 Open Access Article
Open Access ArticleAntimicrobial peptides from Bombyx mori: a splendid immune defense response in silkworms†
Jannatun Nesaa,
Abdul Sadatb,
Danieli F. Buccinic,
Ahmet Kati de,
Amit K. Mandal
de,
Amit K. Mandal *af and
Octavio L. Franco
*af and
Octavio L. Franco *cg
*cg
aChemical Biology Laboratory, Department of Sericulture, Raiganj University, Uttar Dinajpur, 733134, West Bengal, India. E-mail: amitmandal08@gmail.com
bInsect Ecology and Conservation Biology Laboratory, Department of Sericulture, Raiganj University, Uttar Dinajpur, 733134, West Bengal, India
cS-INOVA Biotech, Post-Graduate Program in Biotechnology, Catholic University Dom Bosco, Campo Grande, Mato Grosso Do Sul, Brazil. E-mail: ocfranco@gmail.com
dBiotechnology Department, Institution of Health Science, University of Health Science, Istanbul, Turkey
eMedical Microbiology Department, Faculty of Medicine, Acibadem Mehmet Ali Aydinlar University, Istanbul, Turkey
fCentre for Nanotechnology Sciences, Raiganj University, Uttar Dinajpur, 733134, West Bengal, India
gCenter of Proteomic and Biochemical Analysis, Post Graduate Program in Genomic Sciences and Biotechnology, Catholic University of Brasilia, Brasilia, Brazil
First published on 2nd January 2020
Abstract
Bombyx mori L., a primary producer of silk, is the main tool in the sericulture industry and provides the means of livelihood to a large number of people. Silk cocoon crop losses due to bacterial infection pose a major threat to the sericulture industry. Bombyx mori L., a silkworm of the mulberry type, has a sophisticated inherent innate immune mechanism to combat such invasive pathogens. Among all the components in this defense system, antimicrobial peptides (AMPs) are notable due to their specificity towards the invading pathogens without harming the normal host cells. Bombyx mori L. so far has had AMPs identified that belong to six different families, namely cecropin, defensin, moricin, gloverin, attacin and lebocin, which are produced by the Toll and immune deficiency (IMD) pathways. Their diverse modes of action depend on microbial pathogens and are still under investigation. This review examines the recent progress in understanding the immune defense mechanism of Bombyx mori based on AMPs.
Introduction
Sericulture is one of the prime agriculture-based industries in the world and it continues to flourish worldwide due to an ever-increasing demand for raw silk.1 To fulfil this demand, silkworm rearing practice has undergone drastic changes over time in order to produce higher quality silk.2 At present, the Indian silk industry has a massive impact on global sericulture, holding the second position in the silk yield rankings. The Indian silk industry produced 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 273 metric tons of raw mulberry silk in 2016-17 [3AR 2017]. But this production rate is insufficient to fulfil the national and international demand. In addition, the theoretical capacity in silk production per capita has not yet been reached.1,3,4 To increase the production rate of high quality silk, cultivation of disease-free silkworms is of paramount importance.
273 metric tons of raw mulberry silk in 2016-17 [3AR 2017]. But this production rate is insufficient to fulfil the national and international demand. In addition, the theoretical capacity in silk production per capita has not yet been reached.1,3,4 To increase the production rate of high quality silk, cultivation of disease-free silkworms is of paramount importance.
The high mortality rate of silkworms due to disease is the main obstacle to high rates of silk production.2,5 The leaves that are fed are considered to be the main source of disease-causing infections and, thus, special emphasis is paid to the cleanliness of food and accommodation.2 Production of disease-resistant silkworm varieties has also drawn attention as an alternative approach. As silkworms lack an acquired immune system, they depend solely on their innate defense mechanism to protect themselves from pathogenic diseases.6 Antimicrobial peptides (AMPs) are one of the chief components of the innate immune system in B. mori.7 Upon pathogenic infection, AMPs are rapidly released into the haemolymph of the insect, and then they eliminate pathogens either by disrupting the cell membrane or by intracellular killing of the invading pathogen.8
The present review article aims to understand how various AMPs from the silkworm function against bacterial infections as a part of an immune defense strategy.
Silkworm diseases
Silkworms have been domesticated by the silk industry, and they cannot reproduce on their own. Indeed, reproduction demands human interference in the rearing room, which makes silkworms susceptible to various infectious diseases, such as pebrine, flacherie, grasserie and muscardine.2 All these infectious diseases are caused by microorganisms, including protozoa, bacteria, viruses and fungi (Table 1).3,4| Sl. no. | Disease | Pathogen | Season | Prevalence in India | Symptoms | % loss of cocoons (2016-17) |
|---|---|---|---|---|---|---|
| 1. | Viral | |||||
| Nuclear polyhedrosis | BmNPV | Summer and rainy season | Prevalent | Swelling on inter-segmental region, shining and fragile skin, milky white fluid | 20 | |
| Cytoplasmic polyhedrosis | BmCPV | Prevalent | Translucent cephalo-thoracic region; diarrhoea; retarded growth; milky white midgut; whitish faeces | |||
| Infectious flacherie | BmIFV | Prevalent | Translucent cephalothoraxes; retarded growth; vomiting and diarrhoea | |||
| Densonucleosis | BmDNV1, BmDNV2, BmDNV3 | Prevalent not reported, not reported | Translucent cephalothoraxes; retarded growth; vomiting and diarrhoea. | |||
| 2. | Bacteraemia | |||||
| Bacterial diseases of digestive tract | Streptococcus sp., Staphylococcus sp., Pseudomonas sp | Summer and rainy season | Prevalent | Sluggish movement; retarded growth; transparent cephalo-thoracic region | 30 | |
| Septicaemia | Bacillus sp., Streptococcus sp., Staphylococcus sp., Serratiamarcescens | Prevalent | Sluggish movement low appetite; swollen thorax; shrinkage; vomiting softening and discoloured body | |||
| Toxicosis | Bacillus thringiensis var. Sotto | Prevalent | Sluggish movement; retarded growth; cessation of feeding; vomiting; paralysis and death; corpse stretched and cephalo-thoracic region bent like hook | |||
| 3. | Mycosis | |||||
| White muscardine | Beauveriabassiana | Rainy season | Prevalent | Oily specks on the body surface; larva on death softens, turns hard and later mummifies; mummified larvae appear white | 10 | |
| Green muscardine | Metarhiziumanisopliae, Nomuraearileyi | Prevalent | Large specks with black periphery; mummified larvae green in colour | |||
| Yellow muscardine | Isariafarinosa | Not reported | Large disease specks around stigma and small on skin, mummified larvae yellow | |||
| Red muscardine | Sporosporellauvella, Isariafumosoroseus | Not reported | Develop red patches few hour before death; no external growth | |||
| Orange muscardine | Sterigmatocystis japonica | Not reported | Develop orange patches few hour before death; no external growth | |||
| Aspergillosis | Aspergillusflavus, Aspergillusoryzae | Prevalent | Formation of light yellow coloured spores on surface with dirty brown | |||
| 4. | Protozoan | |||||
| Pebrine | Nosemabombycis | All season | Prevalent | Sluggish larvae with paler, translucent, wrinkled skin | 40 | |
| Nosema sp. M-11 | Prevalent | |||||
| Nosema sp. M-14 | Not reported | |||||
| Vairomorpha sp. M-12 | Prevalent | |||||
| Pleistomorpha sp. M-24 | Not reported | |||||
| Pleistomorpha sp. M-25 | Not reported | |||||
| Pleistomorpha sp. M-27 | Not reported | |||||
| Thelohania sp. M-32 | Not reported | |||||
| Leptomonassp | Not reported | |||||
Gut microflora of Bombyx mori L.
Bacterial microflora in the arthropod gut is closely related to the digestive capability of the host.9,10 The digestive tract of the silkworm is quite simple, without any specialized structures, consisting of three regions: the foregut, midgut and hindgut. The midgut represents the majority of the digestive tract, while the foregut and hindgut constitute smaller portions.11 Silkworms are holometabolous insects and undergo five larval instars (referred to as L1 through L5), before metamorphosing to short-lived reproductive adults. Metamorphosis directly affects the intestinal microbiota. It is reported that the variety in bacterial species strikingly decreases in the adults.12 Kalpana et al. reported the presence of heterotopic bacterial population coupled with different larval stages of B. mori, and they related this finding with mulberry leaves fed to them.2 It was also revealed that there was more gut microflora in the middle region of the digestive tract of the late (fourth and fifth) instar larvae. The highest bacterial population was recorded in the digestive tract of fifth instar larvae (13 × 106 CFU g−1), whereas first instar larvae possess lower bacterial populations (5.7 × 104 CFU g−1) in their gut.2Silkworm gut microflora belongs to the belongs to the genera Bacillus, Corynebacterium, Micrococcus, Enterobacteriaceae, Moraxella, Alcaligenes, Acinetobacter and Aeromonas. Regarding the phenological and physiological properties, the gut microflora is dominated by rod-shaped bacteria rather than cocci, and by Gram positive bacteria rather than Gram negative. All bacterial populations are capable of glucose fermentation, and they mainly produce protease enzymes (in the form of caseinase and gelatinase), followed by amylase and lipase.10 Another study revealed that fifth instar larvae of B. mori possess both culturable facultative anaerobic bacteria and culturable obligatory anaerobic bacteria in the digestive tract suspension, which utilize polysaccharides as a carbon source.10,13 It was reported earlier that wild families of B. morisuch as Acronicta major (Noctuidae) and Diaphania pyloalis (Pyralididae) share the same food niche. Due to the similar foraging behaviour in all these three species, they possess highly assorted but idiosyncratic microbiota.12 The mid gut microbial content of all these three taxa is dominated by Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes phyla (Fig. 1).12
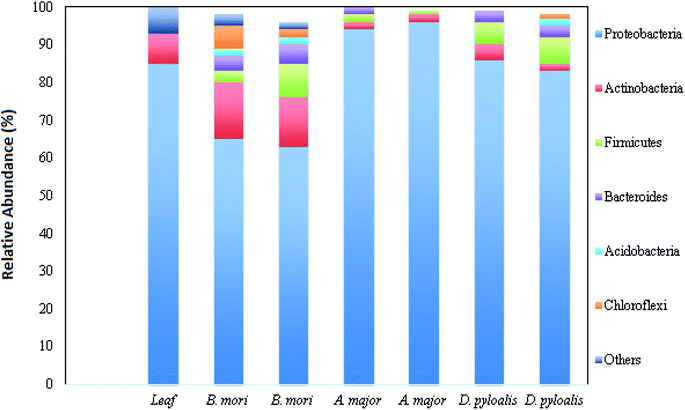 | ||
| Fig. 1 Relative abundance of different bacterial phyla in host samples (this figure has been adapted from Chen et al., 2018 with permission from Springer Nature).12 | ||
Different bacteria coexist in the gut microflora through balanced symbiotic or antagonistic relationships. Antagonism is characterized by synthesis of the antimicrobial products of one microorganism against others that inhabit the same ecological niche.14
Antimicrobial peptides in B. mori L
After the invasion of pathogens through the first line of defense (physical barrier) and their breakthrough into the insect's body, the second line of defense (innate immune system) comes into play.15 The innate immune system in the form of cellular and humoral response prevents spreading and multiplication of the invading pathogens inside the host.16 Cellular response includes hemocyte-mediated actions, such as encapsulation, nodulation and phagocytosis.17 Humoral response leads to production of melanin (melanization), generation of reactive oxygen species (ROS) and stimulation of AMPs production.7AMPs are proteins with low molecular weight that exert a diverse array of defense mechanisms against fungi, bacteria and viruses. These biochemically active elements are the main components of the humoral defense system of various insects. They are the potential and imminent drugs of the future, mainly because their small size enables them to diffuse quickly through the plasma membrane of the target bacterial cells and activate the host's defense mechanism against microbes without developing resistance or toxic effects.18 Different experimental studies during the last few decades have established the presence of six families of AMP from B. mori viz.; cecropin, defensine, moricin, gloverin, lebocin and attacin (Table 2).
| Antimicrobial Peptide | Amino acid sequence of peptide | Activity | Target | Mode of action |
|---|---|---|---|---|
| Cecropin A1 | MNFVRILSFVFALVLALGAVSAAPEPRWKLFKKIEKVGRNVRDGLIKAGPAIAVIGQAKSLGK | Antimicrobial and cytotoxicity | Bacterial and human leukemic cell | Form continuous leaky pore on plasma membrane |
| Cecropin B6 | MNFAKILSFVFALVLALSMTSAAPEPRWKIFKKIEKMGRNIRDGIVKAGPAIEVLGSAKAIGK | |||
| Cecropin C | RWKLFKKIEKVGRNVRDGLIKAGPAIAVIGQAKSL | |||
| Cecropin D | MKISKIFVFVFAIVFATASVSAAPGNFFKDLVSIVLDVSGS | |||
| Cecropin E | MNFSRALFYVFAVFLVCASVMAAPEPRWKIFKKIEKVGQNIRDGIIKAGPAVAVVGQAATIAHGK | |||
| Defensin | MAHQRKSLVIFIFLTVLVFVFALPRDATVFDNQHSEVAIEKSTSKIDSSDVKIPGRIWCEFEEATETAIC | Antimicrobial | Bacteria and fungi | Disrupt bacterial cell membrane. |
| QEHCLPKGYSYGICVSNTCSCI | Hamper cell wall biosynthesis in fungi | |||
| Bmmor | MNILKLFFVFIVAMSLVSCSTAAPAKIPIKAIKTVGKAVGKGLRAINIASTANDVFNFLKPKKRKH | Antimicrobial | Specially gram-positive bacteria and fungi | Increase membrane permeability in target organism |
| Moricin Like A | AKIPIKAIKTVGKAVGKGLRAINIASTANDVFNFLKPKKRKA | |||
| Moricin Like B | MKVFSFFCVVLAMLVLIMGGTSAAPEPKGIGKIIRKGGKVIKHGLTAIGVGAAGHEVYQDSKNSG | |||
| Gloverin1 | MYSKVLLSAALLVCVNAQVSMPPGYAEKYPITSQFSRSVRHPRDIHDFVTWDREMGGGKVFGTLGESDQGLFGKGGYNREFFNDDRGKLTGQAYGTRVLGPGGDSTSYGGRLDWANENAKAAIDLNRQIGGSAGIEASAS | Antimicrobial | Strongly effective against broad spectrum bacteria and viruses. But less effective against yeasts. | Increase membrane permeability in target organism and intra-cellular killing by altering cellular functions |
| GVWDLGKNTHLSAGGVVSKEFGHRRPDVGLQAQITHEW | ||||
| Gloverin2 | MNTNLFYIFATTLVCVNAEVYGPSDYAEDYSISGQSSRRHPRDVTWDKQMGGGKVFGTLGQNDDGLFGKAGYNREIFNDDRGKLTGQAYGTRVLGPGGDSTNYGGRLDWANKNAQATIDLNRQIGGRSGMTASGSGVWDL | |||
| DKNTHFSAGGMVSKEFGHKRPDVGLQAEIRHDW | ||||
| Gloverin3 | MNSKLLFFIATVLVCVNAEVYRSPDYEEEYPIRGLFSKRHPRDVTWDTKMGGGKVFGTLGQNDDGLFGKAGYNREIFNDDRGQLTGQAYGTRVLGPGGDSTNYGGRLDWANKNAQAAIDINRQIGGRSGMTASGSGVWDL | |||
| DKNTHISAGGMVSKEFGHRRPDVGLQAEIRHEW | ||||
| Gloverin4 | MNSKLLYFFATVLVCVNAEVYWEDEEGYPVSGQFSKRHPRDVTWDKQVGGGKVFGTLGQNDDGLFGKAGYNREIFNDDRGKLTGQAYGTRVLGPAGDSTNYGGRLDWANKNAEAAIDINRQIGGRSGMTATGSGVWDLDK | |||
| NTRLSAGGMVSKEFGHRRPDVGVQAEFRHDW | ||||
| Attacin | MSKSVALLLLCACLASGRHVPTRARRQAGSFTVNSDGTSGAALKVPLTGNDKNVLSAIGSADFNDRHKLS | Antimicrobial | Strongly effective against broad spectrum bacteria | Rather than acting on cell wall, attacin hampers plasma membrane synthesis in growing bacterial cell |
| AASAGLALDNVNGHGLSLTGTRIPGFGEQLGVAGKVNLFHNNNHDLSAKAFAIRNSPSAFPNAPNFNTLG | ||||
| GGLDYMFKQKVGASLSAAHSDVINRNDYSAGGKLNLFRSPSSSLDFNAGFKKFDTPFYRSSWEPNVGFSF | ||||
| SKFF | ||||
| Lebocin 1 | DLRFLYPRGKLPVPTPPPFNPKPIYIDMGNRY | Antimicrobial | Bacteria | Form leakage in lipid bilayer of the plasma membrane and showed synergistic effect on cecropin D |
| Lebocin 2 | DLRFLYPRGKLPVPTPPPFNPKPIYIDMGNRY | |||
| Lebocin 3 | DLRFLYPRGKLPVPTPPPFNPKPIYIDMGNRY | |||
| Lebocin 4 | DLRFLYPRGKLPVPTPPPFNPKPIYIDMGNRY |
Cecropin
Cecropin was first isolated from the haemolymph of bacteria-infected giant silk worms (Hyalophoracecropia).19 Later, this AMP was found to be a part of the immune system in Antheraea pernyi and B. mori. Cecropins are cationic peptides that lack a cysteine residue with a strong basic N-terminal linked to a neutral C-terminal by a flexible glycine–proline link. Insects possess three major types of cecropin; viz., cecropin A (37 amino acids), cecropin B (35 amino acids) and cecropinD (37 amino acids). In B. mori, 11 genes are responsible for the production of Bmcecropin, which occurred due to unequal cross-over events of the encoding gene.20 This AMP family is classified into five subtypes (Table 2), namely Bmcec A1 (2 genes), B6 (6 genes), C (1 gene), D (1 gene) and E (1 gene). The presence of six different clustered genes in chromosome 26 which encode amino acids for Bmcec B6 explains the phenomenon of gene duplication for that particular protein.21 Jiggins and Kim suggested that gene duplication increases AMP production to enhance immunity against microbial infections.22Cecropins exhibit a broad range of antimicrobial properties against both Gram-positive and Gram-negative bacteria and fungi.18 Beside this, cecropins and their derivatives (SB-37 and Shiva) act as active suppressors of Trypanosoma and Plasmodium23,24 and also inhibit the proliferation of tumour cell lines.25 Cecropin A causes the lysis of both Gram-positive and Gram-negative bacteria by the following mechanism: at first, Cecropin A binds to the negatively charged membrane lipids via its strongly positively charged N-terminus, and then it induces pore formation by its hydrophobic C-terminus, which then renders the membrane permeable, eventually leading to bacterial death.26 Among all the paralogs, BmcecB6 and BmcecD show the highest antimicrobial activity by inhibiting Bacillus bombysepticus, Bacillus subtilis, Bacillus thuringiensis, Bacillus thuringiensis galleriae, Escherichia coli, Serratia marcescens, Pseudomonas aeruginosa and Ralstoniasolaanacearum.20 Besides, they are found to be active against the human leukaemia cell line.27 While these two paralogs remain inactive only against Staphylococcus aureus and Xanthomonas campestris. Bmcec A1 and BmcecE are active against five tested bacteria, and BmcecC has no inhibition activity against tested bacterial populations (Table 3).20
| Gram-positive bacteria | Gram-negative bacteria | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus subtilis | Bacillus bombysepticus | Bacillus thuringiensis | Bacillus thuringiensisgalleriae | Staphylococcus aureus | Serratia marcescens | Escherichia coli | Psedomonas aeruginosa | Ralstonia solaanacearum | Xanthomonas campestris | |
| a N. B. “+” active, “−” inactive. | ||||||||||
| BmcecA1 | + | − | + | − | − | − | + | + | + | − |
| BmcecB6 | + | + | + | + | − | + | + | + | + | − |
| BmcecC | − | − | − | − | − | − | − | − | − | − |
| BmcecD | + | + | + | + | − | + | + | + | + | − |
| BmcecE | − | − | + | − | − | + | + | + | + | − |
| Bmmor | + | + | + | + | + | + | + | + | + | − |
| BmmorLA | + | + | − | − | + | − | + | − | + | − |
| BmmorLB | − | − | − | − | − | − | − | − | − | − |
| Bmglv1 | − | − | + | + | − | + | + | + | + | + |
| Bmglv2 | − | − | + | + | − | + | + | + | + | + |
| Bmglv3 | − | − | + | + | − | + | + | + | + | + |
| Bmglv4 | − | − | + | + | − | + | + | + | + | + |
Xia et al. reported successful expression of the cecropin XJ gene in E. Coli and its inhibition of Gram-positive and Gram-negative bacteria,28,29 which opens up a new avenue for using cecropin against bacterial infections in silkworm and as a potent drug for broader therapeutic applications.
Defensin
An insect defensin was first isolated from the haemolymph of immunized flesh fly (Saecophoga peregrine).30 They are cationic peptides (Table 2) comprised of six conserved cysteine molecules which are responsible for the formation of three intra-molecular disulfide bonds.31 Phormicins, royalcins, sapecins and spodoptericis are different groups of the insect defensin family which were isolated and characterised from the haemolymph of the order Lepidoptera.32,33 These AMPs are mainly active against Gram-positive bacteria, including Micrococcus luteus, Aerococcus viridians, Bacillus megaterium, Bacillus subtilis, Bacillus thuringiensis, and Staphylococcus aureus.34Wen et al. discovered the BmDefensinA gene from the B. mori genome, which is assumed to be related to insect defensins.35 Complete characterization of this gene revealed that the 5′-upstream region contains three conserved regulatory sequences; viz., NF-kβ binding, IL-6 responsive and GATA element. This BmDefensinA gene from B. mori translates into a large peptide which consists of an N-terminus signal peptide (22 amino acids), a pro-peptide (34 amino acids) and a mature peptide (36 amino acids) with molecular mass of 4 kDa. The mature peptide contains six conserved cysteine motifs, homologous to insect defensins. Unlike insect defensins, this mature BmDefA peptide is a novel anionic defensin with an isoelectric point of 4.2. This AMP is produced in large quantities in haemocytes, head, silk gland, fat body and ovary, and participates in both the defense mechanism and metamorphosis.18 Yoichi et al. isolated another peptide, namely BmDefB, homologous to and insect defensin with six conserved cysteine motifs, but only 27% amino acid similarities to the BmDefA peptide.36 The BmDefB peptide is largely synthesised in fat body and associated with protection against a broad spectrum of bacteria, including Escherichia coli and Bacillus subtilis. The Bm Defensin peptide forms large channels in the plasma membrane of bacterial cells and causes the efflux of cytosolic K+ ions, partial depolarization of plasma membrane, decreased cytosolis of ATP and inhibition of respiration, eventually leading to bacterial cell death.37
Moricin
Moricin belongs to the AMP family that was first isolated from B. mori. Immunised B. mori produce this AMP in the haemolymph, and thus actively suppress the multiplication of Staphylococcus aureus.38 Moricin (encoded by multiple gene family) consists of a cationic peptide chain (42 residues long) formed by an amphipathic α-helix with charged amino acids in the N-terminal half, at an interval of every three to four amino acid residues.39,40 The B. mori genome harbours 12 moricin-coding genes that are synthesized into three subtypes (Table 2) viz., Bmmor (1 gene), moricin-like A (3 genes) and moricin-like B (8 genes).20 All the genes encoded mature moricin, which contains a positively charged C-terminus and an amphipathic alpha-helical N-terminus without any post-translational modification. Lack of this post-translational modification made this moricin able to be synthesized chemically.18This cationic peptide easily attaches to the negatively charged cell surface of bacteria, and the amphipathic α-helical motif forms channels in bacterial plasma membrane that disrupt the ionic balance of that bacterial cell.41 Though it is active against both bacterial and fungal infection, its activity is recorded as higher against Gram-positive bacteria. The α-helical motif of moricin plays a pivotal role in increasing the membrane permeability in order to kill bacterial pathogens.34 Lepidopteran-specific moricin shows a wide array of antibacterial activity, but this peptide from silkworm differs in its spectrum and activity. An in vitro experiment showed that moricin from B. mori (Bmmor), at dosages of 0.625–1.25 μL L−1, exhibited antibacterial activities against B. bombysepticus, B. subtilis, B. thuringiensis, B. thuringiensis galleriae, E. coli, S. marcescens, S. aureus, P. aeruginosa and R. solaanacearum (Rd). Among the moricin family of B. mori, Bmmor exhibits the highest antibacterial toxicity (p < 0.01) followed by moricin-like A subtype. In contrast, moricin-like B subtype remains inactive against those tested bacteria.20
Gloverin
Gloverin was first isolated from pupal haemolymphs of the giant silk moth (Hyalaphora cecropia). Unlike other AMPs, gloverinis made up of glycine-rich (18.5%) amino acids and devoid of cysteine.42 In addition, this AMP is only found in lepidopteran insects, including B. mori and Antheraea mylitta.43,44 Whole genome analysis of B. mori revealed that mulberry silkworm contains four genes (Table 2) which encode Bmglv 1, 2, 3 and 4.20 These peptides are over-expressed in fat body of B. mori after induction with E. coli.45 The BmGlov gene contains an NF-kB like motif that provides a binding site in the upstream region of the gene. The Bmglv 1 gene is the ancestral one and evolved into Bmglv2–4 upon duplication in embryonic stage, indicating that the derived genes have gained embryonic expression and novel function.45,46Experimental work showed that gloverin actively inhibits mutant strains of E. coli (D21, Df21f2 and D22) that possess lipopolysaccharide (LPS) on their cell surface, and it was found to be inactive against smooth LPS containing E. coli strain. The product of the whole gene family significantly suppresses bacterial infection.18 Bmglv family peptides showed an antibacterial effect on B. thuringiensis, B. thuringiensis galleriae, E. coli, S. marcescens, P. aeruginosa and R. Solaanacearum, except against S. aureus, B. bombysepticus and B. subtilis.20
Attacin
Attacin is a peptide chain (Table 2) with a molecular mass of 20–23 kDa and was first isolated and characterised from the pupal haemolymph of bacterial immunized Hyalaphora cecropia.47 This AMP is expressed both in acidic and basic form with isoelectric points ranging from 5.7 to 8.3, due to post-translational modification of two parental pro-peptide sequences. Acidic attacin possesses a high content of aspartic acid, arginine and isoleucine, whereas basic attacin contains a high content of lysine, glutamic acid and tryptophan.18Attacin shows antibacterial activity against E. coli, Stenotrophomonas maltophilia and Acinetobacter calcoaceticus. Unlike cecropin, attacin does not degrade the cell wall of the target bacteria, instead killing the bacteria by acting on growing cells and causing achain formation. Attacin restrains the synthesis of E. coli membrane proteins (OmpA, OmpC, OmpF and LamB), which alters the membrane structure and permeability.48,49
Lebocin
Lebocin is a proline-rich peptide (Table 2) with O-glycosylated threonine (15-Thr) residue isolated from the haemolymph of immunized B. Mori.50 Glycosylation of the threonine residue of this AMP is crucial for its antibacterial activity. Four structurally related lebocin peptides were isolated and characterised, viz., lebocin 1, 2, 3 and 4, which are similar in peptide sequence (Table 2) but differ only in their sugar moiety. An in vitro study revealed that lebocin causes leakage of the lipid bilayer under low ionic conditions, suggesting active rupture of the bacterial membrane.50 But its exact mechanism is still under investigation, due to its low sensitivity and the need for a low ionic condition for its proper functioning. Lebocin 3 has a synergistic effect on cecropin D, as their conjugation greatly increases the antibacterial activity of cecropin D in B. mori.51Molecular mechanism of AMP synthesis via Toll and IMD signalling pathway
In silkworms, fungal or Gram-positive bacterial infection triggers activation of the Toll pathway (Fig. 2), resulting in the systemic production of AMPs.52,53 Cell wall components of invading fungi like β-glucan and, in Gram-positive bacteria in the form of Lysine-type peptidoglycan (Lys-PG), trigger the activation of serine protease (SP) cascades.54 The recognition of both β-glucan and/or Lys-PG may be mediated by Gram-negative binding protein (GNBP) 3 which is an extracellular recognition factor.55 The SP cascade triggers the activation of the Toll receptor ligand, Spatzle (Spz) with the help of the Spatzle-processing enzyme (SPE).56,57 Spz (PRO Spz) is present in the cell membrane of the infected silkworm with hydrophobic C-terminal Spz region in inactive form. SPE induces proteolysis, which leads to conformational changes which expose the determinants that are critical for binding of the Toll receptor.58 After this conformational modification, the Spz-Toll complex binds to an adaptor protein, MyD88, through the intracellular TIR domain.59 Following this, a heterotrimeric complex (MyD88-Tube-Pelle) is formed, leading to phosphorylation and degradation of the IkB factor, cactus.60 In general, cactus is bound to the NF-kB transcription factor(s) like Dorsal and/or Dif, inhibiting its activity and nuclear localization. The activation and nuclear translocation of Dorsal and Dif requires degradation of cactus by the kinase activity of Pelle,61–63 leading to an increase in the synthesis of AMPs.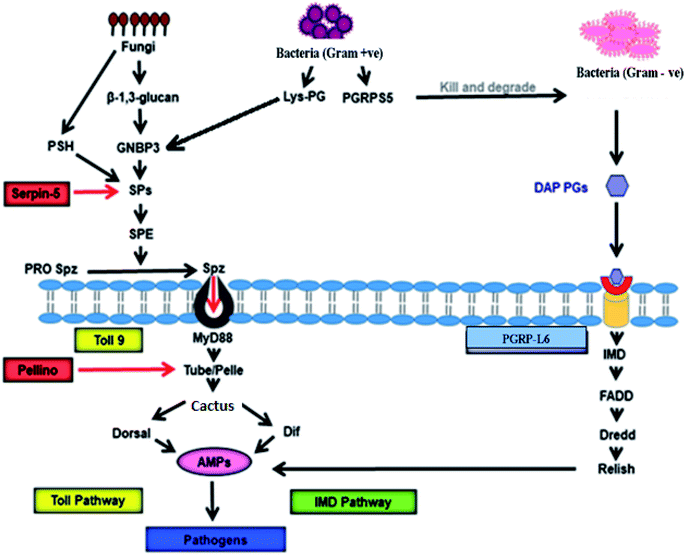 | ||
| Fig. 2 Signalling pathway of antimicrobial activity achieved by AMPs in B. mori. GNBP-3 recognize β-1,3-glucan present on the cell wall of the invading fungi that activate Toll pathway through Toll9, using serine protease cascade, and persuade production of the AMPs that kill invading pathogens. On the other hand, invading Gram-positive bacteria produce Lys-PG, which also activates expression of AMPs via Toll9 using serine protease cascade. PGRPs recognise PGs of the Gram-positive and Gram-negative bacterial cell wall to induce AMP production through IMD pathway. PGRP-L6 in B. mori may act as intracellular receptors that control IMD pathway. GNBP: Gram-Negative Binding protein, PSH: persephone, SP: serine protease, SPE: Spatzle-processing enzyme, pro Spz: pro Spatzle, Spz: Spatzle, PG: peptidoglycan, Lys-PG: Lysis-PG, PGRP: peptidoglycan recognition protein, IMD: immune deficiency, DAP-PG: diaminopimelic acid-PGs, AMPs: antimicrobial peptides.52–65 | ||
Peptidoglycan recognition proteins (PGRPs) were first identified and characterized in the silkworm Bombyx mori.64 These are 17 kDa proteins with a strong affinity towards Peptidoglycans (PGs). The PGRPs are an evolutionarily conserved family of microbial recognition proteins/enzymes found in both insects and mammals.65 The PGRPs consist of a domain with homology to a PG-digesting enzyme known as N-acetylmuramyl-L-alanine amidase, which cleaves the bond between the lactyl group in N-acetylmuramic acid and L-alanine in the stem peptide of PG. PGRPs with amidase activity can degrade Gram-negative bacteria. The amidase activity is restricted to diaminopimelic acid-containing peptidoglycans (DAP-PG), which are most common inthe cell wall of Gram-negative bacteria. However, PGRP binds to Lys-PG without any catalyzing activity in Gram-positive bacteria.66 This catalyzing activity of PGRP probably prevents DAP-PG from activating the Toll pathway and initiates the expression of AMPs via the IMD pathway.65 Infection in Bombyx mori with Gram-negative bacteria simultaneously triggers the IMD pathway (Fig. 2). The intracellular IMD signalling pathway requires the formation of a receptor complex that includes IMD protein, FADD, and the caspase Dredd.67 With the help of caspase activity Dredd, it cleaves Relish (a drosophila NF-kB precursor protein) and causes its phosphorylation. After phosphorylation, the N-terminal NF-kB component of Relish translocates into the nucleus and triggers AMP gene expression several-fold to fight against Gram-positive bacteria.65
Regulation of AMP production in silkworm
AMP production is in direct step with the pathogenic infection, but many intrinsic factors regulate specific AMP production following immune challenges. Insects have rapid and transient activation of immune genes after microbial infection to produce effectors. When microorganisms reach the haemolymph, the recognized invaders occur for the first time due to cellular immune systems and humoral immune reactions.21 Modulation and signalling factors are stimulated, and signal transduction is caused only in specific tissues.21 The genes that encode effectors are activated by cascade signalling.8It was reported earlier that different geographical types of silkworm (Japanese, Chinese, European and Tropical) are characterized by variable susceptibility to infectious pathogens. It was noted that European and Indian strains display the lowest sensitivity to E. mundtii and S. marcescens respectively. Although all four types of silkworm produce AMPs against both pathogens, European and Indian strains regulate their AMP production in a different manner for different pathogens. The European strain produces a specific composition of its AMP cocktail, with a more effective variant cecropin B6 isoform to suppress E. mundtii, while the Indian strain becomes resistant to S. marcescens with its prompt ability to activate the systemic transcription of AMPs. It is suggested that B. mori strains with a distinctive gene pool employ different strategies to fight bacterial infections, whose efficiency appears to be pathogen-dependent.68
Among AMPs, cecropin (Cec) includes five subtypes (A–E): the CecB subtype is active against Gram-positive and Gram-negative bacteria. CecB antimicrobial activity is related to interactions with bacterial membranes and pore formation.69 Further study revealed that B. mori cells, challenged against P. aeruginosa, expressed three classes of AMPs of which cecropin B isoform is of utmost important.70 The membranolytic activity for both CecB (Q53 CecB and E53 CecB) isoforms on the P. aeruginosa outer membrane, followed by permeabilization of the inner membrane and subsequent disintegration of both, causes cytoplasmic content leakage. The Q53 CecB isoform differs from the E53 CecB variant in just one amino acid (glutamic acid replaced by glutamine) and showed the highest antimicrobial and membranolytic activity against P. Aeruginosa.70 The Q53 CecB isoform contains a critical factor in stabilizing the hydrophobic segment that interacts with the bacterial membrane, determining the highest antimicrobial activity of the whole peptide. Studies suggested, in fact, that the same AMP might possess different targets when tested against different pathogens.8
Recombinant DNA technology synthesized the gloverin2 peptide from the BmGlv2 gene in a prokaryotic system, which are more stable (15–82.5 °C) and more active in preventing Gram-negative bacteria by disrupting cell integrity.69
AMPs mediated defense mechanism in silkworm
Silkworm produces ROS, phenoloxidase and AMPs as an array of defensive tools to combat bacterial infection.6,71 Among these three mechanisms, bacterial inhibition by AMPs is the most suitable because of its specificity towards invading pathogens.9 One property of insect innate immunity is the existence of an efficient systemic humoral immune response that fights microbial infections. This defense response consists of rapid and transient production of potent antibacterial and antifungal peptides, acting alone or in synergy when released into the haemolymph. These peptides are produced in the fat body (a functional homologue of the mammalian liver) and some blood cells.72 The AMPs are then secreted into the haemolymph, where they are accumulated at high concentrations and further diffuse throughout the body.73 In insects, production of AMPs was first observed in Drosophila melanogastor. Expressions of AMPs (Fig. 2) are mainly under the control of nuclear factor-kB through Toll and IMD pathways, which was discussed earlier.7,21,65,74–77 Silkworm AMPs show broad spectrum antimicrobial activity in comparison to AMPs from D. melanogaster.28 Oral administration of heat-killed Pseudomonas aeruginosa cells and Serratia marcescens significantly activates production of AMPs against those strains.6 In contrast, oral administration heat-killed S. aureus remains unable to trigger production of AMPs against P. aeruginosa infection. This suggests that the presence of Gram-negative bacteria and fungi in the diet of silkworms activates the immune response against P. aeruginosa infection, but Gram-positive bacteria failed to trigger such responses. It was reported by in another study that S. marcescens and P. aeruginosa are naturally occurring insect pathogens, in contrast to S. aureus.78–80 Thus, during co-evolution, insects may have developed a specific immune-activation strategy against such naturally occurring pathogenic bacteria.In insects, AMP production through the IMD pathway needs the activation of NF-kB under regulation of the I kappa B kinase (IKK) complex.81 Oral administration of heat-killed P. aeruginosa cells into silkworm activates the synthesis of IKKγ (a constituent of the IKK complex). Furthermore, these heat-killed orally administered bacteria, which acquire peptidoglycans, activate an insect cytokine known as paralytic peptide (PP), which leads to synthesis of AMPs through the IMD pathway.6,7,81,82
Defensin, at a dosage of 0.5 mg per animal, actively inhibits pathogenic Staphylococcus aureus bacteria, which are found to be resistant towards methicillin.83 When wounded mice were treated with D2A21, an analogue to cecropin, 100% survival was recorded when compared to that of control.84 Another group of synthetic cecropins (Shiva 11, d5c and Hecate) showed antibacterial activity against pathogens screened from infected contact lenses. Nowadays, these AMPs are used in a lens sterile solution to combat bacterial infections in conjunctiva.85 Silkworm AMPs show antimicrobial activity against a wide range of bacteria, such as Klebsiella pneumoniae, Klebsiella ozaenae, Shigella flexneri, Shigella sonnei, Staphylococcus aureus, Enterococcus faecalis, Staphylococcus epidermidis, Bacillus bombysepticus, Bacillus subtilis, Bacillus thuringiensis, Bacillus thuringiensis galleriae, Escherichia coli, Serratia marcescens, Pseudomonas aeruginosa, Ralstonia solaanacearum, Staphylococcus aureus and Xanthomons campestris.7,20,85 An earlier report showed that genetically engineered peptide Cecropin XJ is active against S. aureus with MIC of 0.4 μM, while other AMPs failed to exhibit this level of inhibition.28 Silkworm gut microflora plays a critical role in the synthesis of such AMPs.14 Silkworms actively suppress the infection against Yersinia pseudotuberculosis, B. bombyseptieus, E. coli, B. subtilis and P. aeruginosa with the help of AMPs that recognize bacterial peptidoglycans. The presence of gut microflora of B. mori induces expression of AMPs in fat body and haemocytes.6,77 The expression of attacin, cecropin, defensin, gloverin, leocin and moricin is accelerated by the oral administration of P. aeruginosa in gut microflora.6 It was also reported that specific proteins such as BmCPT1, BmPGRP-S5, and BmLBP collectively recognize E. coli in the midgut of B. mori to express different AMPs.86 On the other hand, B. bombyseptieus, a Gram-positive bacterium, activates the expression of a group of AMPs such as enbocin (which belongs to the cecropin family), lebocin, attacin, gloverin and moricin in the silkworm gut.87 Artificial feeding of B. mori with attenuated M. luteus leads to the increased production of cecropin D, cecropin E and gloverin 3 in body fat, while BmSerpin-5 reduces the production of these AMPs, down-regulating the Toll pathway by targeting BmHP6 and BmSP21.88 Laboratory experiments showed that recombinant BmSerpin-6, BmSerpin-15 down-regulated the expression of gloverin 2, cecropin D and moricin in fat body and hemocytes of B. mori.88–90 In a separate study it was revealed that Bombyx mori nuclear polyhedrosis virus (BmNPV) infection in silkworm causes highest level of expression of attacin, whereas serpin-5 and cecropin-D exhibited a negative regulatory correlation.91
The literature revealed that AMPs are a key component for immune defense against bacterial infection in silkworm.7,87,92–96 These observations stipulate that production of AMPs in silkworm through the Toll and IMD pathway is absolutely critical and remains unresolved. Hence, there is a need for detailed studies to understand such mechanisms, which will aid in providing more powerful tools against pathogenic multi-drug-resistant microbes.
Conclusion and future prospects97
The present study looks at the molecular mechanism for the synthesis of AMPs in B. mori, which are the crucial effectors of the innate immune system, elucidating their development and their classification. These natural AMPs of B. mori are useful in illustrating the way in which the design of AMP variants for tackling the rising number of multi-drug-resistant infections, as a suitable substitute for conventional antibiotics.Author contributions
J. N., A. S., D. F. B., A. K. wrote the manuscript with the help of A. K. M. and O. L. F.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Pesquisa e Desenvolvimento (CNPq), Fundação de Apoio à Pesquisa do Distrito Federal (FAPDF) and Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul (FUNDECT).References
- S. R. Sarandavar, MS thesis, University of Agricultural Science, Dharwad, 2014.
- S. Kalpana, A. A. M. Hatha and P. Laksmanaperumalsamy, Insect Sci. Its Appl., 1994, 15, 499–502 Search PubMed.
- Annual report, Central Silk Board, Ministry of Textile, Govt. of India, 2017 Search PubMed.
- Annual report, Central Silk Board, Ministry of Textile, Govt. of India, 2016 Search PubMed.
- B. B. Patnaik, D. H. Kim, H. O. Seung, Y. S. Song, N. D. M. Chanh, J. S. Kim, W. Jung, A. K. Saha, B. B. Bindroo and Y. S. Han, PLoS One, 2012, 7, e50900 CrossRef CAS PubMed.
- A. Miyashita, S. Takahashi, K. Ishii, K. Sekimizu and C. Kaito, PLoS One, 2015, 10, e0130486 CrossRef PubMed.
- K. Chen and Z. Lu, Dev. Comp. Immunol., 2018, 83, 3–11 CrossRef CAS PubMed.
- J. A. Hoffmann, Nature, 2003, 426, 33–38 CrossRef CAS PubMed.
- J. M. Harris, L. J. Seiderer and M. I. Lucas, Microb. Ecol., 1991, 21, 277–296 CrossRef CAS PubMed.
- A. A. P. Anand, S. J. Vennison, S. G. Sankar, D. I. G. Prabhu, P. T. Vasan, T. Raghuraman, C. J. Geoffrey and S. E. Vendan, J. Insect Sci., 2010, 10, 1–20 CrossRef PubMed.
- Anatomy of Silkworm Bombyx mori online, https://hbmahesh.weebly.com/uploads/3/4/2/2/3422804/4.anatomy.pdf, accessed June 2019 Search PubMed.
- B. Chen, K. Du, C. Sun, A. Vimalanathan, X. Liang, Y. Li, B. Wang, X. Lu, L. Li and Y. Shao, ISME J., 2018, 12, 2252–2262 CrossRef CAS PubMed.
- V. B. Khyade and R. M. Marathe, Global Journal of Bio-Science and BioTechnology, 2012, 2, 191–200 Search PubMed.
- E. Garcia-Gutierrez, M. J. Mayer, P. D. Cotter and A. Narbad, Gut Microbes, 2019, 10, 1–21 CrossRef CAS PubMed.
- D. Hultmark, Curr. Opin. Immunol., 2003, 15, 12–19 CrossRef CAS PubMed.
- B. Lemaitre and J. Hoffmann, Annu. Rev. Immunol., 2007, 25, 697–743 CrossRef CAS PubMed.
- C. A. Janeway Jr and R. Medzhitov, Annu. Rev. Immunol., 2002, 20, 197–216 CrossRef PubMed.
- S. J. Islam, S. Ezbaruah and J. Kalita, J. Adv. Biol. Biotechnol., 2016, 9, 1–15 CrossRef.
- D. Hultmark, A. Engström, H. Bennich, R. Kapur and H. G. Boman, Eur. J. Biochem., 1982, 127, 207–217 CrossRef CAS PubMed.
- W. Yang, T. Cheng, M. Ye, X. Deng, H. Yi, Y. Huang, X. Tan, D. Han, B. Wang, Z. Xiang, Y. Cao and Q. Xia, PLoS One, 2011, 6, e18109 CrossRef CAS PubMed.
- H. Tanaka, J. Ishibashi, K. Fujita, Y. Nakajima and A. Sagisaka, Insect Biochem. Mol. Biol., 2008, 38, 1087–1110 CrossRef CAS PubMed.
- F. M. Jiggins and K. W. Kim, Genetics, 2005, 171, 1847–1859 CrossRef CAS PubMed.
- S. C. Barr, D. Rose and J. M. Jaynes, J. Parasitol., 1995, 81, 974–978 CrossRef CAS PubMed.
- J. Boisbouvier, A. Prochnicka-Chalufour, A. R. Nieto, J. A. Torres, N. Nanard, M. H. Rodriguez, L. D. Possani and M. Delepierre, Eur. J. Biochem., 1998, 257, 263–273 CrossRef CAS PubMed.
- H. Suttmann, M. Retz, F. Paulsen, J. Harder, U. Zwergel, J. Kamradt, B. Wullich, G. Unteregger and M. Stockle, BMC Urol., 2008, 8, 5–8 CrossRef PubMed.
- S. M. Gregory, A. Cavenaugh, J. Velvet, P. Antje and F. F. A. Paulo, Biophys. J., 2008, 94, 1667–1680 CrossRef CAS PubMed.
- N. S. Parachin and O. L. Franco, Front. Microbiol., 2014, 5, 147 Search PubMed.
- L. Xia, F. Zhang, Z. Liu, J. Ma and J. Yang, Exp. Ther. Med., 2013, 5, 1745–1751 CrossRef CAS PubMed.
- B. J. Shahaji, O. A. Kumar, J. C. Balbhim, D. S. Mallikarjun and T. P. Ramrao, J. Adv. Bioinf. Appl. Res., 2015, 6, 8–22 CAS.
- K. Matsuyama and S. Natori, J. Biol. Chem., 1988, 263, 17112–17116 CAS.
- Y. Xiao, A. L. Hughes, J. Ando, Y. Mastuda, J. F. Cheng, D. Skinner-Noble and G. A. Zhang, BMC Genomics, 2004, 5, 56 CrossRef PubMed.
- S. Fujiwara, J. Imai, M. Fujiwara, T. Yaeshima, T. Kawashima and K. Kobayashi, J. Biol. Chem., 1990, 265, 11333–11337 CAS.
- K. Yamada and S. Natori, Biochem. J., 1993, 291, 275–279 CrossRef CAS PubMed.
- H. Yi, M. Chowdhury, Y. D. Huang and X. Q. Yu, Appl. Microbiol. Biotechnol., 2014, 98, 5807–5822 CrossRef CAS PubMed.
- H. Wen, X. Lan, T. Cheng, N. He, K. Shiomi, Z. Kajiura, Z. Zhou, Q. Xia, Z. Xiang and M. Nakagaki, Mol. Biol. Rep., 2009, 36(4), 711–716 CrossRef CAS PubMed.
- K. Yoichi, T. Hiromitsu, I. Jun, I. Takashi and Y. Inoru, Biosci., Biotechnol., Biochem., 2008, 72, 2353–2361 CrossRef PubMed.
- L. Huang, T. Cheng, P. Xu, D. Cheng, T. Fang and Q. Xia, PLoS One, 2009, 12, e8098 CrossRef PubMed.
- S. Hara and M. Yamakawa, J. Biol. Chem., 1995, 270, 29923–29927 CrossRef CAS PubMed.
- T. Cheng, P. Zhao, C. Liu, P. Xu and Z. Gao, Genomics, 2006, 87, 356–365 CrossRef CAS PubMed.
- H. Tanaka, A. Sagikasa, K. Fujita, S. Furukuwa, J. Ishibashi and M. Yamakawa, Insect Biochem. Mol. Biol., 2012, 42, 474–481 CrossRef CAS PubMed.
- H. Hemmi, J. Ishibashi, S. Haraand and M. Yamakawa, FEBS Lett., 2002, 518, 33–38 CrossRef CAS PubMed.
- A. Axen, A. Carlsson, A. Engström and H. Bennich, Gloverin, Eur. J. Biochem., 1997, 247, 614–619 CrossRef CAS PubMed.
- S. Kawaoka, S. Katsuma, T. Daimon, R. Isono, N. Omuro, K. Mita and T. Shimada, Arch. Insect Biochem. Physiol., 2008, 67, 87–96 CrossRef CAS PubMed.
- A. S. Gandhe, K. P. Arunkumar, S. H. John and J. Nagaraju, BMC Genomics, 2006, 7, 184 CrossRef PubMed.
- N. Mrinal and J. Nagaraju, J. Biol. Chem., 2008, 283, 23376–23387 CrossRef CAS PubMed.
- H. Y. Yi, X. J. Deng, W. Y. Yanga, C. Z. Zhouc, Y. Caoa and X. Q. Yub, Insect Biochem. Mol. Biol., 2013, 43, 612–625 CrossRef CAS PubMed.
- M. Hedengren, K. Borge and D. Hultmark, Biochem. Biophys. Res. Commun., 2000, 279, 574–581 CrossRef CAS PubMed.
- A. Carlsson, P. Engstrom, E. T. Palva and H. Bennich, Infect. Immun., 991, 59, 3040–3045 Search PubMed.
- M. Sugiyama, H. Kuniyoshi, E. Kotani, K. Taniai, K. Kadono-Okuda, Y. Kato, M. Yamamoto, M. Shimabukuro, S. Chowdhury, J. Xu, S. K. Choi, H. Katoaka, A. Suzuki and M. Yamakawa, Insect Biochem. Mol. Biol., 1995, 25, 385–392 CrossRef CAS.
- S. Hara and M. Yamakawa, Biochem. J., 1995, 310, 651–656 CrossRef CAS PubMed.
- M. Kangayam, K. M. Ponnuvel and M. Yamakawa, Curr. Sci., 2002, 83, 447–454 Search PubMed.
- C. Hetru and J. A. Hoffmann, Cold Spring Harbor Perspect. Biol., 2009, 1, a000232 Search PubMed.
- K. Aggarwal and N. Silverman, BMB Rep., 2008, 41, 267–277 CrossRef CAS PubMed.
- L. El Chamy, V. Leclerc, I. Caldelari and J. M. Reichhart, Nat. Immunol., 2008, 9, 1165–1170 CrossRef CAS PubMed.
- M. Gottar, V. Gobert, A. A. Matskevich, J. M. Reichhart, C. Wang, T. M. Butt, M. Belvin, J. A. Hoffmann and D. Ferrandon, Cell, 2006, 127, 1425–1437 CrossRef CAS PubMed.
- D. Morisato and K. V. Anderson, Cell, 1994, 76, 677–688 CrossRef CAS PubMed.
- D. S. Schneider, Y. Jin, D. Morisato and K. V. Anderson, Development, 1994, 120, 1243–1250 CAS.
- C. J. Arnot, N. J. Gay and M. Gangloff, J. Biol. Chem., 2010, 285, 19502–19509 CrossRef CAS PubMed.
- T. Horng and R. Medzhitov, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 12654–12658 CrossRef CAS PubMed.
- H. Sun, B. N. Bristow, G. Qu and S. A. Wasserman, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 12871–12876 CrossRef CAS PubMed.
- L. P. Wu and K. V. Anderson, Nature, 1998, 392, 93–97 CrossRef CAS PubMed.
- P. Towb, A. Bergmann and S. A. Wasserman, Development, 2001, 128, 4729–4736 CAS.
- H. R. Huang, Z. J. Chen, S. Kunes, G. D. Chang and T. Maniatis, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 8322–8327 CrossRef CAS PubMed.
- M. Ochiai and M. Ashida, J. Biol. Chem., 1999, 274, 11854–11858 CrossRef CAS PubMed.
- T. Kaneko and N. Silverman, Cell. Microbiol., 2005, 7, 461–469 CrossRef CAS PubMed.
- C. I. Chang, S. S. Pili-Floury, M. Herve, C. Parquet, Y. Chelliah and B. Lemaitre, PLoS Biol., 2004, 2, E277 CrossRef PubMed.
- S. Hu and X. Yang, J. Biol. Chem., 2000, 275, 30761–30764 CrossRef CAS PubMed.
- O. Romoli, A. Saviane, A. Bozzato, P. D'Antona, G. Tettamanti, A. Squartini, S. Cappellozza and F. Sandrelli, Sci. Rep., 2017, 7(1), 1048 CrossRef PubMed.
- Q. Wang, P. Guo, Z. Wang, H. Liu, Y. Zhang, S. Jiang, W. Han, Q. Xia and P. Zhao, Int. J. Mol. Sci., 2018, 19(8), E2275 CrossRef PubMed.
- O. Romoli, S. Mukherjee, S. A. Mohid, A. Dutta A, A. Montali, E. Franzolin, D. Brady, F. Zito, E. Bergantino, C. Rampazzo, G. Tettamanti, A. Bhunia and F. Sandrelli, ACS Infect. Dis., 2019, 5(7), 1200–1213 CrossRef CAS PubMed.
- Y. Yasuhara, Y. Koizumi, C. Katagiri and M. Ashida, Arch. Biochem. Biophys., 1995, 320, 14–23 CrossRef CAS PubMed.
- D. Ferrandon, Nat. Rev. Immunol., 2007, 7, 862–874 CrossRef CAS PubMed.
- R. A. B. Ezekowitz and J. A. Hoffmann, Innate immunity, Humana Press, Totowa: NJ, 2002 Search PubMed.
- S. Rutschmann, A. Kilinc and D. Ferrandon, J. Immunol., 2002, 168, 1542–1546 CrossRef CAS PubMed.
- M. Yamakawa and H. Tanaka, Dev. Comp. Immunol., 1999, 23, 281–289 CrossRef CAS PubMed.
- Y. Kaneko, S. Furukawa, H. Tanaka and M. Yamakawa, Biosci., Biotechnol., Biochem., 2007, 71, 2233–2241 CrossRef CAS PubMed.
- Y. Kaneko, H. Tanaka, J. Ishibashi, T. Iwasaki and M. Yamakawa, Biosci., Biotechnol., Biochem., 2008, 72, 2353–2361 CrossRef CAS PubMed.
- P. A. Grimont and F. Grimont, Annu. Rev. Microbiol., 1978, 32, 221–248 CrossRef CAS PubMed.
- N. Vodovar, M. Vinals, P. Liehl, A. Basset, J. Degrouard and P. Spellman, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11414–11419 CrossRef CAS PubMed.
- S. M. Braxton, D. W. Onstad, D. E. Dockter, R. Giordano, R. Larsson and R. A. Humber, J. Invertebr. Pathol., 2003, 83, 185–195 CrossRef CAS PubMed.
- B. Lemaitre, Nat. Rev. Immunol., 2004, 4, 521–527 CrossRef CAS PubMed.
- K. Ishii, H. Hamamoto, M. Kamimura, Y. Nakamura, H. Noda and K. Imamura, J. Biol. Chem., 2010, 285, 28635–28642 CrossRef CAS PubMed.
- M. Yamada, K. Nakamura, H. Saido-Sakanaka, A. Asaoka, M. Yamakawa, Y. Yamamoto, Y. Koyama, K. Hikosaka, A. Shimizu and Y. Hirota, J. Vet. Med. Sci., 2005, 67, 1005–1011 CrossRef CAS PubMed.
- W. S. Jang, H. N. Kim, Y. S. Lee, M. H. Nam and I. H. Lee, FEBS Lett., 2002, 521, 81–86 CrossRef CAS PubMed.
- L. B. Sousa, M. J. Mannis, I. R. Schwab, J. Cullor, H. Houstani, W. Smith and J. Jaynes, The use of synthetic cecropin (D5C) in disinfecting contact lens solutions, CLAO J., 1999, 22, 114–117 Search PubMed.
- J. Liang, T. Wang, Z. Xiang and N. He, Insect Biochem. Mol. Biol., 2015, 58, 76–88 CrossRef CAS PubMed.
- L. Huang, T. Cheng, P. Xu, D. Cheng, T. Fang and Q. Xia, PLoS One, 2009, 4, e8098 CrossRef PubMed.
- J. L. Li, L. Ma, Z. Lin, Z. Zou and Z. Q. Lu, Insect Biochem. Mol. Biol., 2016, 73, 27–37 CrossRef CAS PubMed.
- Y. Li, P. Zhao, H. Liu, X. Guo, H. He, R. Zhu, Z. Xiang and Q. Xia, Insect Biochem. Mol. Biol., 2015, 57, 11–19 CrossRef CAS PubMed.
- B. Li, H. Z. Yu, C. J. Ye, Y. Ma, X. Li, T. Fan, F. S. Chen and J. P. Xu, Gene, 2017, 610, 64–70 CrossRef CAS PubMed.
- W. T. Dong, X. D. Ling, L. F. Xiao, J. J. Hu, X. X. Zhao, J. X. Liu and Y. Zhang, Microb. Pathog., 2019, 130, 137–145 CrossRef CAS PubMed.
- K. K. Chen, L. Zhou, F. Chen, Y. Peng and Z. Q. Lu, Dev. Comp. Immunol., 2016, 61, 126–135 CrossRef CAS PubMed.
- S. Wu, X. Zhang, Y. He, J. Shuai, X. Chen and E. Ling, Dev. Comp. Immunol., 2010, 34, 1191–1198 CrossRef CAS PubMed.
- C. Y. Zhou, X. F. Zha, C. Liu, M. J. Han, L. Y. Zhang, P. P. Shi, H. Wang, R. W. Zheng and Q. Y. Xia, Insect Sci., 2016, 23, 502–512 CrossRef CAS PubMed.
- L. Zhi, Ma. Yan, L. Xuan, L. Yi Li and F. Dai, Anim. Biol., 2019, 69(4), 391–410 Search PubMed.
- S. Panthee, A. Paudel, H. Hamamoto and K. Sekimizu, Front. Microbiol., 2017, 8, 373 Search PubMed.
- O. Fleitas and O. L. Franco, Front. Microbiol., 2016, 7, 381 Search PubMed.
Footnote |
| † Dedicated to Prof. Ranadhir Chakraborty, University of North Bengal, on the occasion of his 57th birthday. |
| This journal is © The Royal Society of Chemistry 2020 |
