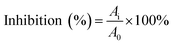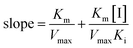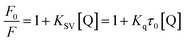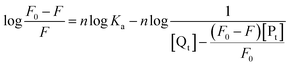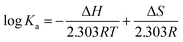Open Access Article
Open Access ArticleExploring the inhibitory mechanism of piceatannol on α-glucosidase relevant to diabetes mellitus†
Lili Jianga,
Zhen Wanga,
Xiaoyu Wanga,
Shujuan Wanga,
Jun Cao*b and
Yong Liu *a
*a
aSchool of Life and Pharmaceutical Sciences, Dalian University of Technology, 2 Dagong Road, Liaodongwan New District, Panjin 124221, China. E-mail: yliu@dlut.edu.cn
bDepartment of Occupational and Environmental Health, Dalian Medical University, No. 9 W. Lvshun South Road, Dalian 116044, China. E-mail: caojunly@163.com
First published on 29th January 2020
Abstract
Due to their association with type 2 diabetes mellitus treatment, α-glucosidase inhibitors have attracted increasing attention of researchers. In this study, we systemically investigated the kinetics and inhibition mechanism of piceatannol on α-glucosidase. Enzyme kinetics analyses showed that piceatannol exhibited strong inhibition on α-glucosidase in a non-competitive manner. Spectroscopy analyses indicated that piceatannol could bind with α-glucosidase to form complexes via high affinity. Further, computational molecular dynamics and molecular docking studies validated that the binding of piceatannol was outside the catalytic site of α-glucosidase, which would induce conformational changes of α-glucosidase and block the entrance of substrate, causing declines in α-glucosidase activities. Our results provide useful information not only for the inhibition mechanism of piceatannol against α-glucosidase but also for a novel target site for developing novel α-glucosidase inhibitors as potential therapeutic agents in the treatment of type 2 diabetes mellitus.
1. Introduction
Diabetes mellitus (DM) is a group of chronic metabolic diseases affecting millions of people worldwide. As estimated by the World Health Organization (WHO), there will be 642 million people that suffer from DM by 2040,1 with type 2 diabetes mellitus (T2DM), accounting for up to 90% of these affected patients.2 Persistent hyperglycemia in diabetes can cause serious complications such as blindness, kidney disease, neuropathy, and cardiovascular disease.3,4 The goal of the treatment of diabetes mellitus involves lowering blood glucose through blocking carbohydrate digestion and absorption.5 α-Glucosidase, which is involved in the cleavage of glucose from disaccharides and oligosaccharides, plays an important role in carbohydrate digestion.6,7 Therefore, the inhibitors of this enzyme can reduce postprandial hyperglycemia, and have been recognized as new anti-diabetic drug candidates. Several oral drugs, such as acarbose, voglibose, and miglitol,8–10 have been used to inhibit the α-glucosidase activity in patients with T2DM. However, these drugs often showed several side effects such as meteorism, abdominal distention, and liver disorders.11 Therefore, much effort has been undertaken to find natural and safer α-glucosidase inhibitors without possibly harmful side effects.Natural products of great structural diversity are considered as a good source for screening α-glucosidase inhibitors.12 In recent years, phenolic compounds have received much attention, and several phenolic compounds have shown significant inhibitory effects against α-glucosidase. Li et al. reported that gallotannin had a strong α-glucosidase inhibitory effect (IC50 = 1.31 μM) in a parabolic mixed-type manner.13 The polyphenol extract of Geranium collinum root was found to show inhibitory activity on α-glucosidase in vitro.14 Moreover, some other phenolic compounds, such as green tea polyphenols,15 water extract of black tea,16 phloretin in apple2 and apigenin in celery,17 have revealed significant inhibition of α-glucosidase activity and alleviated the symptoms of type 2 diabetes.
Piceatannol is a polyphenolic phytochemical, belonging to a natural analog of resveratrol.18 Piceatannol is abundant in various fruits, such as Vaccinium berries (138–422 ng g−1),19 red grapes (374 ng g−1),20 and passion fruit seeds (4.8 mg g−1).21 Piceatannol has a wide spectrum of biological activity, such as vasorelaxant effects, antibacterial activity, antioxidant and pro-oxidant activities.20,22 Piceatannol has also been suggested to be an inhibitor of α-glucosidase.23 However, the effects of piceatannol on α-glucosidase activity have not been fully characterized. Moreover, few studies have reported the specific ligand-binding mechanisms of piceatannol to α-glucosidase molecule.24 Understanding the effect of piceatannol on α-glucosidase activity and its mechanism is important to ensure its safe administration and develop new functional food.
The aim of this study was to investigate the inhibition effect and its possible mechanism of piceatannol on α-glucosidase by using a combination of UV-vis and fluorescence spectral analysis, enzyme kinetic analysis as well as molecular docking methods. This study provides some significant clues to improving the applications of piceatannol in functional food for the prevention and treatment of type 2 diabetes and also suggests a novel target site for developing novel α-glucosidase inhibitors.
2. Materials and methods
2.1 Materials
α-Glucosidase from Saccharomyces cerevisiae (EC 3.2.1.20, 50.3 U mg−1), 4-nitrophenyl-β-D-glucopyranosiduronic acid (pNPG), and acarbose were obtained from Sigma-Aldrich (St, Louis, USA). p-Nitrophenol (pNP) was purchased from Aladdin Biotechnology (Shanghai, China). Piceatannol was purchased from Selleck Chemicals (USA). All other chemicals were of analytical reagent grade, and their aqueous solutions were prepared with freshly ultrapure water.2.2 α-Glucosidase inhibition assay
The α-glucosidase inhibitory activity assay was performed according to the methods described by previous reports with minor modifications.25,26 The enzyme was incubated with varying concentrations of piceatannol in a 100 mM phosphate buffer (pH 6.8) at 37 °C. The substrate reaction mixture contained 0.6 mM pNPG substrate in a 100 mM phosphate buffer (pH 6.8) with and without piceatannol at various concentrations. The enzyme activity assay was conducted following incubation at 37 °C for 7 min.The percentage inhibition of α-glucosidase activity was calculated using the following equation:
2.3 Inhibitory kinetic analysis
Inhibition constant (Ki) values were determined using various concentrations of pNPG in the presence or absence of piceatannol. Ki values were calculated by nonlinear regression using the equations for competitive inhibition (eqn (1)), non-competitive inhibition (eqn (2)), or mixed inhibition (eqn (3)):
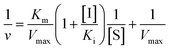 | (1) |
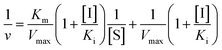 | (2) |
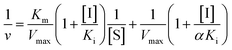 | (3) |
A secondary plot can be acquired from
2.4 Fluorescence spectra measurements
A 2.0 mL substrate reaction mixture containing 6.96 × 10−2 μM α-glucosidase was added to a 1.0 cm quartz cuvette, and then the solution was titrated by successive additions of piceatannol (the final concentrations of piceatannol were varied from 0 to 1.46 μM). These solutions were allowed to stand for 5 min to equilibrate, and then the fluorescence spectra at three temperatures (209, 304 and 310 K) were measured on a Hitachi spectrofluorometer (Model F-7000, Hitachi, Japan) in the wavelength range of 290–500 nm by exciting at 280 nm. Both excitation and emission slit widths were set at 2.5 nm. The appropriate blanks, corresponding to the sodium phosphate buffer (pH 6.8) and the piceatannol, were deducted to correct the fluorescence background.To differentiate the dynamic and static quenching, the quenching effect of piceatannol was further investigated by the Stern–Volmer equation:27
The number of binding sites of piceatannol on α-glucosidase was determined by the following the double-logarithm equation:28
Thermodynamic parameters were calculated by the van't Hoff equation as follows:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka versus 1/T, and the free energy change (ΔG) was estimated from the following relationship:
Ka versus 1/T, and the free energy change (ΔG) was estimated from the following relationship:| ΔG = ΔH − TΔS |
Synchronous fluorescence spectra were measured by maintaining the interval of the excitation wavelength and emission wavelength constant (Δλ = λem − λex) at 15 nm and 60 nm, respectively.29
Because of re-absorption, the fluorescence data were corrected for the absorption of the excitation and emitted light to eliminate the re-absorption and inner filter effects caused by UV absorption, according to the following relationship:30,31
| Fc = Fme(Aex+Aem)/2 |
2.5 Homology modeling and molecular docking studies
A three-dimensional (3D) structure of α-glucosidase was built on the Swiss Modeling server (https://swissmodel.expasy.org/interactive) using homology modeling based on the X-ray structure of the isozyme (PDB ID: 5NN3), which has a high sequence identity and residues similarity to the target protein sequence (GenBank: ABI53718.1).32 The final model was optimized using PROCHECK (http://servicesn.mbi.ucla.edu/SAVES/). ESI Fig. 1 and 2† provide a summary of the homology modelling parameters and the Ramachandran plot analysis of the homology model, respectively.Based on a previous computational study,33 the molecular docking was performed by means of the Autodock version 4.2 program. The homology model was used as the receptor model, the Gasteiger charges and polar hydrogen bonds were added to the macromolecule file. The 3D structures of small molecules were prepared using Chem3D Ultra 8.0. Then, the docking simulations were carried out with the Lamarckian genetic algorithm for 100 docking runs and were extended over the whole receptor protein to find visible combination models. In the process of docking, the remaining parameters were set as default. The lowest energy conformation of the more populated cluster was considered as the most reliable solution.
2.6 Statistical analysis
Data were expressed as means ± standard deviation (n = 3). Data were analyzed by nonlinear regression using the Graphpad Prism 5 software, p < 0.05 was considered statistically significant.3. Results
3.1 Inhibition effect of piceatannol on α-glucosidase activity
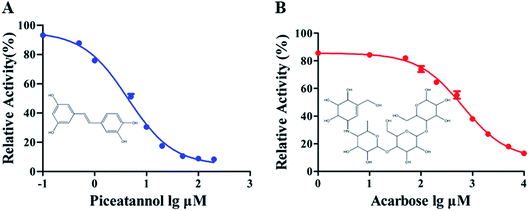
The α-glucosidase inhibitory activity of piceatannol is presented in Fig. 1. The results revealed that piceatannol significantly inhibited α-glucosidase activity in a dose-dependent manner (Fig. 1). The half-maximal inhibitory concentration (IC50) of piceatannol and acarbose, a known inhibitor, against α-glucosidase were calculated as 4.39 ± 0.74 μM and 647.70 ± 1.09 μM, respectively. Piceatannol exerted a more potent inhibitory effect on α-glucosidase activity than that of acarbose.3.2 Kinetics of inhibition
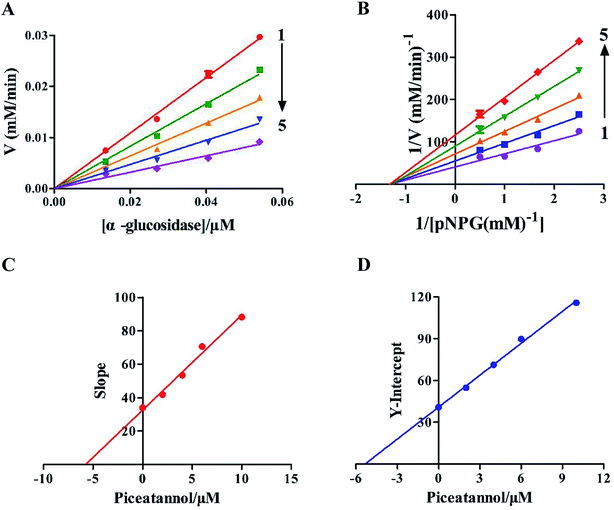
The kinetic experiments were performed to further characterize the inhibition of α-glucosidase activity by piceatannol. Piceatannol strongly inhibited the formation of pNP by α-glucosidase. Plots of the reaction velocity (υ) versus [α-glucosidase] at various concentrations of piceatannol were shown in Fig. 2A. The results showed that all straight lines passed through the origin, whose slopes were dropped with the increase in piceatannol concentration, indicating that the inhibition of α-glucosidase by piceatannol was reversible.Changes in Vmax values were observed on the Lineweaver–Burk plots (Fig. 2B). Obviously, the value of Vmax decreased, whereas −1/Km was constant with the increasing concentrations of inhibitors, and all the lines crossed the negative direction of the horizontal axis. Together the data suggested that the inhibition followed a typical non-competitive mechanism. The value of the inhibition constant (Ki) was calculated to be 5.38 ± 0.24 μM. Furthermore, the secondary plots of the slope and Y-intercept versus [piceatannol] were linear, indicating that one or a class of inhibition sites existed in α-glucosidase with respect to the piceatannol (Fig. 2C and D).
3.3 Quenching of α-glucosidase fluorescence by piceatannol
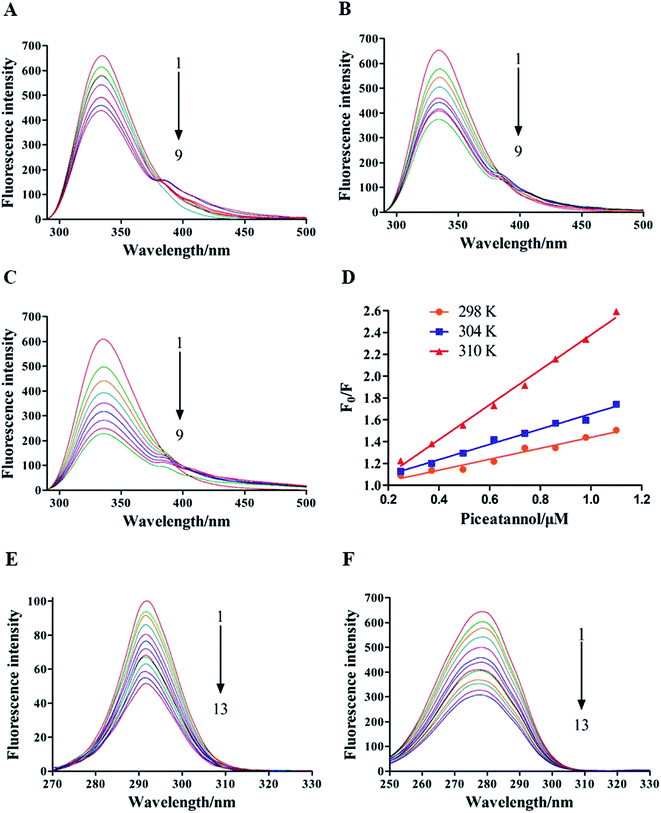
As observed in Fig. 3D, KSV values increased with increasing temperature, suggesting that the quenching of α-glucosidase by piceatannol was a dynamic process. However, the calculated values of Kq (5.02 × 1013, 7.01 × 1013, 16.04 × 1013 L mol−1 s−1 at 298, 304 and 310 K, respectively) were obviously greater than 2 × 1010 L mol−1 s−1, indicating that the fluorescence quenching process was dominated by a static procedure resulting from the piceatannol–α-glucosidase complex formation. These results indicated that the quenching of α-glucosidase by piceatannol was a combination of dynamic and static processes.
As shown in Table 1, the values of n were approximately equal to 1, suggesting that there was one binding site on α-glucosidase for piceatannol, and the result was consistent with that mentioned above. Moreover, in accordance with the alteration in KSV, Ka increased with temperature from 298 K to 310 K. The Ka values were in the order of 105 to 106 L mol−1, suggesting that a high affinity existed between piceatannol and α-glucosidase.
| T (K) | KSV (×105 L mol−1) | Ra | Ka (×105 L mol−1) | n | Rb | ΔH° (kJ mol−1) | ΔG° (kJ mol−1) | ΔS° (J mol−1 K−1) |
|---|---|---|---|---|---|---|---|---|
| 298 | 5.02 ± 0.039 | 0.98 | 4.93 | 1.17 ± 0.057 | 0.99 | 64.55 ± 9.67 | −32.39 ± 9.64 | 325.31 ± 31.80 |
| 304 | 7.01 ± 0.032 | 0.99 | 7.24 | 1.16 ± 0.045 | 1.00 | −34.34 ± 9.64 | ||
| 310 | 16.04 ± 0.049 | 0.99 | 13.54 | 1.32 ± 0.017 | 1.00 | −36.30 ± 9.64 |
3.4 The binding site of piceatannol on α-glucosidase
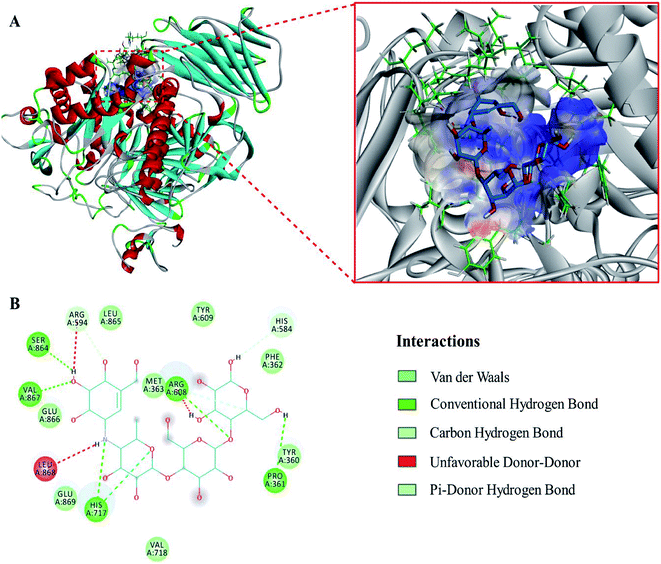
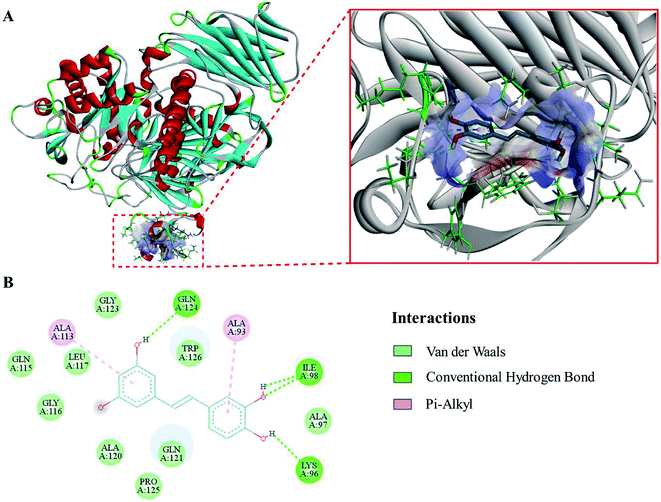
The evaluation results of PROCHECK showed that 91.3%, 8.5% and 0.1% of the residues were located within the most favorable, additionally allowed and generously allowed regions of the Ramachandran plot, respectively. The results indicated that the homology model was suitable for the following molecular docking study.To expound the binding mode of piceatannol on α-glucosidase, molecular docking was carried out. The results showed that acarbose, a known competitive inhibitor, was bound to the active cavity of α-glucosidase and inhibited the catalytic action of α-glucosidase through hydrogen bonds and van der Waals forces (Fig. 4). As shown in Fig. 5, piceatannol was mainly surrounded by amino acid residues Lys 96, Ile 98, Gln 124, Gly 116, Leu 117, Ala 113, Gly 123, Trp 126, Ala 93 and Ala 97 of α-glucosidase, and hydrogen bonds were formed between hydroxyl groups and amino acid residues Lys 96, Ile 98, Gln 124. Furthermore, hydrophobic interactions were formed between piceatannol and the residues Gly 116, Leu 117, Ala 113, Gly 123, Trp 126, Ala 93 and Ala 97 of α-glucosidase, indicating that the hydrophobic interactions were another force in the binding of piceatannol with α-glucosidase. These results indicate that all potential binding sites of piceatannol with α-glucosidase were away from the active cavities of the enzyme, which is also in agreement with the experimental results.
4. Discussion
Our data offer in vitro evidence that piceatannol, a hydroxylated analog of resveratrol, is a potent non-competitive inhibitor of α-glucosidase. Resveratrol with antidiabetic effect has been reported to be a good inhibitor of α-glucosidase.2,34 Interestingly, the observed inhibitory effect against α-glucosidase by piceatannol in the current study is much more potent than that demonstrated by resveratrol,35 which may be structure related. Compared with resveratrol, piceatannol has an additional hydroxy group at the 8′-position. A previous study reported that the inhibition activity on α-glucosidase was increased while adding hydroxyl groups to the B ring of flavones.36 Moreover, a recent study from Liu et al.37 also indicated that the positions of the hydroxyl groups of the polyphenols had an important influence on their interactions with enzymes and the enzyme activities. It is likely that the additional hydroxyl group at this position is conducive to the distribution of the electron cloud, which causes the hydroxyl group to form hydrogen bonds with α-glucosidase.From the fluorescence spectra, an apparent blue shift in λmax was observed on the Trp, suggesting that the interactions between piceatannol and α-glucosidase resulted in significant variations in Trp residues or conformational changes.38 Moreover, the formation of the piceatannol–α-glucosidase complex was an endothermically and entropically increased process, which is frequently regarded as typical evidence for hydrophobic interactions.39 Therefore, we suggest that piceatannol–α-glucosidase complexation could be driven by hydrophobic forces. These assumptions were further verified in our docking study.
According to the quenching theory, the quenching mechanisms of piceatannol included both dynamic and static processes. These phenomena are different from other potent α-glucosidase inhibitors, for example, quercetin, isoquercetin40 and butylisobutyl-phthalate (BIP)41 all of which formed a stable complex. Furthermore, there was a high affinity between piceatannol and α-glucosidase. Therefore, we speculate that the potent inhibitory effect of piceatannol might due to its combination with α-glucosidase and could consequently induce conformational changes in the enzyme. This binding mechanism of piceatannol to α-glucosidase was different from that of other proteins, such as bovine serum albumin (BSA).42
In our docking study, we found that the binding site of piceatannol was at a hydrophobic pocket outside of the catalytic site of α-glucosidase, and the binding could block the entrance of other substrates. This hydrophobic pocket, which contained Lys 96, Ile 98, Gln 124, was different from the binding pocket of acarbose, a known competitive inhibitor.43 These results suggest that piceatannol and acarbose could bind to α-glucosidase at different sites. The docking simulation in our study provides a novel target site outside the catalytic site for developing and designing novel α-glucosidase inhibitors.
5. Conclusions
The present study has systemically investigated the inhibitory effects of piceatannol on α-glucosidase and has provided new insights into the inhibitory and interaction mechanisms of piceatannol on α-glucosidase. This study demonstrates that piceatannol is a vital and potent α-glucosidase inhibitor. Further studies suggest that the interaction between piceatannol and α-glucosidase is a combination of dynamic and static processes, and is mainly driven by hydrophobic interactions. Moreover, the non-competitive inhibition of piceatannol against α-glucosidase may be due to the binding of piceatannol outside the active site of α-glucosidase. These results provide useful information not only for the inhibition mechanism of piceatannol against α-glucosidase, but also provide a basis for further application of piceatannol in novel functional foods or antidiabetic drugs.Abbreviations
| DM | Diabetes mellitus |
| WHO | World health organization |
| T2DM | Type 2 diabetes mellitus |
| pNPG | 4-Nitrophenyl-β-D-glucopyranosiduronic acid |
| pNP | p-Nitrophenol |
| Tyr | Tyrosine |
| Trp | Tryptophan |
| BIP | Butylisobutyl-phthalate |
Authors' contributions
Study conception and design: Yong Liu, Jun Cao. Performing of the research: Lili Jiang, Zhen Wang. Analysis and interpretation of data: Zhen Wang, Xiaoyu Wang, Shujuan Wang. Drafting of manuscript: Lili Jiang, Zhen Wang. Critical revision: Yong Liu, Jun Cao.Conflicts of interest
The authors declare that there is no conflict of interests regarding the publication of this article.Acknowledgements
This study was financially supported by the National Key Research and Development Program of China (2017YFC1702006), the Dalian Science and Technology Innovation Foundation (2018J13SN114).References
- World Health Organization, avaiable from: https://www.who.int/diabetes/global-report, 2016, accessed February 11, 2019.
- A. J. Zhang, A. M. Rimando, C. S. Mizuno and S. T. Mathews, J. Nutr. Biochem., 2017, 47, 86–93 CrossRef CAS PubMed.
- D. M. Nathan, N. Engl. J. Med., 1993, 328, 1676–1685 CrossRef CAS PubMed.
- W. Duckworth, C. Abraira, T. Moritz, D. Reda, N. Emanuele, P. D. Reaven, F. J. Zieve, J. Marks, S. N. Davis, R. Hayward, S. R. Warren, S. Goldman, M. McCarren, M. E. Vitek, W. G. Henderson and G. D. Huang, N. Engl. J. Med., 2009, 360, 129–139 CrossRef CAS PubMed.
- G. S. Chadha and M. E. Morris, AAPS J., 2015, 17, 1464–1474 CrossRef CAS PubMed.
- J. Poongunran, H. K. Perera, L. Jayasinghe, I. T. Fernando, R. Sivakanesan, H. Araya and Y. Fujimoto, Pharm. Biol., 2017, 55, 206–211 CrossRef CAS PubMed.
- S. Kumar, S. Narwal, V. Kumar and O. Prakash, Pharmacogn. Rev., 2011, 5, 19–29 CrossRef CAS PubMed.
- R. K. Singla, R. Singh and A. K. Dubey, Curr. Top. Med. Chem., 2016, 16, 2625–2633 CrossRef CAS PubMed.
- K. Kaku, Expert Opin. Pharmacother., 2014, 15, 1181–1190 CrossRef CAS PubMed.
- L. J. Scott and C. M. Spencer, Drugs, 2000, 59, 521–549 CrossRef CAS PubMed.
- K. R. Rengasamy, M. A. Aderogba, S. O. Amoo, W. A. Stirk and J. Van Staden, Food Chem., 2013, 141, 1412–1415 CrossRef CAS PubMed.
- X. Peng, G. Zhang, Y. Liao and D. Gong, Food Chem., 2016, 190, 207–215 CrossRef CAS PubMed.
- L. M. Yue, J. Lee, L. Zheng, Y. D. Park, Z. M. Ye and J. M. Yang, Int. J. Biol. Macromol., 2017, 103, 829–838 CrossRef CAS PubMed.
- S. Numonov, S. Edirs, K. Bobakulov, M. N. Qureshi, K. Bozorov, F. Sharopov, W. N. Setzer, H. Zhao, M. Habasi, M. Sharofova and H. A. Aisa, Molecules, 2017, 22, 983 CrossRef PubMed.
- X. Yang and F. Kong, J. Sci. Food Agric., 2016, 96, 777–782 CrossRef CAS PubMed.
- D. P. Tong, K. X. Zhu, X. N. Guo, W. Peng and H. M. Zhou, Int. J. Biol. Macromol., 2018, 107, 129–136 CrossRef CAS PubMed.
- L. Zeng, G. Zhang, S. Lin and D. Gong, J. Agric. Food Chem., 2016, 64, 6939–6949 CrossRef CAS PubMed.
- H. Uchida-Maruki, H. Inagaki, R. Ito, I. Kurita, M. Sai and T. Ito, Biol. Pharm. Bull., 2015, 38, 629–633 CrossRef CAS PubMed.
- A. M. Rimando, W. Kalt, J. B. Magee, J. Dewey and J. R. Ballington, J. Agric. Food Chem., 2004, 52, 4713–4719 CrossRef CAS PubMed.
- H. Piotrowska, M. Kucinska and M. Murias, Mutat. Res., 2012, 750, 60–82 CAS.
- J. Kershaw and K. H. Kim, A Review, J. Med. Food, 2017, 20, 427–438 CrossRef CAS PubMed.
- S. Sano, K. Sugiyama, T. Ito, Y. Katano and A. Ishihata, J. Agric. Food Chem., 2011, 59, 6209–6213 CrossRef CAS PubMed.
- M. Minakawa, Y. Miura and K. Yagasaki, Biochem. Biophys. Res. Commun., 2012, 422, 469–475 CrossRef CAS PubMed.
- H. He and Y. H. Lu, J. Agric. Food Chem., 2013, 61, 8110–8119 CrossRef CAS PubMed.
- J. Q. Zhao, Y. M. Wang, Y. L. Yang, Y. Zeng, Q. L. Wang, Y. Shao, L. J. Mei, Y. P. Shi and Y. D. Tao, Food Chem., 2017, 227, 93–101 CrossRef CAS PubMed.
- E. Apostolidis, Y. I. Kwon and K. Shetty, Innovative Food Sci. Emerging Technol., 2007, 8, 46–54 CrossRef CAS.
- A. Bhogale, N. Patel, P. Sarpotdar, J. Mariam, P. M. Dongre, A. Miotello and D. C. Kothari, Colloids Surf., B, 2013, 102, 257–264 CrossRef CAS PubMed.
- S. R. Feroz, S. B. Mohamad, Z. S. D. Bakri, S. N. A. Malek and S. Tayyab, PLoS One, 2013, 8, e76067 CrossRef CAS PubMed.
- Y. Wang, G. Zhang, J. Pan and D. Gong, J. Agric. Food Chem., 2015, 63, 526–534 CrossRef CAS PubMed.
- X. Wu, J. Liu, H. Huang, W. Xue, X. Yao and J. Jin, Int. J. Biol. Macromol., 2011, 49, 343–350 CrossRef CAS PubMed.
- G. Zhang, N. Zhao and L. Wang, J. Lumin., 2011, 131, 880–887 CrossRef CAS.
- G. C. Wang, M. Chen, J. Wang, Y. P. Peng, L. Y. Li, Z. Z. Xie, B. Deng, S. Chen and W. B. Li, Synthesis, biological evaluation and molecular docking studies of chromone hydrazone derivatives as α-glucosidase inhibitors, Bioorg. Med. Chem. Lett., 2017, 27, 2957–2961 CrossRef CAS PubMed.
- X. Li, Z. R. Lü, W. Wang, X. P. Han, J. M. Yang, Y. D. Park, H. M. Zhou, Q. Sheng and J. Lee, Effect of Ba2+ on the activity and structure of α-glucosidase: inhibition kinetics and molecular dynamics simulation, Process Biochem., 2015, 50, 582–588 CrossRef CAS.
- A. M. Rimando, R. Nagmani, D. R. Feller and W. Yokoyama, J. Agric. Food Chem., 2005, 53, 3403–3407 CrossRef CAS PubMed.
- Y. Zhao, M. X. Chen, K. T. Kongstad, A. K. Jager and D. Staerk, J. Agric. Food Chem., 2017, 65, 4421–4427 CrossRef CAS PubMed.
- J. Xiao, G. Kai, K. Yamamoto and X. Chen, Crit. Rev. Food Sci. Nutr., 2013, 53, 818–836 CrossRef CAS PubMed.
- M. Liu, T. T. Liu, Y. B. Shi, Y. N. Zhao, H. Yan, B. Sun, Q. P. Wang, Z. P. Wang and J. Han, Food Funct., 2019, 12, 8182–8194 RSC.
- F. Xiao, M. Gu, Y. Liang, L. Li and Y. Luo, Spectrochim. Acta, Part A, 2014, 118, 1106–1112 CrossRef CAS PubMed.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS PubMed.
- M. Liu, W. Zhang, L. Qiu and X. Lin, J. Biochem., 2011, 149, 27–33 CrossRef CAS PubMed.
- Y. Q. Li, F. C. Zhou, F. Gao, J. S. Bian and F. Shan, J. Agric. Food Chem., 2009, 57, 11463–11468 CrossRef CAS PubMed.
- X. Xu, M. S. Zhao, Q. Q. Han, H. J. Wang, H. M. Zhang and Y. Q. Wang, Spectrochim. Acta, Part A, 2019, 117706 CrossRef PubMed.
- K. Bharatham, N. Bharatham, K. H. Park and K. W. Lee, J. Mol. Graphics Modell., 2008, 26, 1202–1212 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra09028b |
| This journal is © The Royal Society of Chemistry 2020 |