Open Access Article
Open Access ArticleBenzimidazolium salts prevent and disrupt methicillin-resistant Staphylococcus aureus biofilms†
Jérémie Tessier and
Andreea R. Schmitzer *
*
Department of Chemistry, University of Montreal, PO Box 6128, Succursale Centre-Ville Montreal, QC, H3C 3J7, Canada. E-mail: ar.schmitzer@umontreal.ca
First published on 4th March 2020
Abstract
Emergence of resistant bacteria encourages us to develop new antibiotics and strategies to compensate for the different mechanisms of resistance they acquire. One of the defense mechanisms of resistant bacteria is the formation of biofilms. Herein we show that benzimidazolium salts with various flexible or rigid side chains act as strong antibiotic and antibiofilm agents. We show that their antibiofilm activity is due to their capacity to destroy the biofilm matrix and the bacterial cellular membranes. These compounds are able to avoid the formation of biofilms and disperse mature biofilms showing a universal use in the treatment of biofilm-associated infections.
Introduction
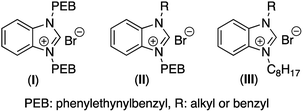
For several years now, researchers have been studying the increase of bacterial resistance,1,2 which plays an increasingly important role in chronic diseases, namely in lung infection and inflammation.3 Deaths from related infections caused by Gram positive resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) are now a major concern in hospital environments.4,5 In addition to developing resistance in their planktonic form, this specific strain of bacteria is known to form biofilms.6 These biofilms are microbial communities attached to a surface and protected from the external environment by a matrix named extracellular polymeric substance (EPS).7 This matrix is composed of polysaccharides, DNA and proteins, surrounded by channels responsible for air, water and nutrient transport into the entire biofilm. This mixture of biopolymers and nutriments provides a viable environment for any microorganism to survive and proliferate. While in the biofilms, bacteria are less sensitive to antibiotics and antimicrobials due to the limited permeation of these compounds into the biofilm and impossibility to reach their site of action. Development of compounds able to destroy the biofilm matrix and possessing bacterial membrane permeation properties and antibacterial properties at the same time seems a good strategy to eradicate these MRSA biofilms. The formation of a biofilm is a multistep process which involves four different stages.8 First, the planktonic bacteria attach to an abiotic or biotic surface. In a second stage, the attachment of the planktonic bacteria to a surface becomes irreversible because of the synthesis of surface proteins, and then bacterial proliferation can occur.9 Subsequent steps involve biofilm maturation and growth, which results in the formation of a bacterial community embedded in the EPS. The detachment of the microorganisms from the biofilm occurs in the last step where planktonic bacteria are released from the biofilm matrix in order to colonize new sites and enlarge the overall structure of the biofilm. Once a biofilm has reach maturity, the bacteria trapped inside the protective layer are less accessible for antibiotics. As the negatively charged extracellular DNA is a major component of these biofilms' matrix,10,11 many new cationic amphiphile agents have been designed to interact with the DNA and achieve a biofilm penetration/destruction. Recent synthesis of cationic polymers,12–14 cationic peptides,15 cationic pillararenes,16 cationic porphyrins,17 cationic dendrimers,18 biomimetic biguanidinylated polyamines,19 and amphiphilic small molecules20 showed interesting results toward the inhibition of biofilm formation and its destruction. Commercially available quaternary ammonium salts and benzalkonium chlorides (BAC) in particular, disrupt the interactions of the extracellular DNA and other components of the bacterial EPS, induce bacterial death, while showing very low toxicity on eukaryotic cells.21 We previously showed that BAC has no activity on MRSA biofilms, even at concentrations higher than its planktonic MIC (330 μg mL−1).22 We previously designed and studied amphiphilic imidazolium and benzimidazolium salts with various flexible or rigid side chains that showed better antibiofilm activities than BAC.21,22 The unique property of benzimidazolium salts is that they can be converted into many structural analogs with completely new properties, for example by only changing the hydrophilic/hydrophobic distribution in the structure.21 The electrostatic interactions with the cellular membranes and their capacity to produce reactive oxygen species are general features that make imidazolium- and benzimidazolium compounds to alter the integrity of microorganisms. Herein, we modified the previously reported bis-phenylethynylbenzyl (bis-PEB) benzimidazolium motif (I) that showed very good planktonic antimicrobial activity, but modest antibiofilm activity,21,22 due to their property to self-assemble in channel-like aggregates. The π-stackable bis-PEB moiety allows the formation of empty channels inside the bacterial membranes which leads to electrolytic imbalance and causes bacterial death.21,22 Once formed inside the biofilm, these channels have low probability to affect the overall electrolytic potential of the isolated biofilm system, which leads to a low or no antibiofilm activity. Benzimidazolium-based mobile synthetic transporters which have very low mobility inside the well packed and organized biofilm matrix also showed only modest antibiofilm activity.23Results and discussion
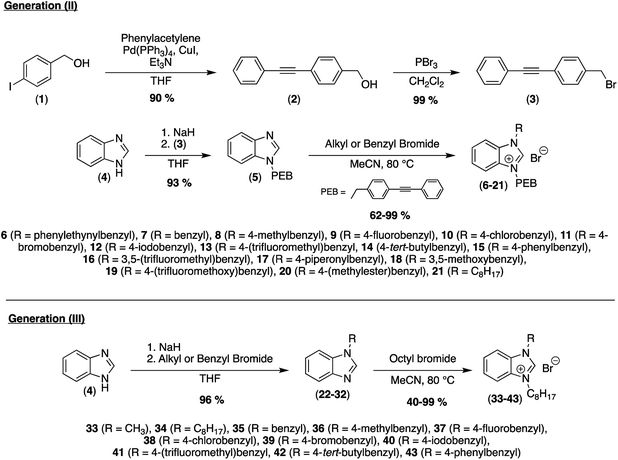
We designed and synthesized new amphiphilic analogues in order to decrease their self-assembling properties and increase their biofilm penetration (Fig. 1) while keeping an overall molecular rigidity. The general synthetic routes for these compounds are summarized in Scheme 1. With this synthetic method, we were able to synthesize a library of compounds bearing different aromatic and aliphatic groups side chains. We substituted one of the highly π-stackable PEB side chain with various benzyl derivative (II) in order to maintain their global hydrophobicity while reducing their channel formation potential.We first investigated the ability of the rigid benzimidazolium salts (II) to kill planktonic bacteria (Table 1). Most of the newly synthesized compounds showed good antimicrobial activity compared to the previously reported generation (I) or commercially available BAC on all the studied bacterial stains. Compounds 11 and 14 possess not only activity on planktonic Gram-positive bacteria (MRSA and VRE) similar to compound 6, but also a 4-fold increased activity on Gram-negative bacteria. Gram-negative bacteria possess an outer membrane acting as an extra protective layer that usually reduces the antibacterial power of synthetic compounds.25
| Antimicrobials | MIC (μg mL−1) | HC50 (μg mL−1) | ||
|---|---|---|---|---|
| MRSA | VRE | E. coli | ||
| a MRSA = methicillin-resistant S. aureus (ATCC 43300), VRE = vancomycin-resistant E. faecium (BAA-2316), E. coli (ATCC 25922), HC50: concentration of the antimicrobials at which 50% of human red blood cells get lysed. | ||||
| BAC | 100 | — | 100 | 95 |
| 6 | 1.25 | 1.25 | 100 | 70 |
| 7 | 5 | 20 | 50 | 200 |
| 8 | 4 | 10 | 20 | 200 |
| 9 | 5 | 10 | 25 | 200 |
| 10 | 2.5 | 5 | 20 | 200 |
| 11 | 1.25 | 5 | 10 | 120 |
| 12 | 2.5 | 5 | 25 | 145 |
| 13 | 2.5 | 5 | 20 | 95 |
| 14 | 1.25 | 2.5 | 25 | 60 |
| 15 | 1.25 | 1.25 | 100 | 70 |
| 16 | 2.5 | 2.5 | 20 | 120 |
| 17 | 5 | 10 | 50 | 150 |
| 18 | 5 | 10 | 50 | 200 |
| 19 | 5 | 5 | 10 | 140 |
| 20 | 5 | 10 | 50 | 200 |
| 21 | 2 | 2.5 | 10 | 50 |
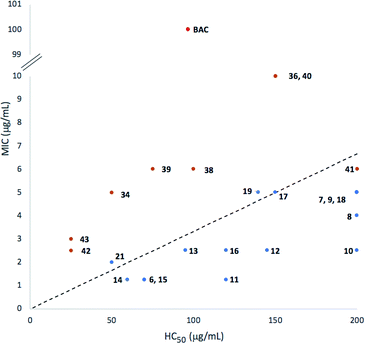
Our new analogues may act on the outer cellular membrane inducing its permeabilization. In addition to their interesting antibacterial activity, these new analogues show low toxicity on red blood cells (Table 1, HC50 values). Indeed, we observed different specificities of these compounds on MRSA, versus red blood cells. We recently showed that the addition of an octyl chain to the benzimidazole fragment of miconazole increased the capacity of these drugs to penetrate phospholipid membranes and increased their antimicrobial activities.24 In this regard we synthesized a semi-rigid analogue (21) bearing one octyl chain and one PEB.
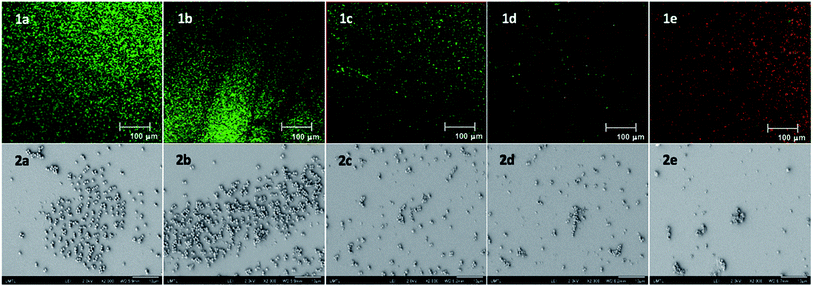
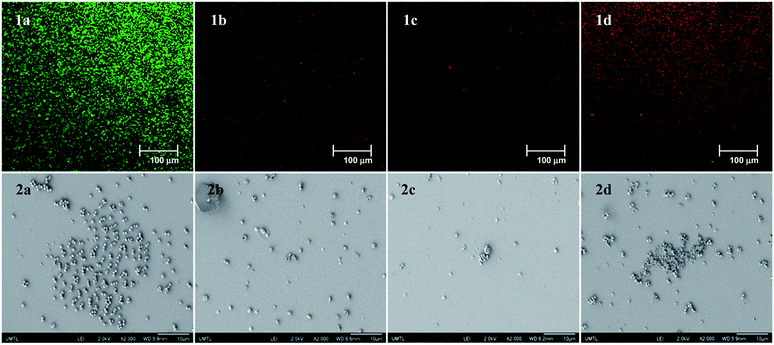
This new compound also showed general antibacterial activity, and low toxicity similar to 6, but also a lower selectivity compared to 11. With these results in hand, we studied the ability of the most active compounds 11, 14, and 21 to prevent the formation of MRSA biofilms using a commercially available fluorescent assay (Baclight Live/Dead).26 Simultaneously, we studied the effect of these analogues on the morphology of the biofilms using scanning electron microscopy (SEM). Using these two techniques, we were able not only to measure the bacterial Live/Dead ratio in the biofilm, but we were also able to observe the effect of those compounds on the spatial organization of the bacterial communities inside the biofilm (Fig. 2).27 Confocal and SEM images of an untreated bacterial biofilms showed the formation of well-defined and homogenous biofilms (Fig. 2(1a and 2a)). Once treated with compound 6 at its MIC, a stripe organization is observed (Fig. 2(1b and 2b)). This change of morphology is the consequence of the bacterial stress response inside the biofilm. This change of morphology is the consequence of the bacterial stress response inside the biofilm. When treated with 6, the bacteria regroup to form a tighter and stronger biofilm, decreasing the ability of the antibiotic to penetrate. As we previously showed, a 20-fold MIC concentration is needed for this type of synthetic transporter to completely inhibit the formation of the biofilm.23 However, once treated with compounds 11 and 14 at their MIC, we observed the formation of overall thinner biofilms (Fig. 2(1c and d, 2c and d)). These fast acting anti-biofilm agents may have an increased capacity to penetrate, weaken and disrupt bacterial biofilms and prevent bacterial stress response. Bacterial biofilm disruption was even more pronounced when the biofilm was treated with the semi-rigid compound 21. As shown by both confocal and SEM images (Fig. 2(1e and 2e)), this antibiofilm agent can disrupt and weaken the bacterial biofilm at its MIC concentration. Not only does this compound have an effect on the thickness and the density of the biofilm, but complete death of the bacterial community can be observed inside the biofilm. These results suggest that the octyl chain can strongly increase the antibiofilm activity and the overall efficiency of those antibacterial agents, acting both on the biofilm matrix and the bacterial membranes.
As compound 21 showed the most promising activity on MRSA biofilms, we designed a new series of analogues bearing octyl chains (Scheme 1, generation III). The decrease of the lipophilic character of an antibiotic often comes with a decrease of its toxicity on red blood cells,28 but these new compounds showed lower specificity for MRSA bacteria versus red blood cells (Table 2).
| Entry | Antimicrobials (–R) | MIC (μg mL−1) | HC50 (μg mL−1) | ||
|---|---|---|---|---|---|
| MRSA | VRE | E. coli | |||
| a MRSA = methicillin-resistant S. aureus (ATCC 43300), VRE = vancomycin-resistant E. faecium (BAA-2316), E. coli (ATCC 25922), HC50: concentration of the antimicrobials at which 50% of human red blood cells get lysed. | |||||
| 33 | Methyl | 400 | 400 | 400 | >200 |
| 34 | Octyl | 5 | 2.5 | 50 | 50 |
| 35 | Benzyl | 50 | 100 | 200 | >200 |
| 36 | 4-Methylbenzyl | 10 | 25 | 200 | 150 |
| 37 | 4-Fluorobenzyl | 25 | 50 | 200 | >200 |
| 38 | 4-Chlorobenzyl | 6 | 15 | 100 | 100 |
| 39 | 4-Bromobenzyl | 6 | 15 | 100 | 75 |
| 40 | 4-Iodobenzyl | 10 | 25 | 100 | 150 |
| 41 | 4-(Trifluoromethyl)benzyl | 6 | 25 | 100 | >200 |
| 42 | 4-tert-Butylbenzyl | 2.5 | 2.5 | 50 | 25 |
| 43 | 4-Phenylbenzyl | 3 | 2.5 | 50 | 25 |
We selected the most actives compounds 34, 39, and 42 and studied their ability to prevent the formation of MRSA biofilms. Once treated with these compounds at their planktonic MICs, we observed a fast and strong inhibition of MRSA biofilm compared to the negative control (Fig. 3). Those amphiphilic salts have the capacity not only to kill bacteria, but also to penetrate and disrupt the biofilm and kill the present bacteria. These compounds are also able to interfere with the biofilm formation and disperse mature biofilms having the potential for universal use in the treatment of biofilm-associated infections.
Conclusions
In regard to the rising MRSA related infections around the world and knowing that the primary defence mechanism of these bacteria is the formation of biofilms, we believe that the inhibition and destruction of those biofilms at low antibiotic concentrations is the key to overcome the challenges brought by the appearance of resistant bacteria. For example, compound 11 showed a 100-fold selectivity to MRSA compared to RBC and a great ability to disrupt mature biofilms (Fig. 4). Herein we show that benzimidazolium salts have antibiotic, antibiofilm activities and good selectivity. Our readily available intermediates and relatively simple synthesis of new analogues make them good candidates for diversification and screening for stronger new activities on bacterial resistant strains.Experimental
1H- and 13C-NMR spectra were recorded on 400 and 75 MHz spectrometers, respectively, in the indicated solvent. Chemical shifts are reported in ppm with internal reference to TMS, and J values are given in hertz. The purity of final compounds used in biological assays was determined by ESI/LC-MS on Quantum TSQ Ultra instruments (C18 (2.1 × 100 mm) column, water +0.1% formic acid). Benzimidazole (4) and 1-methylbenzimidazole (22) were purchase from Sigma-Aldrich and use without further purification. Fresh human red blood cells (blood type O) purchased from Innovative Research in Alserver's solution.(4-Phenylethynyl)benzyl alcohol (2)
To a carefully degassed solution of 4-iodobenzyl alcohol (4.0 g, 17.1 mmol), PPh3 (90 mg, 0.343 mmol), and PdCl2(PPh3)2 (120 mg, 0.171 mmol) in 50 mL of dry THF was added CuI (65 mg, 0.343 mmol) and Et3N (19.1 mL, 137 mmol). The mixture was degassed for another 5 min before phenylacetylene (1.98 mL, 18 mmol) was added dropwise. The reaction was stirred overnight at 50 °C under nitrogen atmosphere. Then, 50 mL of water was added and the aqueous layer was extracted three times with EtOAc. The combined organic fractions were washed with NH4Cl (sat.) where a blue aqueous phase was observed, brine, and dried over Na2SO4. The solvent was removed under reduced pressure and the resulting brownish solid was triturated with hexane until no more apolar (solvent front) product was observed by TLC (40% EtOAc/hexane). Filtration of the resulting solid afforded (4-phenylethynyl)benzyl alcohol (2) as a white solid in 90% isolated yield (3.21 g, 15.4 mmol). 1H NMR (300 MHz, CDCl3) δ 7.54–7.50 (m, 4H), 7.37–7.24 (m, 5H), 4.66 (s, 2H), 2.00 (b, 1H). 13C NMR (75 MHz, CDCl3) δ 140.9, 131.7, 131.5, 128.3, 128.2, 126.7, 123.1, 122.3, 89.3, 89.1, 64.8. HRMS (ESI) calcd for C15H13O+[M + H]+: 209.0961, found 209.0969.(4-Phenylethynyl)benzyl bromide (3)
(4-Phenylethynyl)benzyl alcohol (2) (1.0 g, 4.8 mmol) was dissolved in 20 mL of dry CH2Cl2. The mixture was kept at 0 °C and phosphorus tribromide (0.68 mL, 7.20 mmol) was added dropwise. Then the mixture was stirred 2 hours at 0 °C then water was added and the aqueous phase was extracted 3 times with CH2Cl2. Then, silica gel was added before the solvent was removed under reduced pressure. The product was purified by filtration on silica gel (hexane 100%) to afford (4-phenylethynyl)benzyl bromide (3) as a white solid in 100% isolated yield (1.3 g, 4.8 mmol). 1H NMR (300 MHz, CDCl3) δ 7.54–7.48 (m, 4H), 7.37–7.24 (m, 5H), 4.48 (s, 2H). 13C NMR (75 MHz, CDCl3) δ 137.6, 131.9, 131.5, 129.0, 128.3, 128.2, 123.3, 122.9, 90.2, 88.8, 32.9. HRMS (ESI) calcd for C15H12Br+[M + H]+: 271.0117, found 271.0109.(4-Phenylethynyl)benzyl benzimidazole (5)
To a solution of benzimidazole (869 mg, 7.36 mmol) in THF (30.0 mL) at 0 °C was added sodium hydride (336 mg, 8.4 mmol) portion wise. After stirring for 15 minutes, (4-phenylethynyl)benzyl bromide (3) (1.9 g, 7.00 mmol) was slowly added. The reaction was allowed to warm up to R.T. and stirred until no more starting benzimidazole was observed by TLC (100% EtOAc) (2–3 hours). The reaction was then quenched with water (200 mL) and the resulting precipitate was filtered and washed with water, EtOAc, and hexane to afford (4-phenylethynyl)benzyl benzimidazole (5) as a white solid in 93% yield (2.0 g, 6.5 mmol). 1H NMR (500 MHz, DMSO-d6) δ 7.66–7.51 (m, 7H), 7.44–7.38 (m, 3H), 7.33 (d, J = 8.0 Hz, 2H), 7.21 (d, J = 6.2 Hz, 2H), 5.56 (s, 2H). 13C NMR (126 MHz, DMSO-d6) δ 137.6, 131.7, 131.5, 131.4, 131.3, 128.8, 128.8, 128.8, 127.7, 122.5, 122.1, 121.6, 89.5, 88.9, 47.5. HRMS (ESI) calcd for C22H16N2[M + H]+: 309.1386, found 309.1380.General procedure for generation I and II of benzimidazolium salt (6–21)
To a solution of (4-phenylethynyl)benzyl benzimidazole (5) (135 mg, 0.438 mmol) in MeCN (2 mL) was added the corresponding alkyl or benzyl bromide (1.31 mmol, 3 eq.). The reaction was stirred until LC-MS analysis showed completion of the reaction (24–48 h). MeCN was evaporated and the resulting solid was triturated with EtOAc to remove the excess alkyl or benzyl bromide. The resulting white solid was filtered, washed with hexane, and dried to afford the corresponding benzimidazolium salt (6–21) as a white powder.1,3-Bis(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (6)
Yield 95%. Purity 98.99%. 1H NMR (300 MHz, CDCl3): δ ppm = 9.97 (s, 1H), 7.90–7.96 (m, 1H), 7.52 (m, 14H), 7.38–7.45 (m, 7H), 5.81 (s, 4H). 13C NMR (126 MHz, CDCl3) δ 143.5, 137.3, 131.7, 131.5, 131.4, 131.3, 128.8, 127.6, 122.5, 122.1, 121.1, 89.5, 88.9, 47.5. HRMS (ESI): m/z calcd for C37H27N2[M]+: 499.2169, found 499.2175.3-Benzyl-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (7)
Yield 65%. Purity 100%. 1H NMR (400 MHz, DMSO-d6) δ 10.11 (s, 1H), 7.98 (m, J = 8.8, 6.4, 3.0 Hz, 3H), 7.75–7.51 (m, 12H), 7.50–7.34 (m, 8H), 5.84 (d, J = 15.6 Hz, 4H). 13C NMR (126 MHz, DMSO-d6) δ 143.3, 134.9, 134.4, 132.4, 131.9, 131.6, 129.5, 129.3, 129.2, 129.2, 128.8, 127.3, 123.1, 122.4, 114.5, 114.5, 90.6, 89.2, 50.5, 50.2. HRMS (ESI): m/z calcd for C29H23N2[M]+: 399.1856, found 399.1856.3-(4-Methylbenzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (8)
Yield 65%. Purity 99.48%. 1H NMR (400 MHz, DMSO-d6) δ 10.03 (s, 1H), 7.96 (m, J = 12.2, 6.8, 3.1 Hz, 2H), 7.69–7.60 (m, 4H), 7.60–7.52 (m, 4H), 7.48–7.40 (m, 5H), 7.24 (d, J = 7.9 Hz, 2H), 5.83 (s, 2H), 5.74 (s, 2H), 2.30 (s, 3H).·13C NMR (126 MHz, DMSO-d6) δ 142.7, 138.3, 134.5, 131.9, 131.4, 131.1, 130.8, 129.6, 129.0, 128.8, 128.7, 128.4, 126.9, 122.7, 122.0, 114.1, 114.0, 90.1, 88.7, 49.9, 49.7, 20.7. HRMS (ESI): m/z calcd for C30H25N2[M]+: 413.2001, found 413.2012.3-(4-Fluorobenzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (9)
Yield 79%. Purity 98.65%. 1H NMR (400 MHz, DMSO-d6) δ 10.07 (s, 1H), 8.08–7.90 (m, 2H), 7.68–7.49 (m, 10H), 7.46–7.39 (m, 3H), 7.29 (t, J = 8.9 Hz, 2H), 5.82 (d, J = 17.6 Hz, 4H). 13C NMR (126 MHz, DMSO-d6) δ 163.7, 161.7, 143.3, 134.9, 132.4, 131.9, 131.6, 131.5, 131.4, 131.3, 130.6, 129.5, 129.3, 129.2, 127.3, 123.1, 122.4, 116.5, 116.3, 114.5, 114.5, 90.6, 89.2, 65.4, 62.5, 50.2, 49.8. 19F NMR (471 MHz, DMSO-d6) δ −114.73. HRMS (ESI): m/z calcd for C29H22FN2[M]+: 417.1762, found 417.1760.3-(4-Chlorobenzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (10)
Yield 75%. Purity 96.89%. 1H NMR (400 MHz, DMSO) δ 10.07 (s, 1H), 8.04–7.90 (m, 2H), 7.76–7.49 (m, 12H), 7.47–7.35 (m, 3H), 5.83 (d, J = 12.9 Hz, 4H). 13C NMR (126 MHz, DMSO-d6) δ 143.0, 134.4, 133.5, 132.8, 131.9, 131.4, 131.1, 131.0, 130.4, 129.0, 128.8, 128.7, 126.9, 122.6, 122.0, 114.0, 90.1, 88.7, 49.7, 49.3. HRMS (ESI): m/z calcd for C29H22ClN2[M]+: 433.1466, found 433.1456.3-(4-Bromobenzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (11)
Yield 80%. Purity 99.53%. 1H NMR (400 MHz, DMSO-d6) δ 10.09 (s, 1H), 7.97 (s, 2H), 7.72–7.48 (m, 12H), 7.43 (s, 3H), 5.83 (d, J = 19.5 Hz, 4H). 13C NMR (126 MHz, DMSO) δ 143.0, 134.4, 133.2, 131.9, 131.4, 131.1, 131.0, 130.7, 129.0, 128.8, 128.7, 126.9, 122.6, 122.1, 122.0, 114.0, 90.1, 88.7, 49.7, 49.4. HRMS (ESI): m/z calcd for C29H22CBrN2[M]+: 477.0961, found 477.0959.3-(4-Iodobenzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (12)
Yield 99%. Purity 100%. 1H NMR (400 MHz, DMSO-d6) δ 10.04 (s, 1H), 7.95 (s, 2H), 7.81 (m, J = 6.9 Hz, 2H), 7.70–7.51 (m, 8H), 7.49–7.24 (m, 5H), 5.80 (d, J = 28.5 Hz, 4H). 13C NMR (126 MHz, DMSO) δ 142.9, 137.8, 134.4, 133.5, 131.9, 131.4, 131.1, 131.1, 130.7, 129.0, 128.8, 128.7, 126.9, 122.6, 122.0, 114.0, 95.5, 90.1, 88.7, 49.7, 49.5. HRMS (ESI): m/z calcd for C29H22IN2[M]+: 525.0822, found 525.0804.1-(4-(Phenylethynyl)benzyl)-3-(4-(trifluoromethyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (13)
Yield 65%. Purity 99.57%. 1H NMR (500 MHz, DMSO-d6) δ 10.11 (s, 1H), 8.09–7.90 (m, 2H), 7.83 (d, J = 8.3 Hz, 2H), 7.76 (d, J = 8.2 Hz, 2H), 7.71–7.55 (m, 6H), 7.55 (m, 2H), 7.48–7.37 (m, 3H), 5.91 (d, J = 39.2 Hz, 4H). 13C NMR (126 MHz, DMSO) δ 143.4, 138.7, 134.5, 132.1, 131.6, 131.3, 129.3, 129.2, 129.0, 128.9, 127.1, 127.1, 126.1, 126.0, 122.8, 122.1, 114.2, 114.1, 90.2, 88.8, 49.9, 49.6. 19F NMR (471 MHz, DMSO) δ −63.71. HRMS (ESI): m/z calcd for C30H22F3N2[M]+: 467.1729, found 467.1727.3-(4-(tert-Butyl)benzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (14)
Yield 62%. Purity 99.52%. 1H NMR (500 MHz, DMSO-d6) δ 10.09 (s, 1H), 8.06–8.02 (m, 1H), 7.98–7.93 (m, 1H), 7.68–7.52 (m, 8H), 7.50–7.41 (m, 7H), 5.80 (d, J = 40.9 Hz, 4H), 2.08 (s, 1H), 1.26 (s, 9H). 13C NMR (126 MHz, DMSO) δ 151.3, 142.7, 134.5, 131.9, 131.4, 131.1, 131.1, 131.0, 129.0, 128.8, 128.2, 126.8, 125.8, 122.6, 122.0, 114.1, 114.0, 90.1, 88.7, 49.7, 34.4, 31.0. HRMS (ESI): m/z calcd for C33H31N2[M]+: 455.2482, found 455.2483.3-([1,1′-Biphenyl]-4-ylmethyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (15)
Yield 69%. Purity 100%. 1H NMR (500 MHz, DMSO-d6) δ 10.09 (s, 1H), 8.09–8.03 (m, 1H), 8.00–7.95 (m, 1H), 7.79–7.71 (m, 2H), 7.71–7.52 (m, 12H), 7.51–7.32 (m, 6H), 5.85 (s, 4H). 13C NMR (126 MHz, DMSO) δ 142.9, 140.6, 139.3, 134.4, 133.0, 131.9, 131.4, 131.1, 129.0, 129.0, 128.8, 128.7, 127.8, 127.2, 126.9, 126.7, 122.7, 122.0, 114.1, 114.0, 90.1, 88.7, 49.7. HRMS (ESI): m/z calcd for C35H27N2[M]+: 475.2169, found 455.2159.3-(3,5-Bis(trifluoromethyl)benzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (16)
Yield 68%. Purity 97.62%. 1H NMR (500 MHz, DMSO-d6) δ 10.03 (s, 1H), 8.35 (s, 2H), 8.19 (s, 1H), 8.07–8.01 (m, 1H), 7.99–7.91 (m, 1H), 7.74–7.51 (m, 8H), 7.48–7.39 (m, 3H), 5.97 (s, 2H), 5.84 (s, 2H). 13C NMR (126 MHz, DMSO) δ 143.5, 137.0, 134.4, 131.8, 131.4, 131.1, 131.1, 130.8, 130.6, 129.8, 129.0, 128.8, 128.7, 127.0, 126.9, 122.6, 122.0, 114.1, 113.8, 90.1, 88.7, 49.8, 48.9. 19F NMR (471 MHz, DMSO) δ −62.79. HRMS (ESI): m/z calcd for C31H21F6N2[M]+: 535.1603, found 535.1609.3-(Benzo[d][1,3]dioxol-5-ylmethyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (17)
Yield 87%. Purity 98.85%. 1H NMR (500 MHz, DMSO-d6) δ 10.07 (s, 1H), 8.05–7.99 (m, 1H), 7.99–7.88 (m, 1H), 7.70–7.49 (m, 8H), 7.47–7.36 (m, 3H), 7.20 (d, J = 1.7 Hz, 1H), 7.13 (dd, J = 8.0, 1.8 Hz, 1H), 6.98 (d, J = 8.0 Hz, 1H), 6.04 (s, 2H), 5.84 (s, 2H), 5.69 (s, 2H). 13C NMR (126 MHz, DMSO) δ 147.7, 147.7, 142.7, 134.5, 131.9, 131.4, 131.1, 131.0, 129.0, 128.8, 128.7, 127.2, 126.8, 126.8, 122.7, 122.6, 122.0, 114.1, 113.9, 109.0, 108.6, 101.4, 90.1, 88.7, 50.0, 49.7. HRMS (ESI): m/z calcd for C30H23N2O2[M]+: 443.1754, found 443.1756.3-(3,5-Dimethoxybenzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (18)
Yield 87%. Purity 99.09%. 1H NMR (400 MHz, DMSO-d6) δ 10.15 (s, 1H), 8.15–7.86 (m, 2H), 7.76–7.51 (m, 8H), 7.49–7.41 (m, 3H), 6.75 (s, 2H), 6.51 (s, 1H), 5.87 (s, 2H), 5.72 (s, 2H), 3.74 (s, 6H). 13C NMR (126 MHz, DMSO) δ 160.9, 142.9, 135.9, 134.5, 131.9, 131.4, 131.1, 131.0, 129.0, 128.8, 128.7, 126.8, 122.6, 122.0, 114.1, 114.0, 106.7, 100.0, 90.1, 88.7, 55.4, 50.1, 49.7. HRMS (ESI): m/z calcd for C31H27N2O2[M]+: 459.2067, found 459.2078.1-(4-(Phenylethynyl)benzyl)-3-(4-(trifluoromethoxy)benzyl)-1H-benzo[d]imidazol-3-ium bromide (19)
Yield 75%. Purity 99.63%. 1H NMR (400 MHz, DMSO-d6) δ 10.09 (s, 1H), 8.06–7.89 (m, 2H), 7.72–7.51 (m, 10H), 7.48–7.38 (m, 5H), 5.85 (d, J = 2.7 Hz, 4H). 13C NMR (126 MHz, CDCl3) δ 148.3, 142.9, 134.2, 133.2, 131.7, 131.2, 130.9, 130.9, 130.4, 128.8, 128.6, 128.6, 126.7, 122.5, 121.8, 121.4, 113.9, 113.8, 89.9, 88.5, 49.6, 49.0. 19F NMR (471 MHz, CDCl3) δ −52.03. HRMS (ESI): m/z calcd for C30H22F3N2O[M]+: 483.1679, found 483.1688.3-(4-(Methoxycarbonyl)benzyl)-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (20)
Yield 84%. Purity 100%. 1H NMR (400 MHz, DMSO-d6) δ 10.19 (s, 1H), 8.14–7.85 (m, 4H), 7.78–7.49 (m, 10H), 7.49–7.25 (m, 3H), 5.92 (d, J = 23.2 Hz, 4H), 3.85 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 165.6, 143.0, 139.0, 134.2, 131.7, 131.2, 130.9, 129.6, 129.5, 128.8, 128.6, 128.4, 126.7, 122.5, 121.8, 113.9, 113.8, 89.9, 88.5, 52.1, 49.6, 49.5. HRMS (ESI): m/z calcd for C31H25N2O2[M]+: 457.1911, found 457.1912.3-Octyl-1-(4-(phenylethynyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (21)
Yield 66%. Purity 99.06%. 1H NMR (500 MHz, DMSO-d6) δ 9.97 (s, 1H), 8.27–8.07 (m, 1H), 7.94 (dd, J = 7.3, 1.2 Hz, 1H), 7.76–7.50 (m, 8H), 7.43 (d, J = 2.2 Hz, 3H), 5.82 (s, 2H), 4.51 (t, J = 7.3 Hz, 2H), 2.10–1.73 (m, 2H), 1.41–1.09 (m, 10H), 0.84 (t, J = 7.0 Hz, 3H). 13C NMR (126 MHz, DMSO) δ 142.4, 134.5, 131.8, 131.3, 131.2, 130.7, 128.9, 128.7, 128.5, 126.7, 126.6, 122.5, 121.8, 113.8, 113.7, 90.0, 88.5, 49.5, 46.8, 31.0, 28.4, 28.3, 25.7, 22.0, 13.8. HRMS (ESI): m/z calcd for C30H33N2[M]+: 421.2638, found 421.2645.General procedure for benzimidazole alkylation (23–32)
To a solution of benzimidazole (4) (1.00 g, 8.46 mmol) in THF (25.0 mL) at 0 °C was added sodium hydride (406 mg, 10.2 mmol) portion wise. After stirring for 15 minutes, a solution of the corresponding benzyl or alkyl bromide (8.89 mmol, 1.05 eq.) in THF (5 mL) was slowly added. The reaction was allowed to warm up to R.T. and stirred until no more starting benzimidazole was observed by TLC (100% EtOAc) (2–3 hours). The reaction was then quenched with water and the aqueous layer was extracted three times with EtOAc, dried over Na2SO4 and purified using silica gel chromatography (100% EtOAc) to afford the corresponding alkylated benzimidazole (23–32).1-Octyl-1H-benzo[d]imidazole (23)
Yield 96%, 1H NMR (300 MHz, CDCl3) δ 7.81 (s, 1H), 7.76–7.71 (m, 1H), 7.35–7.31 (m, 1H), 7.25–7.20 (m, 2H), 4.17 (d, 6.3 Hz, 2H), 1.86 (m, 2H), 1.19–1.13 (m, 10H), 0.80 (t, J = 1.8 Hz, 3H) 13C NMR (75 MHz, CDCl3) δ 143.1, 142.6, 133.8, 122.7, 121.2, 120.2, 109.8, 45.3, 31.6, 29.2, 29.3, 26.7, 22.5, 14.0. HRMS (ESI): m/z calcd for C15H22N2[M + H]+: 231.1863, found 231.1867.1-Benzyl-1H-benzo[d]imidazole (24)
Yield 91%, 1H NMR (500 MHz, DMSO-d6) δ 8.41 (s, 1H), 7.68–7.64 (m, 1H), 7.53–7.49 (m, 1H), 7.35–7.25 (m, 5H), 7.23–7.16 (m, 2H), 5.50 (s, 2H). 13C NMR (126 MHz, DMSO) δ 144.25, 143.58, 136.98, 133.67, 128.71, 127.75, 127.40, 122.41, 121.60, 119.51, 110.73, 47.64. HRMS (ESI): m/z calcd for C14H13N2[M + H]+: 209.1073, found 209.1079.1-(4-Methylbenzyl)-1H-benzo[d]imidazole (25)
Yield 40%, 1H NMR (500 MHz, DMSO-d6) δ 8.39 (s, 1H), 7.70–7.60 (m, 1H), 7.55–7.40 (m, 1H), 7.27–7.04 (m, 6H), 5.44 (s, 2H), 2.24 (s, 3H). 13C NMR (126 MHz, DMSO) δ 144.2, 143.6, 137.0, 133.9, 133.6, 129.2, 127.4, 122.3, 121.5, 119.5, 110.7, 47.4, 20.6. HRMS (ESI): m/z calcd for C15H15N2[M + H]+: 223.1230, found 223.1232.1-(4-Fluorobenzyl)-1H-benzo[d]imidazole (26)
Yield 97%, 1H NMR (500 MHz, DMSO-d6) δ 8.44 (s, 1H), 7.71–7.66 (m, 1H), 7.57–7.51 (m, 1H), 7.43–7.36 (m, 2H), 7.27–7.13 (m, 4H), 5.50 (s, 2H). 13C NMR (126 MHz, DMSO) δ 162.59, 160.65, 144.15, 143.62, 133.55, 133.20, 133.17, 129.62, 129.56, 122.42, 121.61, 119.53, 115.58, 115.40, 110.66, 46.87. 19F NMR (471 MHz, DMSO) δ −114.63. HRMS (ESI): m/z calcd for C14H12FN2[M + H]+: 227.0979, found 227.0973.1-(4-Chlorobenzyl)-1H-benzo[d]imidazole (27)
Yield 92%, 1H NMR (500 MHz, DMSO-d6) δ 8.42 (s, 1H), 7.73–7.62 (m, 1H), 7.55–7.47 (m, 1H), 7.43–7.36 (m, 2H), 7.36–7.26 (m, 2H), 7.25–7.15 (m, 2H), 5.50 (s, 2H). 13C NMR (126 MHz, DMSO) δ 144.23, 143.59, 135.98, 133.55, 132.41, 129.30, 128.70, 122.50, 121.69, 119.56, 110.66, 46.90. HRMS (ESI): m/z calcd for C14H12ClN2[M + H]+: 243.0684, found 243.0680.1-(4-Bromobenzyl)-1H-benzo[d]imidazole (28)
Yield 62%, 1H NMR (500 MHz, DMSO-d6) δ 8.42 (s, 1H), 7.71–7.63 (m, 1H), 7.57–7.46 (m, 3H), 7.29–7.24 (m, 2H), 7.24–7.15 (m, 2H), 5.49 (s, 2H). 13C NMR (126 MHz, DMSO) δ 144.2, 143.6, 136.4, 133.6, 131.6, 129.6, 122.5, 121.7, 120.9, 119.6, 110.7, 46.9. HRMS (ESI): m/z calcd for C14H12BrN2[M + H]+: 287.0178, found 287.0173.1-(4-Iodobenzyl)-1H-benzo[d]imidazole (29)
Yield 71%, 1H NMR (400 MHz, DMSO-d6) δ 8.40 (s, 1H), 7.77–7.56 (m, 3H), 7.48 (d, J = 5.7 Hz, 1H), 7.29–6.92 (m, 4H), 5.47 (s, 2H). 13C NMR (126 MHz, DMSO) δ 143.7, 143.1, 135.9, 133.0, 131.1, 129.1, 122.0, 121.2, 120.4, 119.1, 110.2, 46.5. HRMS (ESI): m/z calcd for C14H12IN2[M + H]+: 335.0040, found 335.0041.1-(4-(Trifluoromethyl)benzyl)-1H-benzo[d]imidazole (30)
Yield 96%, 1H NMR (500 MHz, DMSO-d6) δ 8.45 (s, 1H), 7.79–7.60 (m, 3H), 7.56–7.43 (m, 3H), 7.28–7.09 (m, 2H), 5.63 (s, 2H). 13C NMR (126 MHz, DMSO) δ 144.3, 143.6, 141.8, 133.6, 128.0, 125.6, 125.6, 125.6, 122.6, 121.7, 119.6, 110.6, 47.0. 19F NMR (471 MHz, DMSO) δ −64.06. HRMS (ESI): m/z calcd for C15H12F3N2[M + H]+: 277.0947, found 277.0942.1-(4-(tert-Butyl)benzyl)-1H-benzo[d]imidazole (31)
Yield 84%, 1H NMR (500 MHz, DMSO-d6) δ 8.41 (s, 1H), 7.66 (d, J = 7.3 Hz, 1H), 7.55 (d, J = 7.3 Hz, 1H), 7.38–7.32 (m, 2H), 7.26–7.16 (m, 4H), 5.45 (s, 2H), 1.22 (s, 9H). 13C NMR (126 MHz, DMSO) δ 150.1, 134.0, 127.1, 125.4, 122.3, 121.5, 119.5, 110.7, 47.2, 34.2, 31.0. HRMS (ESI): m/z calcd for C18H21N2[M + H]+: 265.1699, found 265.1697.1-([1,1′-Biphenyl]-4-ylmethyl)-1H-benzo[d]imidazole (32)
Yield 44%, 1H NMR (500 MHz, DMSO-d6) δ 8.45 (s, 1H), 7.70–7.54 (m, 6H), 7.47–7.32 (m, 5H), 7.26–7.11 (m, 2H), 5.55 (s, 2H). 13C NMR (126 MHz, DMSO) δ 144.4, 143.8, 139.8, 136.3, 133.8, 129.1, 128.1, 127.7, 127.2, 126.8, 122.6, 121.8, 119.7, 110.9, 47.4. HRMS (ESI): m/z calcd for C20H17N2[M + H]+: 285.1386, found 285.1386.General procedure for generation III of benzimidazolium salt formation (33–43)
To a solution of corresponding alkylated benzimidazole (22–32) (0.438 mmol) in MeCN (2 mL) was added octyl bromide (1.31 mmol, 3 eq.). The reaction was stirred until LC-MS analysis showed completion of the reaction (24–48 h). Another 3 equivalents of octyl bromide were added if the reaction was not completed after 48 h and stirred for an additional 24 h. MeCN was then evaporated and the resulting solid was triturated with EtOAc to remove the excess reagents. The resulting white solid was filtered, washed with hexane, and dried to afford the corresponding benzimidazolium salt (33–43) as a white powder.1-Methyl-3-octyl-1H-benzo[d]imidazol-3-ium bromide (33)
Yield 94%. Purity 99.36%. 1H NMR (500 MHz, CDCl3) δ 11.20 (s, 1H), 7.77–7.73 (m, 1H), 7.71–7.66 (m, 1H), 7.64–7.59 (m, 2H), 4.55–4.48 (m, 2H), 4.26 (s, 3H), 1.98 (q, J = 7.6 Hz, 2H), 1.38–1.31 (m, 2H), 1.31–1.22 (m, 2H), 1.22–1.11 (m, 6H), 0.76 (t, J = 7.0 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 142.72, 131.97, 131.02, 127.28, 127.25, 113.10, 112.90, 47.68, 33.86, 31.58, 29.46, 28.92, 28.91, 26.49, 22.47, 13.97. HRMS (ESI): m/z calcd for C16H25N2[M]+: 245.2012, found 245.2012.1,3-Dioctyl-1H-benzo[d]imidazol-3-ium bromide (34)
Yield 81%. Purity 95.46%. 1H NMR (400 MHz, DMSO-d6) δ 9.88 (s, 1H), 8.11 (dd, J = 6.2, 3.1 Hz, 2H), 7.69 (dd, J = 6.3, 3.1 Hz, 2H), 4.49 (t, J = 7.1 Hz, 4H), 1.98–1.78 (m, 4H), 1.47–1.07 (m, 20H), 0.83 (t, J = 6.7 Hz, 6H). 13C NMR (126 MHz, DMSO) δ 142.07, 131.10, 126.55, 113.74, 46.67, 39.52, 31.14, 28.50, 28.45, 28.39, 25.73, 22.04, 13.93. HRMS (ESI): m/z calcd for C23H39N2[M]+: 343.3113, found 343.3115.1-Benzyl-3-octyl-1H-benzo[d]imidazol-3-ium bromide (35)
Yield 67%. Purity 95.83%. 1H NMR (500 MHz, DMSO-d6) δ 10.03 (s, 1H), 8.13 (dd, J = 7.2, 1.3 Hz, 1H), 7.98 (dd, J = 7.1, 1.3 Hz, 1H), 7.77–7.61 (m, 2H), 7.58–7.48 (m, 2H), 7.47–7.34 (m, 3H), 5.80 (s, 2H), 4.53 (t, J = 7.3 Hz, 2H), 1.98–1.90 (m, 2H), 1.37–1.17 (m, 10H), 0.91–0.82 (m, 3H). 13C NMR (126 MHz, DMSO) δ 142.4, 134.1, 131.3, 130.8, 129.0, 128.7, 128.2, 126.7, 126.6, 113.9, 113.9, 49.8, 46.8, 31.1, 28.5, 28.4, 28.4, 25.8, 22.0, 13.9. HRMS (ESI): m/z calcd for C22H29N2[M]+: 321.2325, found 321.2326.1-(4-Methylbenzyl)-3-octyl-1H-benzo[d]imidazol-3-ium bromide (36)
Yield 99%. Purity 99.69%. 1H NMR (400 MHz, DMSO-d6) δ 10.13 (s, 1H), 8.12 (d, J = 6.9 Hz, 1H), 8.04–7.94 (m, 1H), 7.65 (m, 2H), 7.44 (d, J = 7.9 Hz, 2H), 7.20 (d, J = 7.8 Hz, 2H), 5.76 (s, 2H), 4.53 (t, J = 7.2 Hz, 2H), 2.27 (s, 3H), 1.92 (m, 2H), 1.38–1.13 (m, 10H), 0.83 (t, J = 6.7 Hz, 3H). 13C NMR (126 MHz, DMSO) δ 142.2, 138.1, 131.3, 131.1, 130.7, 129.4, 128.3, 126.6, 126.6, 113.9, 113.9, 49.6, 46.8, 31.1, 28.5, 28.4, 28.4, 25.7, 22.0, 20.7, 13.9. HRMS (ESI): m/z calcd for C23H31N2[M]+: 335.2482, found 335.2473.1-(4-Fluorobenzyl)-3-octyl-1H-benzo[d]imidazol-3-ium bromide (37)
Yield 81%. Purity 100%. 1H NMR (500 MHz, DMSO-d6) δ 10.00 (s, 1H), 8.12 (dd, J = 7.0, 1.7 Hz, 1H), 8.06–7.94 (m, 1H), 7.74–7.57 (m, 4H), 7.35–7.22 (m, 2H), 5.77 (s, 2H), 4.51 (t, J = 7.3 Hz, 2H), 1.93 (p, J = 7.6 Hz, 2H), 1.43–1.18 (m, 10H), 0.84 (t, J = 6.7 Hz, 3H). 13C NMR (126 MHz, DMSO) δ 206.5, 163.1, 161.2, 142.4, 131.3, 130.8, 130.7, 130.3, 130.3, 126.7, 126.7, 115.9, 115.8, 113.9, 113.8, 49.1, 46.8, 31.1, 30.7, 28.5, 28.4, 25.8, 22.0, 13.9. 19F NMR (471 MHz, DMSO) δ −114.68. HRMS (ESI): m/z calcd for C22H28FN2[M]+: 339.2231, found 339.2232.1-(4-Chlorobenzyl)-3-octyl-1H-benzo[d]imidazol-3-ium bromide (38)
Yield 88% Purity 100%. 1H NMR (500 MHz, DMSO-d6) δ 9.99 (s, 1H), 8.21–8.05 (m, 1H), 8.05–7.93 (m, 1H), 7.75–7.64 (m, 2H), 7.57 (d, J = 8.5 Hz, 2H), 7.53–7.39 (m, 2H), 5.79 (s, 2H), 4.51 (t, J = 7.3 Hz, 2H), 1.96–1.89 (m, 2H), 1.37–1.20 (m, 10H), 0.84 (t, J = 7.0 Hz, 3H). 13C NMR (126 MHz, DMSO) δ 142.9, 133.9, 133.5, 131.8, 131.3, 130.8, 129.4, 127.2, 127.1, 114.4, 114.3, 49.6, 47.3, 31.6, 29.0, 28.9, 26.2, 22.5, 14.4. HRMS (ESI): m/z calcd for C22H28ClN2[M]+: 355.1936, found 355.1941.1-(4-Bromobenzyl)-3-octyl-1H-benzo[d]imidazol-3-ium bromide (39)
Yield 93%. Purity 100%. 1H NMR (500 MHz, DMSO-d6) δ 9.88 (s, 1H), 8.12 (dd, J = 7.1, 1.3 Hz, 1H), 7.95 (dd, J = 7.0, 1.3 Hz, 1H), 7.75–7.54 (m, 4H), 7.48 (d, J = 8.5 Hz, 2H), 5.74 (s, 2H), 4.50 (t, J = 7.3 Hz, 2H), 1.92 (d, J = 7.1 Hz, 2H), 1.40–1.15 (m, 10H), 0.85 (t, J = 6.9 Hz, 3H). 13C NMR (126 MHz, DMSO) δ 142.9, 133.9, 132.3, 131.8, 131.2, 131.1, 127.2, 127.1, 122.5, 114.4, 114.3, 49.6, 47.3, 31.6, 29.0, 28.9, 26.2, 22.5, 14.4. HRMS (ESI): m/z calcd for C22H28BrN2[M]+: 399.1430, found 399.1424.1-(4-Iodobenzyl)-3-octyl-1H-benzo[d]imidazol-3-ium bromide (40)
Yield 46%. Purity 97.73%. 1H NMR (500 MHz, DMSO-d6) δ 9.89 (s, 1H), 8.11 (dd, J = 7.3, 1.3 Hz, 1H), 7.94 (dd, J = 7.2, 1.2 Hz, 1H), 7.86–7.73 (m, 2H), 7.73–7.55 (m, 2H), 7.32 (d, J = 8.4 Hz, 2H), 5.72 (s, 2H), 4.50 (t, J = 7.3 Hz, 2H), 1.98–1.84 (m, 2H), 1.40–1.14 (m, 10H), 0.84 (d, J = 6.9 Hz, 3H). 13C NMR (126 MHz, DMSO) δ 142.5, 133.4, 131.8, 131.3, 130.8, 130.6, 126.7, 126.6, 122.0, 113.9, 113.8, 49.1, 46.8, 31.1, 28.5, 28.4, 25.8, 22.0, 13.9. HRMS (ESI): m/z calcd for C22H28IN2[M]+: 447.1292, found 447.1281.3-Octyl-1-(4-(trifluoromethyl)benzyl)-1H-benzo[d]imidazol-3-ium bromide (41)
Yield 71%. Purity 100%. 1H NMR (400 MHz, DMSO-d6) δ 10.05 (s, 1H), 8.14 (d, J = 7.5 Hz, 1H), 7.97 (d, J = 7.5 Hz, 1H), 7.83–7.58 (m, 6H), 5.92 (s, 2H), 4.52 (t, J = 7.2 Hz, 2H), 2.10–1.81 (m, 2H), 1.27 (d, J = 35.6 Hz, 10H), 0.84 (t, J = 6.6 Hz, 3H). 13C NMR (126 MHz, DMSO) δ 143.1, 139.2, 131.8, 131.3, 129.5, 127.3, 127.2, 126.3, 126.3, 114.4, 114.2, 49.7, 47.4, 31.6, 29.0, 28.9, 26.2, 22.5, 14.4. 19F NMR (471 MHz, DMSO) δ −63.87. HRMS (ESI): m/z calcd for C23H28F3N2[M]+: 389.2199, found 389.2206.1-(4-(tert-Butyl)benzyl)-3-octyl-1H-benzo[d]imidazol-3-ium bromide (42)
Yield 83%. Purity 98.83%. 1H NMR (400 MHz, DMSO-d6) δ 10.00 (s, 1H), 8.18–8.08 (m, 1H), 8.08–7.96 (m, 1H), 7.74–7.62 (m, 2H), 7.54–7.37 (m, 4H), 5.73 (s, 2H), 4.51 (t, J = 7.2 Hz, 2H), 1.96–1.76 (m, 2H), 1.24 (s, 19H), 0.96–0.67 (m, 3H). 13C NMR (126 MHz, DMSO) δ 151.2, 142.3, 131.3, 131.2, 130.9, 128.0, 126.7, 126.6, 125.7, 113.9, 49.5, 46.8, 34.4, 31.1, 31.0, 28.5, 28.4, 25.8, 22.0, 13.9. HRMS (ESI): m/z calcd for C26H37N2[M]+: 377.2951, found 377.2954.1-([1,1′-Biphenyl]-4-ylmethyl)-3-octyl-1H-benzo[d]imidazol-3-ium bromide (43)
Yield 97%. Purity 98.66%. 1H NMR (500 MHz, DMSO-d6) δ 10.02 (s, 1H), 8.12 (dd, J = 6.5, 2.4 Hz, 1H), 8.03 (dd, J = 6.5, 2.4 Hz, 1H), 7.74–7.60 (m, 8H), 7.46 (t, J = 7.6 Hz, 2H), 7.41–7.32 (m, 1H), 5.82 (s, 2H), 4.52 (t, J = 7.3 Hz, 2H), 2.02–1.83 (m, 2H), 1.47–1.18 (m, 10H), 0.91–0.66 (m, 3H). 13C NMR (126 MHz, DMSO) δ 142.9, 141.0, 139.8, 133.7, 131.8, 131.4, 129.5, 129.4, 128.2, 127.7, 127.2, 127.2, 114.4, 50.0, 47.3, 31.6, 29.0, 28.9, 26.3, 22.5, 14.4. HRMS (ESI): m/z calcd for C28H33N2[M]+: 397.2638, found 397.2638.Bacterial strains, culture conditions, and viability assays
MICs were determined in 96-well microtiter plates. Assays were conducted in Luria Broth (LB) medium at 37 °C in triplicate. Bacterial cell density (OD at 600 nm) was measured using a Fischer Scientific cell density meter model 40. UV-vis and fluorescence spectroscopy experiments were performed on a Tecan Infinite M200 microplate reader.Hemolytic assay
Red blood cells in Alserver's solution were centrifuged 10 min at 300 × g, washed 3 times with PBS buffer and resuspended in PBS at 2% v/v. In a 96 wells plate was added 195 μL of red blood cells solution and 5 μL of compound in DMSO, and the plate was incubated with light agitation for 1 h at 37 °C. The plate was then centrifuged for 10 min at 300 × g and 50 μL of the supernatant of each well was transferred to another plate. Absorbance was measured at λ = 405 nm.Biofilm inhibition assay
S. aureus cells were incubated in LB medium at 37 °C for 5 h and rediluted in LB medium to the desired final concentration (OD600 nm = 0.1–0.15). S. aureus biofilms were labeled with Live/Dead stain after 12 h incubation with antibiotics in growth media (LB broth). Negative control: DMSO (final concentration not exceeding 10% volume). Positive control: 70% ethanol.Biofilm staining and confocal laser scanning microscopy (LSM) analysis
Biofilms stained with FilmTracer™ LIVE/DEAD® Biofilm viability kit (Molecular Probes, Life Technologies Ltd.). Briefly, a working solution of fluorescent stains was prepared by adding 1 μL of SYTO® 9 stain and 1 μL of PI stain to 1 mL of filter-sterilized water. Two hundred μL of staining solution were deposited on each well of a chambered coverglass (8-well), after 15 min at room temperature in the dark, samples were washed with sterile saline (0.9% NaCl) from base of the support material. Then, biofilms were examined with a confocal laser microscope (Leica model TCS SP5; Leica Microsystems CMS GmbH, Mannheim, Germany) using a 20× dry objective (HC PL FLUOTAR 20.0× 0.50 DRY). A 488 nm laser line was used to excite SYTO® 9, while the fluorescent emission was detected from 500 to 540 nm. PI was sequentially excited with 561 nm laser line and its fluorescent emission was detected from 600 to 695 nm.Scanning electron microscopy
Bacterial samples were applied to titanium surfaces for 12 hours in order to form biofilms, following by a fixation for 1 h at 4 °C in 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M phosphate buffer (PB), pH 7.3, and subsequently rinsed three times with PB. Following fixation, samples were incubated for 1 hour in 1% osmium tetroxide at RT and then dehydrated through an ethanol series (30%, 50%, 70%, 90%, 95% and two times 100%) followed by drying using a Critical Point Drier CPD300 (Leica Biosystems, Concord, ON, Canada). A JEOL JSM-7400F (JEOL Ltd, Tokyo, Japan) field-emission scanning electron microscope (FE-SEM) operated at 1.5 kV was used to image the samples.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Université de Montréal for financial support. We thank J. N. Pelletier and K. J. Wilkinson from the Département de Chimie-Université de Montréal for access to their laboratories and instruments. We thank A. Fouillen (PhD) from the electron microscopy platform of the University of Montreal for the SEM preparation and analysis of the samples.References
- E. Peterson and P. Kaur, Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens, Front. Microbiol., 2018, 9, 2928 CrossRef PubMed
.
- S. B. Zaman, M. A. Hussain, R. Nye, V. Mehta, K. T. Mamun and N. Hossain, A review on antibiotic resistance: alarm bells are ringing, Cureus, 2017, 9, 1403 Search PubMed
.
- A. A. Pragman, J. P. Berger and B. J. Williams, Understanding persistent bacterial lung infections: clinical implications informed by the biology of the microbiota and biofilms, Clin. Pulm. Med., 2016, 23, 57–66 CrossRef PubMed
.
- S. L. Andie, H. de Lencastre, J. Garau, J. Kluytmans and S. Malhotra-Kumar, Andreas Peschel & Stephan Harbarth. Methicillin-resistant Staphylococcus aureus, Nat. Rev. Dis. Primers, 2018, 4, 18033 CrossRef PubMed
.
- S. Lakhundi and K. Zhang, Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology, Clin. Microbiol. Rev., 2018, 31, 1–103 CrossRef PubMed
.
- K. M. Craft, J. M. Nguyen, L. J. Berg and S. D. Townsend, Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype, MedChemComm, 2019, 10, 1231–1241 RSC
.
- H. C. Flemming, T. R. Neu and D. J. Wozniak, The EPS matrix: the “house of biofilm cells”, J. Bacteriol., 2007, 189, 7945–7947 CrossRef CAS PubMed
.
- Y. Z. Nira Rabin, C. Opoku-Temeng, Y. Du, E. Bonsu and H. O. Sintim, Biofilm formation mechanisms and targets for developing antibiofilm agents, Future Med. Chem., 2015, 7, 493–512 CrossRef PubMed
.
- K. H. Dakheel, R. Abdul Rahim, V. K. Neela, J. R. Al-Obaidi, T. G. Hun and K. Yusoff, Methicillin-resistant Staphylococcus aureus biofilms and their influence on cacterial adhesion and cohesion, BioMed Res. Int., 2016, 1, 1–14 Search PubMed
.
- N. K. Archer, M. J. Mazaitis, J. W. Costerton, J. G. Leid, M. E. Powers and M. E. Shirtliff, Virulence, 2011, 2, 445 CrossRef PubMed
.
- S. Sugimoto, F. Sato, R. Miyakawa, A. Chiba, S. Onodera, S. Hori and Y. Mizunoe, Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus, Sci. Rep., 2018, 8, 2254 CrossRef PubMed
.
- D. Borisova, E. Haladjova, M. Kyulavska, P. Petrov, S. Pispas, S. Stoitsova and T. Paunova-Krasteva, Application of cationic polymer micelles for the dispersal of bacterial biofilms, Eng. Life Sci., 2018, 18, 943–948 CrossRef CAS
.
- Z. Jing, K. Xiu and Y. Sun, Amide-based cationic polymeric N-halamines: synthesis, characterization and antimicrobial and biofilm-binding properties, Ind. Eng. Chem. Res., 2019, 58, 6218–6225 CrossRef CAS
.
- P. Zhang, S. Li, H. Chen, X. Wang, L. Liu, F. Lv and S. Wang, Biofilm inhibition and elimination regulated by cationic conjugated polymers, ACS Appl. Mater. Interfaces, 2017, 9, 16933–16938 CrossRef CAS PubMed
.
- B. C. Koppen, P. P. G. Mulder, L. de Boer, M. Riool, J. W. Drijfhout and S. A. J. Zaat, Synergistic microbicidal effect of cationic antimicrobial peptides and teicoplanin against planktonic and biofilm-encased Staphylococcus aureus, Int. J. Antimicrob. Agents, 2019, 53, 143–151 CrossRef CAS PubMed
.
- R. Joseph, A. Naugolny, M. Feldman, I. M. Herzog, M. Fridman and Y. Cohen, Cationic Pillararenes Potently Inhibit Biofilm Formation without Affecting Bacterial Growth and Viability, J. Am. Chem. Soc., 2016, 138, 754–757 CrossRef CAS PubMed
.
- T. L. Collins, E. A. Markus, D. J. Hassett and J. B. Robinson, The effect of a cationic porphyrin on Pseudomonas aeruginosa biofilms, Curr. Microbiol., 2010, 61, 411–416 CrossRef CAS PubMed
.
- A. Barrios-Gumiel, J. Sanchez-Nieves, J. Perez-Serrano, R. Gomez and F. J. de la Mata, PEGylated AgNP covered with cationic carbosilane dendrons to enhance antibacterial and inhibition of biofilm properties, Int. J. Pharm., 2019, 569, 118591 CrossRef CAS PubMed
.
- T. Bottcher, I. Kolodkin-Gal, R. Kolter, R. Losick and J. Clardy, Synthesis and activity of biomimetic biofilm disruptors, J. Am. Chem. Soc., 2013, 135, 2927–2930 CrossRef CAS PubMed
.
- J. Hoque, M. M. Konai, S. Gonuguntla, G. B. Manjunath, S. Samaddar, V. Yarlagadda and J. Haldar, Membrane Active Small Molecules Show Selective Broad Spectrum Antibacterial Activity with No Detectable Resistance and Eradicate Biofilms, J. Med. Chem., 2015, 58, 5486–5500 CrossRef CAS PubMed
.
- C. R. Elie, G. David and A. R. Schmitzer, Strong antibacterial properties of anion transporters: a result of depolarization and weakening of the bacterial membrane, J. Med. Chem., 2015, 58, 2358–2366 CrossRef CAS PubMed
.
- C. R. Elie, A. Hebert, M. Charbonneau, A. Haiun and A. R. Schmitzer, Benzimidazolium-based synthetic chloride and calcium transporters in bacterial membranes, Org. Biomol. Chem., 2013, 11, 923–928 RSC
.
- J. Tessier, M. Lecluse, J. Gravel and A. R. Schmitzer, Antimicrobial and Antibiofilm Activity of Disubstituted Bis-benzimidazolium Salts, ChemMedChem, 2018, 13, 2567–2572 CrossRef CAS PubMed
.
- J. Tessier, M. Golmohamadi, K. J. Wilkinson and A. R. Schmitzer, Anti-staphylococcal biofilm activity of miconazoctylium bromide, Org. Biomol. Chem., 2018, 16, 4288–4294 RSC
.
- M. K. Alexander, A. Miu, A. Oh, M. Reichelt, H. Ho, C. Chalouni, S. Labadie, L. Wang, J. Liang, N. N. Nickerson, H. Hu, L. Yu, M. Du, D. Yan, S. Park, J. Kim, M. Xu, B. D. Sellers, H. E. Purkey, N. J. Skelton, M. F. T. Koehler, J. Payandeh, V. Verma, Y. Xu, C. M. Koth and M. Nishiyama, Disrupting Gram-Negative Bacterial Outer Membrane Biosynthesis through Inhibition of the Lipopolysaccharide Transporter MsbA, Antimicrob. Agents Chemother., 2018, 62, 1–7 CrossRef PubMed
.
- M. Berney, F. Hammes, F. Bosshard, H. U. Weilenmann and T. Egli, Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry, Appl. Environ. Microbiol., 2007, 73, 3283–3290 CrossRef CAS PubMed
.
- L. C. Gomes and F. J. Mergulhao, SEM Analysis of Surface Impact on Biofilm Antibiotic Treatment, Scanning, 2017, 2017, 1–7 CrossRef PubMed
.
- A. Punia, K. Lee, E. He, S. Mukherjee, A. Mancuso, P. Banerjee and N. L. Yang, Effect of relative arrangement of cationic and lipophilic moieties on hemolytic and antibacterial activities of PEGylated polyacrylates, Int. J. Mol. Sci., 2015, 16, 23867–23880 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra00738b |
| This journal is © The Royal Society of Chemistry 2020 |