Open Access Article
Open Access ArticleDNA density-dependent uptake of DNA origami-based two-or three-dimensional nanostructures by immune cells†
Tatsuoki
Maezawa
 a,
Shozo
Ohtsuki
a,
Shozo
Ohtsuki
 a,
Kumi
Hidaka
b,
Hiroshi
Sugiyama
a,
Kumi
Hidaka
b,
Hiroshi
Sugiyama
 bc,
Masayuki
Endo
bc,
Masayuki
Endo
 bc,
Yuki
Takahashi
bc,
Yuki
Takahashi
 a,
Yoshinobu
Takakura
a and
Makiya
Nishikawa
a,
Yoshinobu
Takakura
a and
Makiya
Nishikawa
 *ad
*ad
aDepartment of Biopharmaceutics and Drug Metabolism, Graduate School of Pharmaceutical Sciences, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan
bDepartment of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan
cInstitute for Integrated Cell-Material Sciences, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan
dLaboratory of Biopharmaceutics, Faculty of Pharmaceutical Sciences, Tokyo University of Science, Noda, Chiba 278-8510, Japan. E-mail: makiya@rs.tus.ac.jp
First published on 19th June 2020
Abstract
DNA nanostructures are expected to be applied for targeted drug delivery to immune cells. However, the structural properties of DNA nanostructures required for the delivery have not fully been elucidated. In this study, we focused on the DNA density that can be important for the their recognition and uptake by immune cells. To examine this, DNA nanostructures with almost identical molecular weights and structural flexibility, but with different shapes and DNA densities, were designed using DNA origami technology. We compared the following five types of DNA nanostructures, all of which consisted of ten DNA helices using an identical circular, single-stranded scaffold and staples. Rec180 had a rectangular-shaped, almost flat structure. Rec90, Rec50 and Rec0 were bent forms of Rec180 at the center by 90, 50 or 0 degrees, respectively. Rec50/50 has two bends of 50 degrees each so that the both ends stick together to form a triangular prism shape. The fluctuation, or flexibility, of these DNA nanostructures under solution conditions was estimated using CanDo software. The DNA density estimated from the average distance between any two of the ten DNA helices in the DNA nanostructures was different among them; Rec50, Rec0 and Rec50/50 had a higher density than Rec180 and Rec90. Agarose gel electrophoresis and atomic force microscopy showed that all of the nanostructures were prepared with high yield. Flow cytometry analysis revealed that the uptake of DNA nanostructures by murine macrophage-like RAW264.7 cells was higher for those with higher DNA density than those with low density. There was a positive correlation between the density and cellular uptake. These results indicate that DNA nanostructures with high DNA density are suitable for delivery to immune cells.
Introduction
In 1982, Seeman presented the concept of DNA nanostructures, which are composed of only DNA, and this is the birth of DNA nanotechnology.1 In 1998, Winfree et al. annealed a double-crossover DNA tile and constructed a crystal using the tile as the structural unit.2 With this achievement, he demonstrated that DNA nanostructures can be constructed by utilizing hybridization between complementary sequences. At that time, however, it was difficult to control the size and form of DNA nanostructures. Therefore, the number of designable DNA nanostructures was limited. DNA origami technology was devised by Rothemund in 2006. In this technology, a long, single-strand DNA, a scaffold, and short “staple” DNAs complementary to parts of the scaffold are annealed into a targeted shape by self-assembly.3 This technology enables the preparation of various, perhaps arbitrary, DNA nanostructures, and many researchers accomplished the construction of various DNA nanostructures.4–6 In addition, versatile functions were added to DNA nanostructures by adding various molecules such as proteins and photo-responsive functional groups.7,8DNA nanostructures are expected to be used in many fields, and are expected to play important roles in drug delivery, because DNA is biodegradable and biocompatible, which are suitable properties for delivery systems. Some researchers succeeded in drug delivery by using DNA nanostructures in vitro and in vivo. Schuller et al. demonstrated that DNA containing an unmethylated cytosine-phosphate-guanine (CpG) motif connected to a tubular DNA nanostructure activated immune cells with higher efficiency than single-strand CpG DNA because of its accelerated cellular uptake.9 Jiang et al. loaded doxorubicin into tubular and triangular DNA nanostructures and demonstrated an enhanced delivery of the anticancer agent to human breast cancer-derived MCF7 cells.10 Douglas et al. developed a box-shaped DNA nanostructure sealed with aptamers against the platelet-derived growth factor (PDGF) and showed that it selectively delivered a conjugated antibody to human CD3 cells that expressed PDGF.11
To develop DNA nanostructures that are suitable for drug delivery, it is important to elucidate the relationship between the structural properties of DNA nanostructures and their cellular uptake. However, despite many studies on the applications of DNA nanostructures to drug delivery, little information is available about the structural properties of DNA nanostructures suitable for cellular uptake.
We developed various DNA nanostructures including polypod-like structured DNA, or polypodna,12 polypodna-based DNA dendrimers,13 tetrahedral and tetragonal DNAs,14 and rectangular DNA nanostructures.15 By comparing their uptake by immune cells, it was found that DNA nanostructures with more complicated structures tended to be more effectively taken up by immune cells than simpler ones. In addition, this structure-dependent uptake was almost specific to dendritic cells and macrophages, but was hardly applied to fibroblasts and endothelial cells.16 However, in these studies, multiple factors of DNA nanostructures, such as molecular weight and DNA density, were different among them. These factors could affect the cellular uptake of DNA nanostructures, which could make it difficult to elucidate the factors determining their cellular uptake.
In this study, we focused on the DNA density of DNA nanostructures and designed five types of DNA which have different DNA densities for one structure. We also paid attention to the molecular weight and structural flexibility, and designed DNA nanostructures with nearly identical molecular weights and structural flexibility to minimize the possible influence of these parameters. To this end, we designed five types of two- or three-dimensional DNA nanostructures using an identical circular, single-stranded scaffold and staples, each of which was bent at different angles to endow them with different DNA densities. By evaluating their structural properties and cellular uptake by murine macrophage-like RAW264.7 cells, we elucidated the relationship between the DNA density of DNA nanostructures and their uptake by the cells.
Materials and methods
Chemicals
RPMI1640 medium was purchased from Nissui Pharmaceutical Co., Ltd (Tokyo, Japan). Fetal bovine serum (FBS) and Opti-modified Eagle's medium (Opti-MEM) were obtained from Thermo Fisher Scientific (Waltham, MA, USA). All of the other chemicals were of the highest grade available and used without further purification.Cell culture
Murine macrophage-like RAW264.7 cells were obtained from ATCC (Manassas, VA, USA) and cultured in RPMI1640 medium supplemented with 10% heat-inactivated FBS, 0.2% NaHCO3, 100 units per ml of penicillin, 100 μg ml−1 of streptomycin and 2 mM L-glutamine at 37 °C in humidified air containing 5% CO2. The cells were then seeded on 96-well culture plates at a density of 5 × 04 cells per well and cultured for 24 h prior to use.Scaffold
To construct DNA nanostructures with an optimal size for cellular uptake, a scaffold 1801 bases long developed in a previous study15 was used. The sequence of the scaffold is as follows:5′-CTAGATACAAATGTGGTATGGAATTCAGTCAATATGTTCACCCCAAAAAAGCTGTTTGTTAACTTGCCAACCTCATTCTAAAATGTATATAGAAGCCCAAAAGACAATAACAAAAATATTCTTGTAGAACAAAATGGGAAAGAATGTTCCACTAAATATCAAGATTTAGAGCAAAGCATGAGATGTGTGGGGATAGACAGTGAGGCTGATAAAATAGAGTAGAGCTCAGAAACAGACCCATTGATATATGTAAGTGACCTATGAAAAAAATATGGCATTTTACAATGGGAAAATGATGATCTTTTTCTTTTTTAGAAAAACAGGGAAATATATTTATATGTAAAAAATAAAAGGGAACCCATATGTCATACCATACACACAAAAAAATTCCAGTGAATTATAAGTCTAAATGGAGAAGGCAAAACTTTAAATCTTTTAGAAAATAATATAGAAGCATGCCATCAAGACTTCAGTGTAGAGAAAAATTTCTTATGACTCAAAGTCCTAACCACAAAGAAAAGATTGTTAATTAGATTGCATGAATATTAAGACTTATTTTTAAAATTAAAAAACCATTAAGAAAAGTCAGGCCATAGAATGACAGAAAATATTTGCAACACCCCAGTAAAGAGAATTGTAATATGCAGATTATAAAAAGAAGTCTTACAAATCAGTAAAAAATAAAACTAGACAAAAATTTGAACAGATGAAAGAGAAACTCTAAATAATCATTACACATGAGAAACTCAATCTCAGAAATCAGAGAACTATCATTGCATATACACTAAATTAGAGAAATATTAAAAGGCTAAGTAACATCTGTGGCTTAATTAAAACAGGTAGTTGACAATTAAACATTGGCATAGTATATCTGCATAGTATAATACAACTCACTATAGGAGGGCCATCATGGCCAAGTTGACCAGTGCTGTCCCAGTGCTCACAGCCAGGGATGTGGCTGGAGCTGTTGAGTTCTGGACTGACAGGTTGGGGTTCTCCAGAGATTTTGTGGAGGATGACTTTGCAGGTGTGGTCAGAGATGATGTCACCCTGTTCATCTCAGCAGTCCAGGACCAGGTGGTGCCTGACAACACCCTGGCTTGGGTGTGGGTGAGAGGACTGGATGAGCTGTATGCTGAGTGGAGTGAGGTGGTCTCCACCAACTTCAGGGATGCCAGTGGCCCTGCCATGACAGAGATTGGAGAGCAGCCCTGGGGGAGAGAGTTTGCCCTGAGAGACCCAGCAGGCAACTGTGTGCACTTTGTGGCAGAGGAGCAGGACTGAGGATAACCTAGGAAACCTTAAAACCTTTAAAAGCCTTATATATTCTTTTTTTTCTTATAAAACTTAAAACCTTAGAGGCTATTTAAGTTGCTGATTTATATTAATTTTATTGTTCAAACATGAGAGCTTAGTACATGAAACATGAGAGCTTAGTACATTAGCCATGAGAGCTTAGTACATTAGCCATGAGGGTTTAGTTCATTAAACATGAGAGCTTAGTACATTAAACATGAGAGCTTAGTACATACTATCAACAGGTTGAACTGCTGATCTTAATTAAAATTATCTCTAAGGCATGTGAACTGGCTGTCTTGGTTTTCATCTGTACTTCATCTGCTACCTCTGTGACCTGAAACATATTTATAATTCCATTAAGCTGTGCATATGATAGATTTATCATATGTATTTTCCTTAAAGGATTTTTGTAAGAACTAATTGAATTGATACCTGTAAAGTCTTTATCACACTACCCAATAAATAATAAATCTCTTTGTTCAGCTCTCTGTTTCGCTGAGGT-3′.
Staples
Staples, short single-strand DNA 16–50 bases long, were purchased from Eurofin Genomics Japan K.K. (Tokyo, Japan). Ten staples (two staples for each DNA nanostructure) labeled with Alexa Fluor488 at the 5′-end were purchased from Japan BioService Co.,Ltd (Saitama, Japan). Sequences of staples used in DNA nanostructures are listed in ESI, Table S1.†Design of DNA nanostructures
DNA nanostructures were designed using caDNAno, computer-aided design software of two- and three-dimensional DNA nanostructures as previously reported.2Fluctuation or flexibility of DNA nanostructures
The JavaScript Object Notation files of the five DNA nanostructures were prepared using caDNAno and analyzed in CanDo, a DNA nanostructure's model generating system.3 In CanDo analysis, the DNA nanostructure is divided into hundreds of parts, and all conformations speculated to exist in solution at room temperature are calculated. Based on this calculation, the root mean square fluctuation (RMSF) of each part is determined as a parameter of fluctuation or flexibility.DNA density of DNA nanostructures
The average distance between two of the DNA helices composing the DNA nanostructures was used as an index of DNA density of DNA nanostructures.Preparation of DNA nanostructures
Each structure was prepared by mixing equimolar amounts of the scaffold and 50 staples. The scaffold and the staples dissolved in a TE-Mg2+ buffer (20 mM Tris-HCl, 1 mM ethylenediaminetetraacetic acid (EDTA), and 10 mM magnesium chloride, pH 7.3) were mixed at a final concentration of 60 nM for each DNA. When preparing fluorescently labeled DNA nanostructures, staples labeled with Alexa Fluor488 were mixed instead of unlabeled staples. The mixtures were then heated to 85 °C and gradually cooled down to 4 °C using a thermal cycler (PC-818S Program Temp Control System; ASTEC Co., Ltd, Fukuoka, Japan). The products were analyzed by agarose gel electrophoresis in 0.5× TBE buffer containing 10 mM potassium (45 mM Tris borate, 1 mM EDTA, supplemented with 10 mM MgCl2) at 4 °C. The gel was stained with SYBR Gold (Thermo Fisher Scientific) and observed using a LAS3000 (Fujifilm, Tokyo, Japan).AFM imaging of DNA nanostructures
Atomic force microscopy (AFM) images of each structure were obtained using an AFM system (Nano Live Vision, RIBM, Tsukuba, Japan) using a silicon nitride cantilever (BL-AC10EGS; Olympus, Tokyo, Japan). In brief, a DNA sample was adsorbed on a freshly cleaved mica plate pretreated with 0.1% aqueous 3-aminopropyltriethoxysilane for 5 min at room temperature and then washed three times with a buffer solution containing 20 mM Tris and 10 mM MgCl2. Scanning was performed in the same buffer solution.Flow cytometry
Alexa Fluor488-labeled single-stranded DNA (ssDNA) and five DNA nanostructures were diluted in 50 μl Opti-MEM to 3 nM. They were added to RAW264.7 cells on a 96-well plate at a density of 5 × 104 cells per well and incubated for 4 h at 37 °C or 4 °C. RAW264.7 cells were washed three times with PBS and harvested. Thereafter, the fluorescence intensity of cells was determined by flow cytometry (Gallios Flow Cytometer, BD Biosciences, NJ, USA) using Kaluza software (version 1.0, BD Biosciences), and the mean fluorescence intensity (MFI) was calculated.Statistical analysis
Differences were statistically evaluated by one-way analysis of variance (ANOVA) followed by the Tukey–Kramer test for multiple comparisons. A p value less than 0.05 was considered statistically significant.Results and discussion
Design of DNA nanostructures
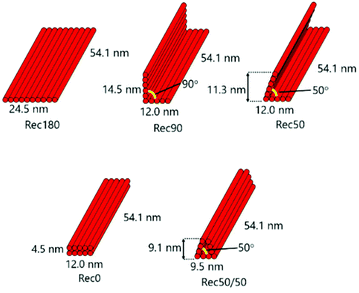
We firstly designed Rec180, an almost flat, rectangular-shaped DNA nanostructure, as reported previously.15 Assuming the helical diameter of DNA, the interhelical gap, and the distance between bases in double-strand DNA were 2, 0.5 and 0.34 nm, respectively, Rec180 has 24.5 nm width (10 helix), 54.1 nm height (160 base), and 2 nm depth (1 helix). There are single-stranded loops on both sides of Rec180 so that the interaction between the DNA nanostructures through π–π stacking hardly occurs. Then, based on the design of Rec180, four DNA nanostructures were designed. Rec180 was bent at the center by 90, 50 or 0 degrees to obtain Rec90, Rec50 and Rec0, respectively, or bent twice by 50 degrees each to obtain Rec50/50. Schematic illustrations of these DNA nanostructures are shown in Fig. 1. They are constructed using the same 1801-base long scaffold and 50 staples, and the total base number per nanostructure was identical to that of Rec180. Therefore, the molecular weights of the five DNA nanostructures designed were almost identical.Structural flexibility of DNA nanostructures
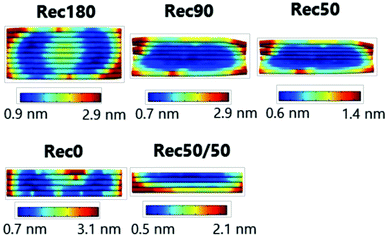
The proportion of the single- and double-stranded parts is important for root mean square fluctuation (RMSF) or structural flexibility of DNA nanostructures. Actually, reducing the number of staples and increasing single-stranded parts result in larger RMSF values. Fig. 2 shows the results of CanDo analysis of each DNA nanostructure. Both sides of the DNA nanostructures were single stranded. Therefore, the RMSF values of the sides were relatively high, indicating that these parts are flexible. On the other hand, the central part of the DNA nanostructures, where there are many crossovers between double helices, had low RMSF values, indicating that this part is a rigid structure. These characteristics were common to all the DNA nanostructures prepared. This could be due to the fact that they were prepared using an identical scaffold and 50 staples. A slightly higher RMSF value of the central part of Rec180 could be explained by the fact that this part has few crossovers (ESI,†Fig. 1). Our previous study showed that DNA nanostructures prepared with different numbers of staples possessed different structural flexibilities depending on the number and position of the staples.15 Therefore, these results suggest that the flexibility is comparable among the DNA nanostructures designed.DNA density of DNA nanostructures
Fig. 3a shows the calculation method for the average distance between DNA helices and the average distance between the DNA helices of each nanostructure. Each DNA nanostructure consists of 10 helices, so that there are 45 combinations of the two helices in total. The average distances of the five DNA nanostructures calculated were 10.7, 7.1, 6.2, 5.0, and 5.1 nm for Rec180, Rec90, Rec50, Rec0, and Rec50/50, respectively (Fig. 3b).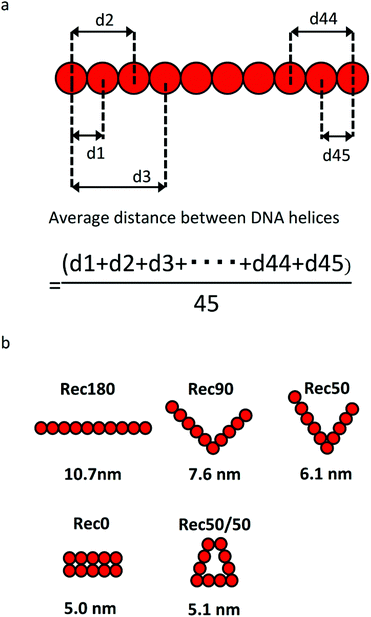 | ||
| Fig. 3 (a) Calculation method of the average distance between DNA helices. (b) Arrangement of helices and the average distance between DNA helices of each DNA nanostructure. | ||
Agarose gel electrophoresis of DNA nanostructures
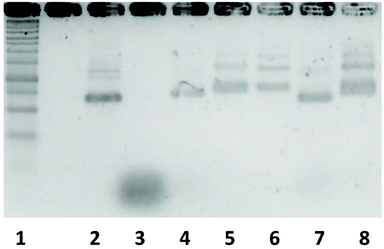
Fig. 4 shows the agarose gel electrophoresis analysis of the five DNA nanostructures. The bands of the DNA nanostructures were slightly higher than those of the scaffold, and the band of the staples was hardly detected in all the DNA nanostructures. These results indicate that the scaffold and the staples hybridize to form each DNA nanostructure.AFM observation of DNA nanostructures
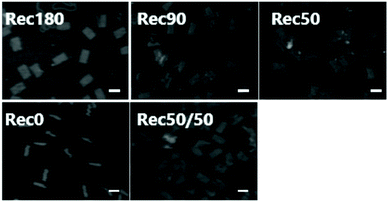
Fig. 5 shows the AFM images of the five DNA nanostructures. These images were obtained after the adsorption of the DNA nanostructure onto mica plates. Therefore, both the two- and three-dimensional DNA nanostructures should be observed as two-dimensional shapes. Rec0 was observed as a rectangle with about 12.5 nm width and about 50 nm height. Other DNA nanostructures were also observed as rectangular shapes with about 25 nm width and about 50 nm height. Some collapsed or unfolded structures were observed in the images, but their percentage was low in all the DNA nanostructures. These results of the electrophoresis and AFM imaging indicate that all the DNA nanostructures are constructed as designed with high yield.Cellular uptake of DNA nanostructures
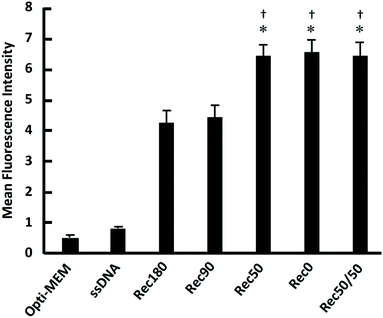
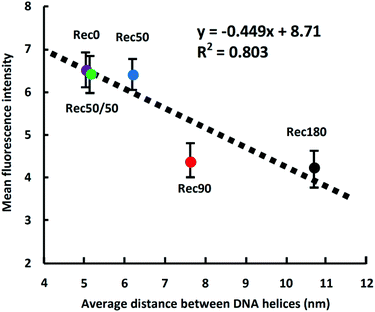
Fig. 6 shows the uptake of the DNA nanostructures by RAW264.7 cells. When RAW264.7 cells were incubated at 37 °C, all the DNA nanostructures were taken up by the cells much more efficiently than single-stranded DNA. The uptake of Rec50/50 was the highest. The DNA nanostructures with high DNA density tended to be taken up more efficiently than those with low density. When RAW264.7 cells were incubated at 4 °C, cellular uptake of all DNA nanostructures was comparable (data not shown). Fig. 7 shows the relationship between the average distance between DNA helices and the uptake by RAW264.7 cells of the DNA nanostructures. There was a negative correlation between these two, suggesting that the DNA density is a key parameter determining the cellular uptake when the molecular weight and structural flexibility were comparable to one another.Most studies on the DNA origami technology use M13mp18 (p7249) single-stranded DNA of 7249 bases as a scaffold. It is because the source of a long single-stranded DNA other than this is limited. However, the size of M13mp18 is relatively large, and M13mp18-based DNA nanostructures are generally larger than 100 nm in length. Taking into consideration that the upper size limit of endocytotic uptake is around 100 nm,17 shorter single-stranded DNAs are suitable as a scaffold to evaluate the relationship between the structural properties and cellular uptake of DNA nanostructures. Therefore, we used a scaffold of 1801 bases which was previously constructed,15 and prepared five DNA nanostructures with a suitable size for cellular uptake. The sizes of these DNA nanostructures are comparable to the size of nanosized drug delivery systems, such as polymeric micelles and liposomes.
TLR9 receptor-positive immune cells would release proinflammatory cytokines upon stimulation with CpG DNA, and these cytokines could affect the cellular uptake of compounds, including DNA nanostructures. Therefore, the DNA nanostructures used should contain a few or no CpG motifs. pCpG-mcs, a CpG-free plasmid DNA, was used as the template for the preparation of the scaffold to accomplish this.
In this study, DNA nanostructures with high DNA density were more efficiently taken up by RAW264.7 cells than those with low DNA density. In contrast, little difference was observed in the MFI values at 4 °C among the DNA nanostructures. These findings suggest that the DNA density of DNA nanostructures affects the cellular uptake, especially the rate of endocytosis. The cellular uptakes of Rec50, Rec0, and Rec50/50 were comparable to one another, although the average distance between DNA helices was slightly different. These results may suggest that there could be an upper limit in the cellular uptake efficacy when the DNA density exceeds a certain value. In general, larger DNA nanostructures are more efficiently taken up by smaller ones. The results of the present study, however, show that the DNA density is another important parameter than the size, because the smaller ones, i.e., Rec50/50 and Rec0, were most efficiently taken up by the cells.
There are several reports about the receptors involved in the cellular uptake of DNA and DNA nanostructures. In these studies, scavenger receptors,18 membrane-anchored protein 1,19 receptors for advanced glycation end product,20 membrane-associated nucleic acid-binding protein (MNAB),21 mannose receptor C type 122 and lymphocyte antigen 75 (DEC-205)23 are reported to be important for DNA uptake. We demonstrated that the transfection of human macrophage scavenger receptor 1 to HEK-Blue HTLR9 cells greatly increased the uptake of both DNA and DNA nanostructures.24 It was also demonstrated that the uptake of complicated DNA nanostructures was more efficient than that of simple ones. Therefore, it is expected that, by using the evaluation method utilized in these studies, the relationship between the interaction of DNA nanostructures with receptors and their structural properties such as DNA density can be elucidated.
Conclusion
In conclusion, the present study suggests that the cellular uptake of DNA nanostructures with high DNA density tends to be more efficient than that of DNA nanostructures with low DNA density. This result will provide useful information for the development of DNA nanostructure-based delivery systems for nucleic acid medicines to immune cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by JST CREST Grant Number JPMJCR1521, Japan, and Grants-in-Aid for Scientific Research (B) (23390010 and 26293008) from the Japan Society for the Promotion of Science.References
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- E. Winfree, F. Liu, L. A. Wenzler and N. C. Seeman, Nature, 1998, 394, 539–544 CrossRef CAS PubMed.
- P. W. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- R. Veneziano, S. Ratanalert, K. Zhang, F. Zhang, H. Yan, W. Chiu and M. Bathe, Science, 2016, 352, 1534 CrossRef CAS PubMed.
- D. Han, S. Pal, J. Nangreave, Z. Deng, Y. Liu and H. Yan, Science, 2011, 332, 342–346 CrossRef CAS PubMed.
- H. Dietz, S. M. Douglas and W. M. Shih, Science, 2009, 325, 725–730 CrossRef CAS PubMed.
- T. Masubuchi, M. Endo, R. Iizuka, A. Iguchi, D. H. Yoon, T. Sekiguchi, H. Qi, R. Iinuma, Y. Miyazono, S. Shoji, T. Funatsu, H. Sugiyama, Y. Harada, T. Ueda and H. Tadakuma, Nat. Nanotechnol., 2018, 13, 933–940 CrossRef CAS PubMed.
- E. M. Willner, Y. Kamada, Y. Suzuki, T. Emura, K. Hidaka, H. Dietz, H. Sugiyama and M. Endo, Angew. Chem., Int. Ed., 2017, 56, 15324–15328 CrossRef CAS PubMed.
- V. J. Schuller, S. Heidegger, N. Sandholzer, P. C. Nickels, N. A. Suhartha, S. Endres, C. Bourquin and T. Liedl, ACS Nano, 2011, 5, 9696–9702 CrossRef PubMed.
- Q. Jiang, C. Song, J. Nangreave, X. Liu, L. Lin, D. Qiu, Z. G. Wang, G. Zou, X. Liang, H. Yan and B. Ding, J. Am. Chem. Soc., 2012, 134, 13396–13403 CrossRef CAS PubMed.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS PubMed.
- K. Mohri, M. Nishikawa, N. Takahashi, T. Shiomi, N. Matsuoka, K. Ogawa, M. Endo, K. Hidaka, H. Sugiyama, Y. Takahashi and Y. Takakura, ACS Nano, 2012, 6, 5931–5940 CrossRef CAS PubMed.
- K. Mohri, E. Kusuki, S. Ohtsuki, N. Takahashi, M. Endo, K. Hidaka, H. Sugiyama, Y. Takahashi, Y. Takakura and M. Nishikawa, Biomacromolecules, 2015, 16, 1095–1101 CrossRef CAS PubMed.
- S. Ohtsuki, N. Matsuzaki, K. Mohri, M. Endo, T. Emura, K. Hidaka, H. Sugiyama, Y. Takahashi, K. Ishiyama, N. Kadowaki, Y. Takakura and M. Nishikawa, Nucleic Acid Ther., 2015, 25, 245–253 CrossRef CAS PubMed.
- S. Ohtsuki, Y. Shiba, T. Maezawa, K. Hidaka, H. Sugiyama, M. Endo, Y. Takahashi, Y. Takakura and M. Nishikawa, Nanoscale, 2019, 11, 23416–23422 RSC.
- K. Mohri, K. Nagata, S. Ohtsuki, S. Toyama, M. Nonomura, Y. Takahashi, Y. Takakura, M. Nishikawa and S. Sakuma, Pharm. Res., 2017, 34, 2362–2370 CrossRef CAS PubMed.
- R. A. Petros and J. M. DeSimone, Nat. Rev. Drug Discovery, 2010, 9, 615–627 CrossRef CAS PubMed.
- Y. Kimura, K. Sonehara, E. Kuramoto, T. Makino, S. Yamamoto, T. Yamamoto, T. Kataoka and T. Tokunaga, J. Biochem., 1994, 116, 991–994 CrossRef CAS PubMed.
- L. Benimetskaya, J. D. Loike, Z. Khaled, G. Loike, S. C. Silverstein, L. Cao, J. E. L. Khoury, T. Q. Cai and C. A. Stein, Nat. Med., 1997, 3, 414–420 CrossRef CAS PubMed.
- C. M. Sirois, T. Jin, A. L. Miller, D. Bertheloot, H. Nakamura, G. L. Horvath, A. Mian, J. Jiang, J. Schrum, L. Bossaller, K. Pelka, N. Garbi, Y. Brewah, J. Tian, C. Chang, P. S. Chowdhury, G. P. Sims, R. Kolbeck, A. J. Coyle, A. A. Humbles, T. S. Xiao and E. Latz, J. Exp. Med., 2013, 210, 2447–2463 CrossRef CAS PubMed.
- D. C. Siess, C. T. Vedder, L. S. Merkens, T. Tanaka, A. C. Freed, S. L. McCoy, M. C. Heinrich, M. E. Deffebach, R. M. Bennett and S. H. Hefeneider, J. Biol. Chem., 2000, 275, 33655–33662 CrossRef CAS PubMed.
- A. P. Moseman, E. A. Moseman, S. Schworer, I. Smirnova, T. Volkova, U. von Andrian and A. Poltorak, J. Immunol., 2013, 191, 5615–5624 CrossRef CAS PubMed.
- M. H. Lahoud, F. Ahmet, J. G. Zhang, S. Meuter, A. N. Policheni, S. Kitsoulis, C. N. Lee, M. O'Keeffe, L. C. Sullivan, A. G. Brooks, R. Berry, J. Rossjohn, J. D. Mintern, J. Vega-Ramos, J. A. Villadangos, N. A. Nicola, M. C. Nussenzweig, K. J. Stacey, K. Shortman, W. R. Heath and I. Caminschi, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16270–16275 CrossRef CAS PubMed.
- S. Ohtsuki, Y. Takahashi, T. Inoue, Y. Takakura and M. Nishikawa, Sci. Rep., 2017, 7, 13661 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr02361b |
| This journal is © The Royal Society of Chemistry 2020 |