Open Access Article
Open Access Article3D multiphoton lithography using biocompatible polymers with specific mechanical properties†
Boris
Buchroithner
a,
Delara
Hartmann
d,
Sandra
Mayr
a,
Yoo Jin
Oh
c,
Dmitry
Sivun
 b,
Andreas
Karner
b,
Andreas
Karner
 a,
Bianca
Buchegger
b,
Thomas
Griesser
d,
Peter
Hinterdorfer
c,
Thomas A.
Klar
b and
Jaroslaw
Jacak
a,
Bianca
Buchegger
b,
Thomas
Griesser
d,
Peter
Hinterdorfer
c,
Thomas A.
Klar
b and
Jaroslaw
Jacak
 *a
*a
aUniversity of Applied Sciences Upper Austria, School of Medical Engineering and Applied Social Sciences, Garnison Str. 21, 4020 Linz, Austria. E-mail: jaroslaw.jacak@fh-linz.at
bInstitute of Applied Physics and Linz Institute of Technology LIT, Johannes Kepler University Linz, Altenberger Str. 69, 4040 Linz, Austria
cInstitute of Biophysics, Johannes Kepler University Linz, Gruberstr. 40, 4020 Linz, Austria
dChair of Chemistry of Polymeric Materials, Montanuniversitaet Leoben, Otto–Glöckel Str. 2, 8700 Leoben, Austria
First published on 9th May 2020
Abstract
The fabrication of two- and three-dimensional scaffolds mimicking the extracellular matrix and providing cell stimulation is of high importance in biology and material science. We show two new, biocompatible polymers, which can be 3D structured via multiphoton lithography, and determine their mechanical properties. Atomic force microscopy analysis of structures with sub-micron feature sizes reveals Young's modulus values in the 100 MPa range. Assessment of biocompatibility of the new resins was done by cultivating human umbilical vein endothelial cells on two-dimensionally structured substrates for four days. The cell density and presence of apoptotic cells has been quantified.
Introduction
Mimicking the mechanical properties of an extracellular matrix is of paramount importance in tissue engineering. The bone formation process or neural tissue reconstruction is especially sensitive to the mechanical, topographical and chemical properties of the substrates.1,2 The matrix closely interacts with the embedded cells and has a direct impact on cell fate.3,4 For the best possible mimicking of a tissue scaffold with controllable topography,3 tuning of the mechanical and chemical properties is required.4,5 All these properties help to guide cell growth and tissue formation, improving cell adhesion and enhancing cell survival.3–8 Consequently, great efforts have been made to study the impact of cell-matrix interactions at a cellular level.5 For a better understanding of these interactions, however, there is still a lack of biocompatible materials with determined mechanical properties, which can be structured in 3D with feature sizes ranging from micro to nanoscale. Multiphoton lithography (MPL) and related methods9 are the only techniques capable of 3D structuring with a resolution in the sub-micrometer regime. MPL outperforms technologies like stereolithography and electrospinning typically used for tissue engineering in regard to the 3D resolution.5,6,10,11 3D structuring of biocompatible polymers helps to better control the scaffolds elastic properties and to adapt the structure for the tissue environment.In the past, MPL fabricated 3D nanostructures were used for tissue engineering in vitro1,2 and in vivo,12 production of metamaterials with flexible mechanical properties,13 fabrication of responsive and programmable materials,14 quantification of forces applied by living cells onto the environment,11,15 or for structuring soft gelatine-based scaffolds and biocompatible 3D hydrogel woodpiles.16–20 Most of these applications require a precise knowledge about the mechanical properties of the material. Mechanical properties (such as Young's modulus (E), stiffness, etc.) of the specific polymers can be investigated via atomic force microscopy (AFM). For Young's modulus analysis, the cantilever tip is used as a nanoindenter, and the deformation of the specimen at a given force is recorded.21,22
In this contribution, we present two biocompatible resins, which can be structured via 3D MPL, and characterize their mechanical properties. The overall performance of these new resins was compared to two resins, which are commonly used in MPL. We show that the new photoresists enable 3D structuring with high aspect ratios and feature sizes in the sub-micrometer regime. For biocompatibility testing, endothelial cells were cultured on polymeric structures for several days. On both resins, closely connected human umbilical vein endothelial cells (HUVECs) with typical endothelial morphology formed a confluent monolayer that covered the glass surface and polymer structures. The increased cell density and low amount of apoptotic cells indicate high biocompatibility of new photoresists.
Materials and methods
Lithography setup
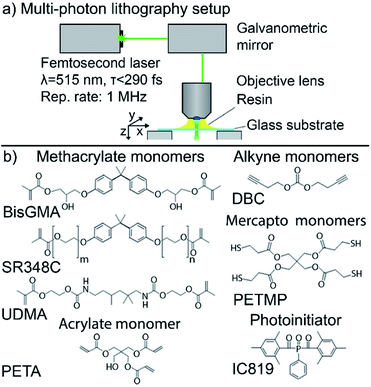
A scheme of the Workshop of Photonics (WOP, Lithuania) MPL setup is given in Fig. 1a. The two-photon photoinitiators were excited with 515 nm ultra-short laser pulses (CARBIDE, 1 MHz repetition rate, 290 fs pulse duration, Light Conversion, Lithuania). All stated intensities are peak intensities calculated from power measured in front of the objective lens (Zeiss, 63× magnification, NA 1.25), transmission of the objective, and diffraction-limited spot area. The writing was performed in a dip-in configuration.23 A 3-axis stage (AEROTECH Nanopositioner, USA) was used for sample motion. Optionally, faster lateral writing is performed by a galvanometric mirror scanner (AEROTECH, USA).Resin formulation
The pentaerythritol triacrylate (PETA) and the hybrid OrmoComp monomers (OrmoComp® without photoinitiator) were purchased from Sigma-Aldrich (Germany) and micro resist technology (Germany), respectively. The photoresist BisSR consists of 29% of Bis-GMA (Esschem Europe, England) and 71% ethoxylated bisphenol A dimethacrylate (SR348C) (SARTOMER, France), both used as received and stirred with a magnetic stirrer until homogeneously mixed. The resin M10 consists of 18.7% pentaerythritol tetra(3-mercaptopropionate) (PETMP), 6.4% di(but-1-yne-4-yl)carbonate (DBC) and 74.7% 1,6-bis-[2-methacryloyloxyethoxycarbonylamino]-2,4,4-trimethylhexane (UDMA) and 2‰ of propylgallat as stabilizer. DBC was synthesized as described elsewhere.24 The schemes of the chemical structures are shown in Fig. 1b. Bis(2,4,-trimethylbenzoyl)-phenylphosphineoxide (IC819) (Sigma-Aldrich, Germany) was used as photoinitiator. 2 wt% of IC819 were used for M10 and 1 wt% for BisSR, OrmoComp®, and PETA, respectively. The photo-initiator was gently mixed into the resin for 10 hours using a magnetic stirrer. All resin formulations were stored at 4 °C.3D structuring
For stability testing, 3D grids with a height of 90 μm and a side length of 30 μm were manufactured. Thereby the thickness of the horizontal bars was set to 1.5 μm (by multiple scans with overlap) with a horizontal periodicity of 8 μm. To reduce optical aberrations, the structures were written in the dip-in configuration.23 The intensity was set to maximize writing speed, but still lower than the damage threshold of the resins. For the structuring of PETA, OrmoComp®, and M10, an intensity (peak intensity at the focal plane) of 0.43 TW cm−2 was used, while for BisSR an intensity of 0.48 TW cm−2 was applied. In the case of M10, each voxel is exposed three times (triple voxel exposure). All structures were written with a speed of 15 mm s−1. For development, OrmoComp® and M10 structures were rinsed with acetone. PETA and BisSR structures were rinsed with ethanol.The samples for AFM nanoindentation experiments were prepared as follows: OrmoComp® support-bars of 3 μm height were written in advance for all four tested resin formulations in the dip-in configuration. The spacing between the support bars is 75 μm. Thereafter, 35 OrmoComp® lines with a distance of 2 μm to the glass surface and 5 μm to each other were written in between two support-bars. The starting intensity for the first line was 0.074 TW cm−2 and the incremental intensity step for each following line was 0.074 TW cm−2 (writing speed 5 mm s−1). For the PETA, BisSR, and M10, the same procedure and parameters were applied. A lower writing speed of 1 mm s−1 was used for M10, to achieve similar feature sizes. The PETA and BisSR resin was developed with ethanol and the OrmoComp® and M10 with acetone.
Young's modulus characterization
The elasticity measurements were performed with an atomic force microscope (JPK Nano Wizard, Germany) using the QI™ mode. The AFM is mounted on top of an Olympus IX71 inverted optical microscope. The preparation of the sample is shown in detail in Fig. 2. Cantilever and sample were immersed in deionized water and thermally equilibrated for 1 h. The Young's modulus was measured with a qp-BioAC-CI cantilever (NanoAndMore, Germany; tip radius = 30 nm; spring constant glass = 0.3 N m−1). Before each measurement, the exact cantilever spring constant and the sensitivity were determined on the substrate adjacent to the polymer lines using the contact based calibration method (JPK, Germany). Force mapping was done using the JPK QI™ mode (400 points per polymer line profile, performed in the central region of the polymer line). The outline of the polymer line is excluded for a constant tip-sample contact area (see ESI, Fig. S1†). The indentation force was constant at a value of 2 nN to ensure nonlinear deformation of the polymer sample. The interaction between a rigid sphere (as an approximation for AFM tip) and an elastic surface (polymer lines) is described best by the Hertz model, thus to determine Young's modulus from the force distance curves.25E was determined by averaging over 400 data points in three technical replicas.Cell culture
Primary HUVEC cells were a kind gift from Prof. J. Breuss (Medical University of Vienna, Austria). The cells were cultured in M199 (Carl Roth, Karlsruhe, Germany) supplemented with 10% BCS (Sigma-Aldrich, Austria), LVES (Life Technologies, USA), GlutaMAX (100x, Life Technologies, USA) and penicillin–streptomycin (100 IU ml−1 penicillin 100 μg ml−1 streptomycin) at 37 °C and 5% CO2.Biocompatibility testing
The endothelial cells (primary HUVEC, passage 6) were seeded onto glass slides with 2D polymer structures (2 grids with a size of 100 × 100 × 15 μm, grid constants 25 μm and 2 grids with a size of 90 × 90 × 15 μm, grid constants 30 μm), coated with 0.2% gelatine in phosphate-buffered saline (Carl Roth, Austria) at a density of 105 cells per cm2. Medium exchange was performed on a daily basis. Four days post seeding, the cells were washed with pre-warmed PBS and fixed with 4% paraformaldehyde (PFA, Sigma-Aldrich) for 15 minutes at room temperature. Subsequently, cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 30 minutes at room-temperature. Microscopic images were taken using a Zeiss Axiovert 200 (ZEN 2 blue edition) using a 10x A-PLAN (NA 0.25) and 20x LD PLAN-Neofluar (NA 0.4) objective lens. A bare glass surface in the surrounding of the grids was used as negative control. This allows taking the local cell density for each sample into account. For quantification, a cell density ratio was used, which is defined as ratio of cell density on the polymer grid (N = 4) to cell density on a negative control. Additionally, a Caspase-Glo® 3/7 assay (Promega, WI, USA) is used for the detection of apoptosis or secondary necrosis.26 By adding the kit reagent (containing DEVD substrate and luciferase) to the supernatant, the DEVD substrate is cleaved by caspases releasing the substrate aminoluciferin. Upon cleavage by luciferase phosphorescence light is detected by a photometer.Results
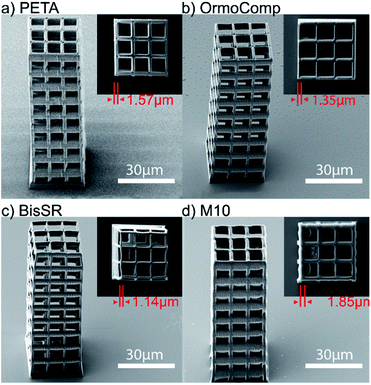
Multiphoton structuring was performed using a 515 nm femtosecond laser (see details of setup in the Material and methods section). The first resin, BisSR, consists of bisphenol-A-glycidyl dimethacrylate (Bis-GMA) and ethoxylated bisphenol A dimethacrylate (SR348C). The second resin, named M10, is a mixture of monomers carrying methacrylate-, mercapto- and alkyne reactive groups. Thereby, the added mercapto- and alkyne monomers lead to delayed gelation inducing both a higher monomer conversion and a lower shrinkage stress.27 The chemical structures of the monomers are shown in Fig. 1b. In order to be able to create large scale structures that can withstand strong mechanical forces (e.g. re-immersion into water), a high (about 1 wt%), but tolerable photoinitiator concentration was chosen.28 These novel resins were compared to two established resin formulations, namely OrmoComp® and pentaerythritol triacrylate (PETA), both containing 1 wt% IC819. To compare the mechanical stability of the polymers, freestanding scaffolds with a height of 90 μm and a side length of 30 μm were fabricated (Fig. 3a–d).The thickness of the horizontal bars was set to 1.5 μm and the distance between each bar was set to 8 μm. For the PETA and OrmoComp® structuring, an intensity of Iex = 0.43 TW cm−2 was used, resulting in a lateral feature size of lf = 1.57 μm and lf = 1.35 μm, respectively. For the BisSR structure, Iex = 0.48 TW cm−2 results in lf = 1.14 μm and for the M10 structure Iex = 0.43 TW cm−2 yields lf = 1.85 μm. For the structuring of M10, each voxel was triple exposed to enhance the mechanical stability, which most probably caused the deviation of the bar thickness (higher value) compared to the designed one. On the other hand, the difference in the feature size observed for BisSR (lower value) relative to the set value is most likely introduced by shrinkage during the polymer development processes. The results show that these formulations allow creating large-scale 3D structures similar to those written with the established OrmoComp® and PETA formulations.
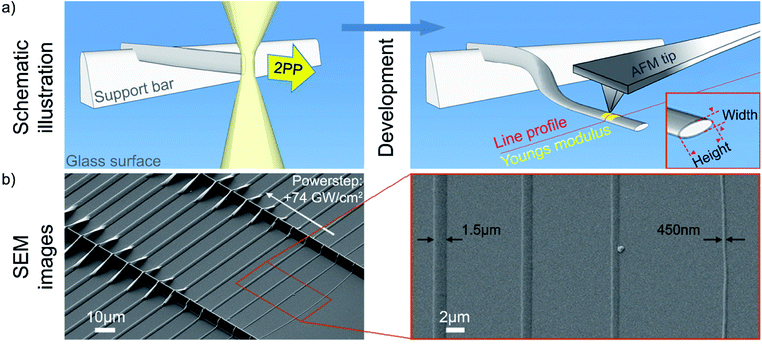
Kunik et al. have shown that the writing parameters strongly depend on the surface proximity.29 In particular, the polymerization at a surface starts by forming polymer islands directly at the interface. Such a heterogeneous 2D polymer growth has an impact on the mechanical properties of structures. In order to exclude such impact, 3D structured hanging lines with various feature sizes were produced (Fig. 2a). In order to characterize the mechanical properties of structured polymer lines at a nanoscopic scale, an atomic force microscopy was used. However, determining the Young's modulus of the 3D hanging nanostructures is a challenging task, which would require a complicated force measurement protocol to exclude the effects of bending and deforming of hanging nanostructures under the pressure of the AFM cantilever. Therefore, we produced micro- and nano-sized lines with length of 75 μm at 2 μm above the glass surface (hanging 3D structure). Due to the large length and small distance to a glass substrate, the written lines were attaching to the glass surface upon development. In addition to that, lines were tipped (rotated) by 90 degrees (see Fig. 2a). Consequently, the side surface of the polymer line was lying flat on the glass surface (Fig. 2a and b). Being supported by a substrate, the standard nanoindentation protocols could be applied for the Young's modulus measurements. Overall, such production method ensures the homogeneity of written nanostructures, and simplifies the determination of their mechanical properties. The line dimensions were tuned by adjusting the writing intensity, in increments of 0.074 TW cm−2 from line to line. In detail, 35 polymer lines, 2 μm above the surface and spaced 5 μm apart of each other were written. Fig. 2b shows scanning electron microscopy (SEM) images of such lines.
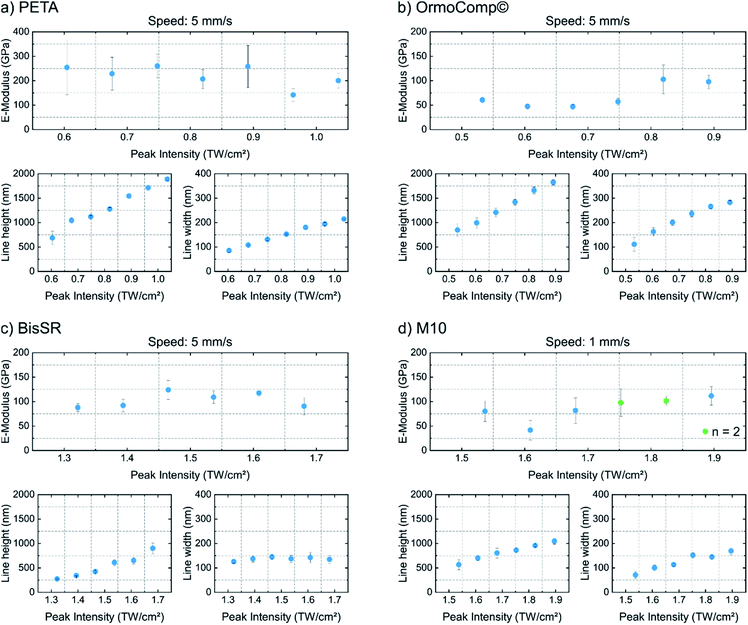
For probing the Young's moduli and line profiles, qp-BioAC-CI cantilevers were used. The exact cantilever spring constant was determined according to the Material and methods section. The Hertz model was used to retrieve the Young's modulus from the measured force distance curves, obtained from three technical replicas (see Fig. 4).
Fig. 4 shows the results of the Young's modulus analysis of the 3D written structures, each consisting of one of the four polymers. The PETA resin formed the stiffest lines with an average E-modulus of 221 ± 40 MPa and the developed lines did not significantly depend on the writing intensity. The heterogeneities of the measured E-values could be induced by a heterogeneous softer gel phase around the core of the polymer,27 introduced during the development process. The smallest developed PETA-line measured w = 86 nm in width and h = 684 nm in height. In contrast, OrmoComp® structures are softer and show an E-modulus increase in the range of 47 ± 5 MPa to 102 ± 29 MPa with increase of feature size (Fig. 4b). For the smallest lines w = 110 nm and h = 846 nm is measured. The tendency to higher E-modulus with increasing structure size is broken by the smallest line (first point in Fig. 4b), which shows an E-modulus 15 MPa higher than the following two (larger) lines. This is probably an artefact, introduced by too high indentation depth compared to the width of the line. In such case the measured E-modulus is shifting30 towards the E-modulus of the substrate (Eglass = 50 GPa).22 Similar to PETA, the BisSR resin does not show a writing intensity dependent increase of the E-modulus (Fig. 4c, average E-modulus of 103 ± 14 MPa). The smallest lines (Fig. 4c), have w = 126 nm and h = 272 nm and E = 88 ± 9 MPa. The Young's modulus of M10 shows an intensity dependent increase from 41 ± 21 MPa to 120 ± 20 MPa. As already observed in OrmoComp®, the Young's modulus of the smallest polymer line is influenced by the stiffness of the glass (Fig. 4d). Here it is worth to mention, that the observed feature sizes yield a high aspect ratio, which cannot be attributed to the typical point spread function of a laser beam. Aspect ratios are found to be 8.7 (on average) for PETA lines, 6.4 for OrmoComp lines, 6.2 for M10 lines, and from 2 to 6.6 for BisSR lines. Similar results were also shown by other studies31,32 and were attributed to self-focusing, self-trapping, or dispersion effects, leading to polymerization beyond the resolution limit.33
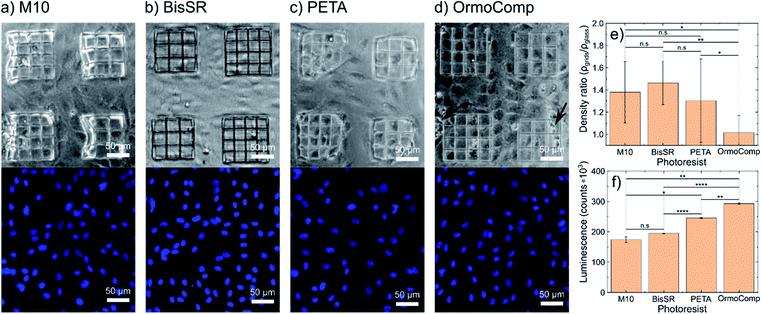
To assess the biocompatibility of the photoresists, human umbilical vein endothelial cells (105 HUVEC per cm2) were seeded on 2D polymer gratings. After four days of cultivation, phase-contrast images were taken for cell morphology comparison and cells were fixed and stained with 4′,6-diamidino-2-phenylindole (DAPI) and CD31-FITC for visualization of the cell nuclei distribution (Fig. 5) and junctions (Fig. S3†) of the cells on the grids, respectively. Fig. 5 shows phase-contrast and fluorescence microscopy images of HUVECs on 2D lithography structures written with M10, BisSR, PETA, and OrmoComp® on a glass surface (see Fig. 5a–d). A plain glass substrate near to the polymeric grids was used as a negative control. For each photoresist, four scaffolds with grid constants of 25 and 30 μm were structured. Biocompatibility was assessed by judgment of the cell density (indicating cell growth/cell division), presence of apoptotic cells, and uniform distribution of cell nuclei on the polymeric grids compared to the adjacent glass surface. Cell density ratios for all four tested polymers are presented in Fig. 5e. As can be seen, the OrmoComp® grid exhibits almost the same density of cells as a bare glass substrate. Grids made of PETA show on average 30% higher density compared to the negative control. The biocompatible resins M10 and BisSR show 38% and 46% increase of the cell density, respectively. The significance of the cell density ratio was tested by nonparametric Kruskal–Wallis multiple comparisons test. Both M10 and BisSR grids show a significant (p < 0.05) increase in the cell density ration compared to the grids made from OrmoComp. Additionally, the biocompatibility of M10 and BisSR was confirmed by an in vitro assay (Caspase-Glo® 3/7, Promega, WI, USA). Extracellular active effector caspases (DEVD-aminofluoromethylcoumarine cleavage activity) are detected by luminescence increase. The luminescence is proportional to the amount of the extracellular caspase-3 and caspase-7, correlating in turn to the amount of apoptotic/secondary necrotic cells. The highest biocompatibility (defined here as low amount of released caspase enzymes) is observed for the polymers M10 and BisSR (174![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 065 ± 9062 and 194
065 ± 9062 and 194![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 494 ± 1154 counts) (Fig. 5f). No significant difference between these two samples were observed (2-way ANOVA, Tukey's multiple comparisons test). The cells cultivated on PETA grids show a significant higher amount of extracellular caspases (245
494 ± 1154 counts) (Fig. 5f). No significant difference between these two samples were observed (2-way ANOVA, Tukey's multiple comparisons test). The cells cultivated on PETA grids show a significant higher amount of extracellular caspases (245![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 403 ± 1353 counts; p < 0.0001 compared to BisSR). Cells grown on OrmoComp exhibit the highest number of extracellular caspases (293
403 ± 1353 counts; p < 0.0001 compared to BisSR). Cells grown on OrmoComp exhibit the highest number of extracellular caspases (293![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 016 ± 2307 counts; p < 0.01 compared to PETA). Values are presented as mean ± S.D. Results are derived from a single experiment in technical triplicates. In general, the formation of a confluent monolayer of cells on BisSR and M10 photoresists indicates a good biocompatibility. The uniform distribution of cell nuclei on the glass surface and the M10 grids indicates the material's non-toxicity. For BisSR, an enhanced affinity of the cells to the polymer lines was observed (Fig. 5b). Our biocompatibility results are encouraging and should be validated on wider level of cell types and conditions. However, such a task is beyond the scope of this manuscript but hopefully will trigger further research in this direction.
016 ± 2307 counts; p < 0.01 compared to PETA). Values are presented as mean ± S.D. Results are derived from a single experiment in technical triplicates. In general, the formation of a confluent monolayer of cells on BisSR and M10 photoresists indicates a good biocompatibility. The uniform distribution of cell nuclei on the glass surface and the M10 grids indicates the material's non-toxicity. For BisSR, an enhanced affinity of the cells to the polymer lines was observed (Fig. 5b). Our biocompatibility results are encouraging and should be validated on wider level of cell types and conditions. However, such a task is beyond the scope of this manuscript but hopefully will trigger further research in this direction.
Conclusion
In this contribution, we introduced two new biocompatible resin formulations BisSR and M10. We characterized their 3D MPL writing capability and mechanical properties. The Young's modulus of single polymeric lines was determined as a function of illumination intensity. The measured Young's modulus of BisSR lines was around 80 MPa and independent from writing intensity. In contrast, an increase in the writing intensity for M10 lines leads to the rise of Young's modulus from 40 to 120 MPa. The M10 resin was designed to provide reduced shrinkage stress and high monomer conversion.27 In comparison, 3D structures made of the BisSR experience around 9% of post-polymerization shrinkage. Furthermore, we were able to structure the polymers in a sub-micrometer range. The results indicate that, M10 lines with even sub-100 nm thickness withstand development. Analysis of human endothelial cells on structured polymeric grids indicates a better biocompatibility of the two new photoresists BisSR and M10 compared to common MPL photoresists. The biocompatibility of these materials has been exclusively shown for in vitro cell cultures. However, we believe that our new materials would be suitable for tissue engineering in vivo12 and here determined mechanical parameters of new photoresists would help in the implementation of mechanical metamaterials13 and reversibly deformable microstructures.14Author contributions
J. J. conceived the project and carried out most of the writing. The cooperation regarding the M10 formulation was initiated by T. G. and T. K. Bo. B. performed the main experimental work and analyzed the data. Y. O., D. S., Bi. B., helped with the experimental work and data analysis P. H., T. K. helped with the experimental design. A. K, S. M. helped with the experimental work. D. H., T. G. provided the M10 polymer. All authors contributed to writing and editing the manuscript.Funding sources
This work was funded by the Interreg project ATCZ14 and the Austria Science Fund (FWF) project P 31827-B21, the European Fund for Regional Development (EFRE, IWB2020) and the Federal State of Upper Austria.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Heidi Piglmayer-Brezina, Bernhard Fragner and Alfred Nimmervoll for technical support.References
- A. R. Amini, C. T. Laurencin and S. P. Nukavarapu, Crit. Rev. Biomed. Eng., 2012, 40, 363–408 CrossRef PubMed.
- L. Binan, C. Tendey, G. De Crescenzo, R. El Ayoubi, A. Ajji and M. Jolicoeur, Biomaterials, 2014, 35, 664–674 CrossRef CAS PubMed.
- H. N. Kim, A. Jiao, N. S. Hwang, M. S. Kim, D. H. Kang, D.-H. Kim and K.-Y. Suh, Adv. Drug Delivery Rev., 2013, 65, 536–558 CrossRef CAS PubMed.
- E. Tamjid, A. Simchi, J. W. C. Dunlop, P. Fratzl, R. Bagheri and M. Vossoughi, J. Biomed. Mater. Res., Part A, 2013, 101, 2796–2807 CrossRef PubMed.
- I. Armentano, M. Dottori, E. Fortunati, S. Mattioli and J. M. Kenny, Polym. Degrad. Stab., 2010, 95, 2126–2146 CrossRef CAS.
- M. Jafari, Z. Paknejad, M. R. Rad, S. R. Motamedian, M. J. Eghbal, N. Nadjmi and A. Khojasteh, J. Biomed. Mater. Res., Part B, 2017, 105, 431–459 CrossRef CAS PubMed.
- B. S. Kim and D. J. Mooney, Trends Biotechnol., 1998, 16, 224–230 CrossRef CAS PubMed.
- B. Guillotin, A. Souquet, S. Catros, M. Duocastella, B. Pippenger, S. Bellance, R. Bareille, M. Rémy, L. Bordenave, J. Amédée and F. Guillemot, Biomaterials, 2010, 31, 7250–7256 CrossRef CAS PubMed.
- A. Ovsianikov, M. Malinauskas, S. Schlie, B. Chichkov, S. Gittard, R. Narayan, M. Löbler, K. Sternberg, K.-P. Schmitz and A. Haverich, Acta Biomater., 2011, 7, 967–974 CrossRef CAS PubMed.
- J. Stampfl, S. Baudis, C. Heller, R. Liska, A. Neumeister, R. Kling, A. Ostendorf and M. Spitzbart, J. Micromech. Microeng., 2008, 18, 125014 CrossRef.
- J. Heitz, C. Plamadeala, M. Wiesbauer, P. Freudenthaler, R. Wollhofen, J. Jacak, T. A. Klar, B. Magnus, D. Köstner, A. Weth, W. Baumgartner and R. Marksteiner, J. Biomed. Mater. Res., Part A, 2017, 105, 891–899 CrossRef CAS PubMed.
- J. Maciulaitis, S. Rekštytė, M. Bratchikov, R. Gudas, M. Malinauskas, A. Pockevicius, A. Usas, A. Rimkunas, V. Jankauskaite, V. Grigaliunas and R. Maciulaitis, Appl. Surf. Sci., 2019, 487, 692–702 CrossRef CAS.
- T. Bückmann, M. Thiel, M. Kadic, R. Schittny and M. Wegener, Nat. Commun., 2014, 5, 1–6 Search PubMed.
- S. Rekštyte, D. Paipulas, M. Malinauskas and V. Mizeikis, Nanotechnology, 2017, 28, 124001 CrossRef PubMed.
- F. Klein, T. Striebel, J. Fischer, Z. Jiang, C. M. Franz, G. von Freymann, M. Wegener and M. Bastmeyer, Adv. Mater., 2010, 22, 868–871 CrossRef CAS PubMed.
- L. Brigo, A. Urciuolo, S. Giulitti, G. Della Giustina, M. Tromayer, R. Liska, N. Elvassore and G. Brusatin, Acta Biomater., 2017, 55, 373–384 CrossRef CAS PubMed.
- X.-H. Qin, P. Gruber, M. Markovic, B. Plochberger, E. Klotzsch, J. Stampfl, A. Ovsianikov and R. Liska, Polym. Chem., 2014, 5, 6523–6533 RSC.
- G. D. Giustina, S. Giulitti, L. Brigo, M. Zanatta, M. Tromayer, R. Liska, N. Elvassore and G. Brusatin, Macromol. Rapid Commun., 2017, 38, 1600570 CrossRef PubMed.
- M. Gebinoga, J. Katzmann, U. Fernekorn, J. Hampl, F. Weise, M. Klett, A. Läffert, T. A. Klar and A. Schober, Eng. Life Sci., 2013, 13, 368–375 CrossRef CAS.
- A. Selimis, V. Mironov and M. Farsari, Microelectron. Eng., 2015, 132, 83–89 CrossRef CAS.
- S. Tomkoria, K. Masuda and J. Mao, Proc. Inst. Mech. Eng., Part H, 2007, 221, 467–473 CrossRef CAS PubMed.
- A. Vinckier and G. Semenza, FEBS Lett., 1998, 430, 12–16 CrossRef CAS.
- T. Bückmann, N. Stenger, M. Kadic, J. Kaschke, A. Frölich, T. Kennerknecht, C. Eberl, M. Thiel and M. Wegener, Adv. Mater., 2012, 24, 2710–2714 CrossRef PubMed.
- A. Oesterreicher, C. Gorsche, S. Ayalur-Karunakaran, A. Moser, M. Edler, G. Pinter, S. Schlögl, R. Liska and T. Griesser, Macromol. Rapid Commun., 2016, 37, 1701–1706 CrossRef CAS.
- J. Zemła, J. Danilkiewicz, B. Orzechowska, J. Pabijan, S. Seweryn and M. Lekka, Semin. Cell Dev. Biol., 2018, 73, 115–124 CrossRef.
- H. Hentze, X. Y. Lin, M. S. K. Choi and A. G. Porter, Cell Death Differ., 2003, 10, 956–968 CrossRef CAS PubMed.
- S. Ye, N. B. Cramer, I. R. Smith, K. R. Voigt and C. N. Bowman, Macromolecules, 2011, 44, 9084–9090 CrossRef CAS PubMed.
- A. Accardo, M.-C. Blatché, R. Courson, I. Loubinoux, C. Vieu and L. Malaquin, Biomedical Physics & Engineering Express, 2018, 4, 027009 Search PubMed.
- D. Kunik, S. J. Ludueña, S. Costantino and O. E. Martínez, J. Microsc., 2008, 229, 540–544 CrossRef CAS PubMed.
- J. Menčík, D. Munz, E. Quandt, E. R. Weppelmann and M. V. Swain, J. Mater. Res., 1997, 12, 2475–2484 CrossRef.
- W. Haske, V. W. Chen, J. M. Hales, W. Dong, S. Barlow, S. R. Marder and J. W. Perry, Opt. Express, 2007, 15, 3426–3436 CrossRef CAS PubMed.
- C. A. Leatherdale and R. J. DeVoe, in Nonlinear Optical Transmission and Multiphoton Processes in Organics, International Society for Optics and Photonics, 2003, vol. 5211, pp. 112–123 Search PubMed.
- A. S. Kewitsch and A. Yariv, Opt. Lett., 1996, 21, 24–26 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: AFM analysis of polymer lines and biocompatibility control measurements. See DOI: 10.1039/d0na00154f |
| This journal is © The Royal Society of Chemistry 2020 |