. C. Fang, E. H. Sargent and S. O. Kelley, Nat. Nanotechnol., 2009, 4, 844–848 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00905a |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |