Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Upregulation of epithelial metallothioneins by metal-rich ultrafine particulate matter from an underground railway
Matthew
Loxham
 *abcd,
Jeongmin
Woo
a,
Akul
Singhania†
a,
Natalie P.
Smithers
a,
Alison
Yeomans
e,
Graham
Packham
e,
Alina M.
Crainic
f,
Richard B.
Cook
f,
Flemming R.
Cassee
gh,
Christopher H.
Woelk‡
*abcd,
Jeongmin
Woo
a,
Akul
Singhania†
a,
Natalie P.
Smithers
a,
Alison
Yeomans
e,
Graham
Packham
e,
Alina M.
Crainic
f,
Richard B.
Cook
f,
Flemming R.
Cassee
gh,
Christopher H.
Woelk‡
 a and
Donna E.
Davies
abc
a and
Donna E.
Davies
abc
aSchool of Clinical and Experimental Sciences, University of Southampton Faculty of Medicine, University Hospital Southampton, Tremona Road, Southampton, UK SO16 6YD. E-mail: m.loxham@soton.ac.uk
bNIHR Southampton Biomedical Research Centre, University Hospital Southampton, Tremona Road, Southampton, UK SO16 6YD
cInstitute for Life Sciences, Highfield Campus, University of Southampton, Southampton, UK SO17 1BJ
dSouthampton Marine and Maritime Institute, University of Southampton, Boldrewood Innovation Campus, Southampton, UK SO16 7QF
eCancer Research UK Centre, Cancer Sciences, University of Southampton Faculty of Medicine, University Hospital Southampton, Southampton, UK SO16 6YD
fNational Centre for Advanced Tribology (nCATS), Mechanical Engineering, Faculty of Engineering and Physical Sciences, University of Southampton, Southampton, UK SO17 1BJ
gCentre for Sustainability, Environment, and Health, National Institute for Public Health and the Environment (RIVM), Bilthoven, The Netherlands
hInstitute for Risk Assessment Sciences (IRAS), Utrecht University, Utrecht, The Netherlands
First published on 16th April 2020
Abstract
Airborne particulate matter (PM) is a leading cause of mortality and morbidity. However, understanding of the range and mechanisms of effects of PM components is poor. PM generated in underground railways is rich in metals, especially iron. In the ultrafine (UFPM; <0.1 μm diameter) fraction, the combination of small size and metal enrichment poses an unknown health risk. This study aimed to analyse transcriptomic responses to underground UFPM in primary bronchial epithelial cells (PBECs), a key site of PM deposition. The oxidation state of iron in UFPM from an underground station was determined by X-ray absorption near edge structure (XANES) spectroscopy. Antioxidant response was assayed using a reporter cell line transfected with an antioxidant response element (ARE)-luciferase construct. Differentiated PBECs were exposed to UFPM for 6 h or 24 h for RNA-Seq and RT-qPCR analysis. XANES showed predominance of redox-active Fe3O4, with ROS generation confirmed by induction of ARE-luciferase expression. 6 h exposure of PBECs to UFPM identified 52 differentially expressed genes (DEGs), especially associated with epithelial maintenance, whereas 24 h exposure yielded 23 DEGs, particularly involved with redox homeostasis and metal binding. At both timepoints, there was upregulation of members of the metallothionein family, low molecular weight proteins with antioxidant activity whose main function is binding and homeostasis of zinc and copper ions, but not iron ions. This upregulation was partially inhibited by metal chelation or ROS scavenging. These data suggest differential regulation of responses to metal-rich UFPM depending on exposure period, and highlight novel pathways and markers of PM exposure, with the role of metallothioneins warranting further investigation.
Significance to metallomicsUnderground railways contain airborne particulate matter (PM) markedly more concentrated and richer in transition metals than urban PM, upon which most knowledge of the effects of PM is based. Here, we show that ultrafine PM (<0.1 μm diameter) from an underground railway station elicits time-dependent changes in the transcriptome of mucociliary primary bronchial epithelial cell cultures, with the initial response seemingly related to maintenance of the epithelial barrier, but later focused on the metal-rich nature of the PM and oxidative stress. Notably, we observe significant upregulation of multiple genes coding for cysteine-rich metal-binding metallothioneins, potentially indicative of an important role in cellular defences. |
Introduction
Exposure to airborne particulate matter (PM) is one of the leading causes of increased mortality and morbidity worldwide, with 9 million deaths per year worldwide attributed to exposure to fine particulate matter (PM2.5, <2.5 μm aerodynamic diameter), although this number has been growing as the exposure-response function for PM is better refined.1–3 There are established associations between PM exposure and development/exacerbation of respiratory and cardiovascular diseases,1,4 as well as emerging evidence for roles in the development of type 2 diabetes and worsening cognitive performance,5,6 along with reduced cognitive function.7 In order to reduce exposure to PM, national and international legislation sets limits on ambient PM concentrations. However, although these limits are defined on a mass concentration basis (i.e. μg m−3), it is unlikely that this is the sole best metric of understanding PM effect.8 As PM diameter decreases, it may pose a greater risk to health on account of a greater ability to enter the lower airways, cells, and circulation, as well as possessing an increased surface area/volume ratio, allowing a greater interaction with the surrounding milieu.A key mechanism by which PM exerts its effects is thought to be through the generation of reactive oxygen species (ROS), which react with biomolecules such as lipids, proteins, and DNA. In vitro studies have shown that redox-active metals in PM can generate ROS and deplete antioxidants, an important initial defence mechanism against oxidative attack, and also elicit inflammatory responses from airway cells.9,10 Furthermore, some studies have suggested a correlation between oxidative stress elicited by PM, and the magnitude of the cellular response,11 suggesting there is a need to understand better the effects of PM from different sources.12
Underground railway systems are used by tens of millions of people daily, and have been shown to have airborne PM mass concentrations several times higher than above ground.13 Underground PM is generally rich in metals (often >50% by mass), especially iron, derived principally from wear of wheels and rails, and electrical arcing. This also applies to the ultrafine fractions (UFPM/PM0.1; <0.1 μm aerodynamic diameter), whose levels are not legally regulated, yet have the potential to be toxic on the basis of both size and composition.14 We have previously shown that exposure of primary bronchial epithelial cells (PBECs) to iron-rich underground UFPM induces ROS generation and inflammatory cytokine release more potently than the fine or coarse fractions, and that this UFPM can penetrate the mucous barrier to enter cells, suggesting potential for longer term toxicity.15 However, there is also a lack of consistent evidence for clinically significant effects on persons exposed to underground railway PM, although this is based on a relatively small body of research.16
Cellular endpoints studied in toxicological analyses tend to focus on generalised markers of PM toxicity including inflammatory mediator release, DNA damage, and cell death. While this allows direct comparison between different PM samples, these responses may neglect other endpoints relevant to composition-related effects of PM. By evaluating the transcriptomic response of cells exposed to PM, RNA-Seq offers the potential to study responses comprehensively in an unbiased, hypothesis-free approach.17 Thus, the aim of this study was to characterise changes in the transcriptome of differentiated mucociliary cultures of PBECs following exposure to underground UFPM whose composition is unlike ambient PM, yet to which millions of people are regularly exposed, leading to potential health risks.18 This study highlights a number of previously unobserved gene perturbations in response to UFPM, including novel observations of widespread and significant upregulation of metallothionein (MT) gene expression.
Methods
PM collection and analysis
Quasi-UFPM (PM0.18; mean airborne concentration 44 μg m−3) was collected as a suspension in ultrapure water at a European mainline underground railway station, using a Versatile Aerosol Concentration Enrichment System (VACES) as described previously.14 PM was gamma irradiated (1250 Gy, 10 h) with a Gammacell 1000 cell irradiator (Atomic Energy of Canada Ltd, Ontario, Canada) prior to use.14PM chemical analysis
PM composition was analysed by inductively coupled plasma mass spectrometry and ion chromatography as described previously.14 PM lipopolysaccharide (LPS) concentration was determined using a Limulus amoebocyte lysate assay (Lonza, Slough, UK) as described previously.15X-ray absorption near edge structure (XANES) spectroscopy of underground railway UFPM
UFPM iron oxidation state was determined using XANES (Beamline I08, Diamond Light Source synchrotron, United Kingdom), with L2,3-edge spectra generated between 700 and 732 eV, at increments of 0.2 eV between 706–728 eV, and 0.5 eV outside this range. Mantis software (2nd Look Consulting, Hong Kong) was used for background correction and aXis2000 (http://unicorn.mcmaster.ca/aXis2000.html) for spectra assembly. For UFPM samples, individual particle spectra across two fields of view (total n = 13) were extracted using the lasso function. For standards of Fe, Fe2O3, and Fe3O4 nanopowders (Sigma Aldrich, Gillingham, UK), average spectra of particles in a single field of view were calculated using the Mantis cluster analysis function. Spectra were plotted using Origin (OriginLab Corporation, USA) with baseline correction and peak heights normalised to 1.Determination of antioxidant response element (ARE) activation in response to UFPM exposure
MCF7 breast cancer epithelial cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with foetal calf serum (10%), L-glutamine (2 mM) and penicillin and streptomycin (50 units per ml, 50 μg ml−1, respectively) as previously described.19 MCF7 cells were transiently transfected with a Nuclear Factor, Erythroid 2 Like 2 (Nrf2)-binding anti-oxidant response element (ARE) upstream of a luciferase reporter sequence,20 using TransFast transfection reagent (Promega, Southampton UK) according to the manufacturer's instructions, with mutated and non-transfected controls. Cells were then trypsinised, seeded in white 96-well plates at 2000 cells per well, and incubated for 4 h to adhere. Cells were exposed to 75 μl UFPM (1.3, 2.5, and 6.3 μg cm−2, equating to 5, 10, and 25 μg ml−1) in serum-free DMEM, or PM-free PBS-adjusted serum-free DMEM alone as a control, for 24 h. Response was assessed using BrightGlo reagent (Promega) according to the manufacturer's instructions.Challenge of primary bronchial epithelial cells with UFPM
PBECs from a total of 18 healthy volunteers (7M/11F, mean age 33 ± 17 year) were established from bronchial brushings obtained at bronchoscopy, with prior ethical approval (Southampton Local Research Ethics Committee Rec. No. 05/Q1702/165, code MRC0268) and written informed donor consent obtained. PBECs were cultured on 0.4 μm pore Transwell inserts at air–liquid interface (ALI), as described previously;21 and used when transepithelial electrical resistance was >330 Ω cm2. PBECs were made quiescent for 24 h prior to use, in bronchial epithelial basal medium (BEBM) containing 1% 100× insulin/transferrin/selenium solution (Sigma Aldrich) and 1 mg ml−1 bovine serum albumin (BSA),22 then exposed apically to 75 μl UFPM suspension at 25 μg ml−1 (5.6 μg cm−2), or 75 μl PBS-adjusted PM-free control, alone or supplemented with desferrioxamine (DFX; 200 μM) or N-acetylcysteine (NAC; 20 mM).15 Following exposure, cells were lysed with 200 μl QIAzol lysis reagent (Qiagen, Hilden, Germany).RNA-Seq
RNA was extracted from cell lysates using miRNeasy spin columns (Qiagen) with on-column DNase, according to manufacturer's instructions; RNA integrity was determined using an Agilent 2100 Bioanalyser (Agilent Technologies UK Ltd, Stockport, UK), with average RNA integrity number (RIN) of 9.9 ± 0.1. RNA-Seq paired end libraries were prepared using an Illumina TruSeq™ stranded mRNA Sample Preparation Kit (Illumina Inc., Cambridge, UK). Flow cells were used for paired end (2 × 50 bp) sequencing on an Illumina HiSeq 2000 with 50 million reads per sample in FASTQ format used for mapping and alignment. Raw paired-end sequencing data quality was assessed using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/), with low quality reads removed. Paired-end reads were aligned to human reference genome GRCh37/hg19 (hg19), using Tophat v2.0.923 with the parameters “-p 16 –library-type fr-firststrand –m 0” (allowing no mismatches). Mappings were converted to gene specific read count values using HTSeq-count v0.5.424 with the default parameters except “-s reverse”, yielding read count values for 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 368 RefSeq annotated genes. Further processing in edgeR v3.4.225 to exclude genes with <2 read counts per million in 50% of samples yielded 13
368 RefSeq annotated genes. Further processing in edgeR v3.4.225 to exclude genes with <2 read counts per million in 50% of samples yielded 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 234 genes which were normalised using the trimmed mean of M-values (TMM) method26 and transformed to log2-counts/million (Log2CPM) using edgeR voom.
234 genes which were normalised using the trimmed mean of M-values (TMM) method26 and transformed to log2-counts/million (Log2CPM) using edgeR voom.
Differentially expressed gene (DEG) paired analysis
After fitting for negative binomial distribution model on TMM normalized data, differentially expressed genes (DEGs) between UFPM-treated and control groups were identified by generalized linear model (GLM) likelihood ratio test using edgeR.25 Genes were identified as significantly differentially expressed with nominal p-value <0.01 and absolute fold change (FC) >1.5. Fold change was calculated in edgeR as the log2 of geometric mean of intensities; a positive and a negative fold change represents genes that were expressed to a greater or lesser extent, respectively, in UFPM-treated vs. control cultures.Gene ontology (GO) and pathway analysis
GO terms associated with biological processes significantly over-represented for DEGs (hypergeometric test with false discovery rate [FDR] corrected p-value (q-value) <0.05) were identified with the ToppGene web tool (https://toppgene.cchmc.org).27,28 Significant GO terms were visualized in semantic similarity-based scatterplots generated by REVIGO (Reduce + Visualise Gene Ontology), using default parameters.29 REVIGO scatterplots were generated with the allowed similarity metric set to medium and the semantic similarity measure set to SimRel. Nodes representing GO terms were depicted in two dimensional space derived by applying multidimensional scaling to a matrix of semantic similarities between GO terms.RT-qPCR analysis
For RT-qPCR analysis, cells from independent donors different to those used in the RNA-Seq analysis, and exposed according to the above protocol, were lysed with 200 μl Trizol lysis reagent (Life Technologies, Paisley, UK). RNA was extracted by standard phenol-chloroform protocol, and reverse transcribed using Precision Reverse Transcription kit (PrimerDesign, Southampton, UK). Probe-based qPCR was performed for cytochrome p450 1A1 (CYP1A1; sense 5′-CAAGGTGTTAAGTGAGAAGGTG-3′, antisense 5′-AGCAGGATAGCCAGGAAGAG-3′; CYP1A1; PrimerDesign) and metallothionein 1G (MT1G; TaqMan assay ID Hs02578922_gH; Applied Biosystems, Paisley, UK) according to the manufacturer's instructions. FC in gene expression relative to PM-free time-matched controls was calculated using the ΔΔCt method, with ubiquitin C (UBC) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as housekeeping genes through a probe-based duplex primer mix (PrimerDesign).Statistical analysis
Statistical analyses for transcriptomic data are provided above. For other datasets, normally distributed data (assessed by Shapiro-Wilk test) were analysed by paired t-test or Bonferroni-corrected one-way repeated measures ANOVA; non-normal data were analysed by Wilcoxon test or Dunn's-corrected Friedman test. Significance threshold was p < 0.05. Data were analysed and plotted using GraphPad Prism 7.03 (GraphPad Software Inc., San Diego, USA).Results
Composition of underground railway UFPM
As previously published, the predominant metal in underground railway UFPM was iron.14Fig. 1 shows the concentrations of all elements comprising at least 0.1 mg g−1 of the mass of the underground railway UFPM. Predominant sources of these are likely wear and mechanical activity relating to steel components (especially iron, manganese, zinc, chromium, titanium, and nickel), electrical components (copper, magnesium, and tin), brakes (barium and copper), and crustal sources which may also include ballast wear (magnesium, calcium, aluminium, sodium, potassium, and titanium). LPS concentration was 6.7 EU mg−1 PM.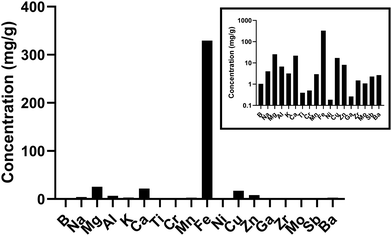 | ||
| Fig. 1 Concentrations of elements in underground railway UFPM. UFPM used in this study was analysed by ICP-MS. Only elements comprising at least 0.1 mg g−1 are shown. Main graph shows concentrations on arithmetic axis. Inset shows same data on logarithmic axis. Data previously published in different format.14 | ||
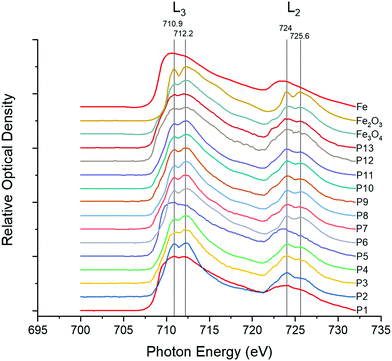
Oxidation state and redox activity of iron in underground railway UFPM
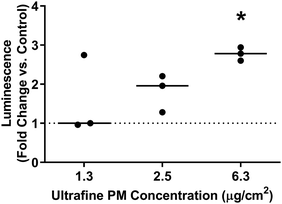
Given the predominance of iron in underground railway UFPM, and in order to know more about potential involvement in oxidative stress, XANES was used to determine iron oxidation state. In 11 of 13 individual underground ultrafine particles analysed, iron existed predominantly as an oxide (Fig. 2). Furthermore, the magnitude of the depression between the L3 peaks and between the L2 peaks in each pair indicated that particles were predominantly composed of Fe(II,III) oxide (i.e. Fe3O4), rather than Fe(III) oxide (Fe2O3). For 2 particles (P1 and P5), the spectrum indicated predominance of metallic iron. Given that Fe(II,III) is able to act as a source of electrons to generate oxidative stress, whereas Fe(III) is not, regulation of the cellular antioxidant response was studied through activation of an ARE-reporter construct. Activation of ARE by Nrf2 is associated with maintenance of cellular redox status through increased transcription of genes related to antioxidant defence, xenobiotic metabolism, and repair of oxidative damage, all associated with PM exposure.30 MCF7 cells were transfected with an ARE-luciferase reporter construct and then exposed to UFPM at 1.3, 2.5, or 6.3 μg cm−2 (5, 10, 25 μg ml−1). After 24 h exposure, there was a UFPM concentration-dependent increase in luminescence, reaching significance with 6.3 μg cm−2 UFPM (Fig. 3), suggesting that underground UFPM is able to induce ARE-dependent gene transcription.Global changes in gene expression after exposure to underground railway UFPM
In order to study effects of UFPM on the airway epithelium, RNA-Seq was performed on PBEC ALI cultures exposed to 5.6 μg cm−2 (25 μg ml−1) underground UFPM for 6 h or 24 h. The concentration and timepoints were chosen based on published work15 and the ARE-luciferase data. Significantly modulated genes were determined by t-test, with significance threshold set at p < 0.01. This analysis showed 52 genes whose expression was altered after 6 h exposure to underground UFPM (13 up-, 39 downregulated; Table 1), while after 24 h exposure, 23 genes were modulated by exposure to underground UFPM (17 up-, 6 downregulated). Notably, there was little overlap between the two timepoints, with only 2 genes (MT1G, HMOX1) upregulated at both timepoints, and 1 gene (TFRC) downregulated at both. Heatmaps of gene modulation at 6 h and 24 h (Fig. 4) indicated a broadly similar response of DEGs across all three donors.| Gene | FC | p-Value | Full name | Entrez gene code |
|---|---|---|---|---|
| Upregulated following 6 h UFPM exposure | ||||
| CYP1A1* | 3.77 | 1.5 × 10−6 | Cytochrome P450 1A1 | 1543 |
| MT1G | 3.37 | 4.1 × 10−3 | Metallothionein 1G | 4495 |
| CYP1B1 | 2.60 | 1.9 × 10−4 | Cytochrome P450 1B1 | 1545 |
| LOC344887 | 1.94 | 9.9 × 10−3 | NmrA-like redox sensor 2, pseudogene | 344887 |
| SLC16A6 | 1.77 | 2.9 × 10−4 | Solute carrier family 16, member 6 | 9120 |
| SCGB1A1 | 1.71 | 2.4 × 10−3 | Secretoglobin, CCSP | 7356 |
| SHISA2 | 1.69 | 8.2 × 10−3 | Transmembrane protein 46/Shisa family member 2 | 387914 |
| TMED7* | 1.68 | 3.2 × 10−6 | Transmembrane P24 trafficking protein 7 | 51014 |
| HMOX1 | 1.65 | 2.0 × 10−4 | Haem oxygenase 1 | 3162 |
| ADARB2 | 1.59 | 5.4 × 10−3 | Adenosine deaminase, RNA-specific B2 (inactive) | 105 |
| CENPQ | 1.54 | 2.5 × 10−3 | Centromere protein Q | 55166 |
| CRISP3 | 1.53 | 7.3 × 10−3 | Cysteine rich secretory protein 3 | 10321 |
| SLMO2 | 1.51 | 1.5 × 10−4 | Slowmo homologue 2/PRELI domain containing 3B | 51012 |
| Downregulated following 6 h UFPM exposure | ||||
| FN1 | 0.25 | 3.8 × 10−4 | Fibronectin 1 | 2335 |
| COL1A2 | 0.33 | 3.6 × 10−3 | Collagen type 1 alpha 2 chain | 1278 |
| CCL5 | 0.38 | 3.1 × 10−3 | Regulated upon activation normal T cell expressed and secreted (RANTES) | 6352 |
| MMP10 | 0.40 | 9.0 × 10−3 | Matrix metalloprotease 10 | 4319 |
| FLG | 0.43 | 1.9 × 10−3 | Filaggrin | 2312 |
| CALML3 | 0.48 | 1.2 × 10−3 | Calmodulin like 3 | 810 |
| MN1 | 0.49 | 4.2 × 10−3 | MN1 proto-oncogene, transcriptional regulator/meningioma 1 | 4330 |
| SERPINA3 | 0.50 | 9.5 × 10−3 | Serpin family A member 3/serpin peptidase inhibitor, clade A (alpha-1 antiprotease, antitrypsin), member 3 | 12 |
| PTPRD | 0.50 | 1.5 × 10−4 | Protein tyrosine phosphatase receptor type D | 5789 |
| SERPINE1 | 0.50 | 5.7 × 10−3 | Serpin family E member 1/plasminogen activator inhibitor-1 | 5054 |
| ECM1 | 0.51 | 5.3 × 10−3 | Extracellular matrix protein 1 | 1893 |
| UCN2 | 0.52 | 2.8 × 10−3 | Urocortin 2 | 90226 |
| HSPA6 | 0.54 | 1.7 × 10−3 | Heat shock protein family A (HSP70) member 6 | 3310 |
| FBN2 | 0.54 | 2.8 × 10−4 | Fibrillin 2 | 2201 |
| SERPINE2 | 0.54 | 4.0 × 10−3 | Serpin family E member 2/serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 | 5270 |
| LAMA3 | 0.57 | 3.2 × 10−3 | Laminin subunit alpha 3 | 3909 |
| ADAMTS15 | 0.57 | 5.5 × 10−4 | ADAM metallopeptidase with thrombospondin type 1 motif 15 | 170689 |
| VIM | 0.60 | 3.6 × 10−3 | Vimentin | 7431 |
| TENM2 | 0.60 | 1.2 × 10−4 | Tenascin M2/teneurin transmembrane protein 2 | 57451 |
| TFRC* | 0.61 | 3.6 × 10−5 | Transferrin receptor | 7037 |
| MMP2* | 0.62 | 1.1 × 10−6 | Matrix metalloprotease 2 | 4313 |
| WFDC5 | 0.62 | 6.6 × 10−3 | WAP four-disulphide core domain 5/WAP1 | 149708 |
| MXRA5* | 0.62 | 4.1 × 10−5 | Matrix-remodelling associated 5/adlikan | 25878 |
| SDK2 | 0.63 | 2.6 × 10−3 | Sidekick cell adhesion molecule 2 | 54549 |
| TNC | 0.63 | 9.7 × 10−3 | Tenascin C | 3371 |
| FOSB | 0.63 | 3.0 × 10−3 | FosB Proto-oncogene, AP-1 transcription factor subunit/FBJ murine osteosarcoma viral oncogene homolog B | 2354 |
| B3GNT8 | 0.64 | 4.5 × 10−3 | UDP-GlcNAc:BetaGal beta-1,3-N-acetylglucosaminyltransferase 8 | 374907 |
| TLL2 | 0.64 | 3.0 × 10−3 | Tolloid-like 2 | 7093 |
| ZNF488 | 0.64 | 2.2 × 10−3 | Zinc finger protein 488 | 118738 |
| ADAMTSL4 | 0.64 | 3.3 × 10−3 | ADAMTS Like 4 | 54507 |
| FAT2 | 0.65 | 2.0 × 10−3 | FAT atypical cadherin 2 | 2196 |
| TMEM217 | 0.65 | 7.1 × 10−3 | Transmembrane protein 217 | 221468 |
| EPHB3 | 0.65 | 7.4 × 10−5 | Ephrin receptor B3 | 2049 |
| COL7A1* | 0.65 | 1.5 × 10−5 | Collagen type VII alpha 1 chain | 1294 |
| AMOTL1* | 0.65 | 6.3 × 10−6 | Angiomotin like 1 | 154810 |
| CEL | 0.65 | 1.5 × 10−4 | Carboxyl ester lipase | 1056 |
| ABCA1* | 0.66 | 2.2 × 10−6 | ATP binding cassette subfamily A member 1 | 19 |
| TGFB1* | 0.66 | 1.0 × 10−5 | Transforming growth factor beta 1 | 7040 |
| FAM83A | 0.66 | 4.4 × 10−4 | Family with sequence similarity 83 member A | 84985 |
| Upregulated following 24 h UFPM exposure | ||||
| MT1G* | 24.02 | 1.2 × 10−11 | Metallothionein 1G | 4495 |
| MT1H* | 21.36 | 1.8 × 10−8 | Metallothionein 1H | 4496 |
| MT1M* | 15.50 | 2.6 × 10−14 | Metallothionein 1M | 4499 |
| HMOX1* | 5.56 | 1.2 × 10−34 | Haemoxygenase-1 | 3162 |
| NRCAM* | 2.56 | 4.6 × 10−6 | Neuronal cell adhesion molecule | 4897 |
| MT1F | 2.17 | 1.6 × 10−4 | Metallothionein 1F | 4494 |
| AKR1B10 | 2.03 | 2.6 × 10−4 | Aldo-keto reductase family 1, member B10 | 57016 |
| MT1E | 1.87 | 9.8 × 10−4 | Metallothionein 1E | 4493 |
| SLC7A11 | 1.74 | 5.1 × 10−4 | Solute carrier family 7 member 11/anionic amino acid transporter light chain, Xc− system, member 11 | 23657 |
| TRIM16L | 1.72 | 9.3 × 10−3 | Tripartite motif containing 16-like | 147166 |
| FMNL3 | 1.71 | 5.1 × 10−3 | Formin-like 3 | 91010 |
| CHST2 | 1.66 | 1.7 × 10−3 | Carbohydrate sulfotransferase 2 | 9435 |
| PLAT | 1.60 | 5.4 × 10−3 | Plasminogen activator. Tissue type | 5327 |
| ABCC3 | 1.58 | 2.8 × 10−4 | ATP-binding cassette, sub-Family C member 3/multidrug resistance-associated protein 3 | 8714 |
| MT2A | 1.58 | 5.4 × 10−4 | Metallothionein 2A | 4502 |
| ME1 | 1.57 | 4.3 × 10−4 | Malic enzyme 1 | 4199 |
| GCLM* | 1.51 | 4.7 × 10−6 | Glutamate-cysteine ligase modifier subunit | 2730 |
| Downregulated following 24 h UFPM exposure | ||||
| RSAD2 | 0.40 | 1.6 × 10−3 | Radical S-adenosyl methionine domain containing 2/viperin | 91543 |
| HERC5 | 0.51 | 5.6 × 10−3 | HECT And RLD domain containing E3 ubiquitin protein ligase 5 | 51191 |
| IFIT1 | 0.54 | 4.4 × 10−3 | Interferon-induced protein with tetratricopeptide repeats 1 | 3434 |
| TFRC* | 0.57 | 2.0 × 10−6 | Transferrin receptor | 7037 |
| EPSTI1 | 0.58 | 2.7 × 10−3 | Epithelial stromal interaction 1 | 94240 |
| GIMAP2 | 0.65 | 7.8 × 10−3 | GTPase, IMAP family member 2 | 26157 |
Validation of RNA-Seq data by RT-qPCR
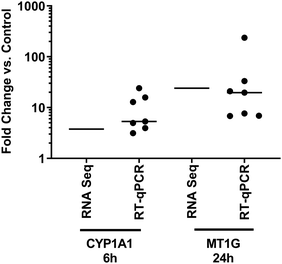
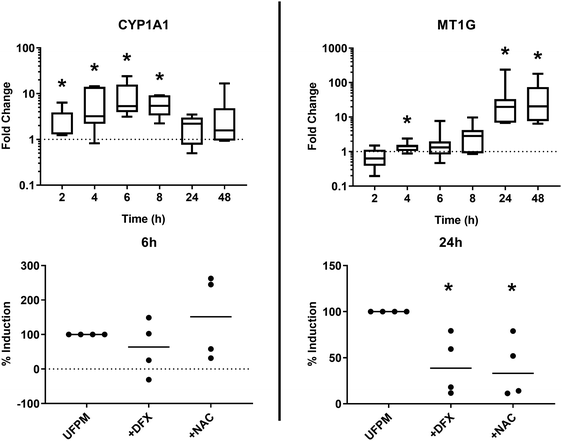
Two genes were selected for validation of the RNA-Seq dataset at timepoints where they were significantly modulated, based on temporal response and lack of obvious functional or regulatory overlap – CYP1A1 (RNA-Seq 6 h FC 3.76, p = 1.46 × 10−6), and MT1G (RNA-Seq 24 h FC 24.1, p = 1.23 × 10−11). Validation was performed using PBECs from 7 donors, distinct from those used for the RNA-Seq studies, but with identical exposure protocols. We contend that this validation in a separate set of donor cultures means that the responses observed are more likely to be reflective of a larger population, rather than simply of those donors used in the RNA-Seq experiments. Median expression changes (vs. UFPM-free controls) were: CYP1A1, 6 h FC 5.32 (IQR 3.95–15.8, p = 0.016); MT1G, 24 h FC 19.7 (IQR 6.92–33.2, p = 0.016) (Fig. 5). Furthermore, we had previously observed upregulation of HMOX1 at 24 h through RT-qPCR (median RNA-Seq 24 h FC 5.54, p = 1.18 × 10−34).15 Thus, upregulation of CYP1A1 at 6 h and of MT1G at 24 h was supported by these validation experiments, along with previous validation of HMOX1 upregulation.Pathway perturbations
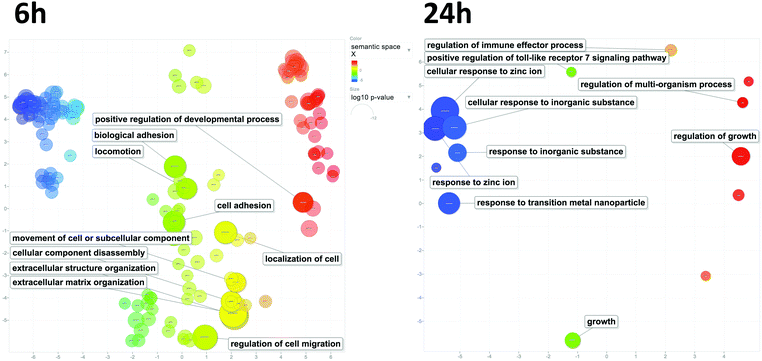
To assess the biological processes perturbed in cells exposed to underground UFPM, DEG sets for 6 h and 24 h exposures were subjected to GO analysis using ToppGene, and the resulting GO terms were visualised in REVIGO (Fig. 6). After 6 h, there were significant changes in expression of genes relevant to epithelial cell function, such as cell adhesion and localisation, and maintenance of external epithelial environment, including extracellular matrix (ECM) organisation and structure organisation. Conversely, after 24 h, there was clear evidence of a response specific to metal-rich underground UFPM, with terms including responses to transition metal nanoparticle, cellular response to inorganic substance, and response to zinc ion. These particle content-specific changes are evidenced especially by upregulation of members of the metallothionein family (Fig. 7). Furthermore, terms relating to antioxidant defences, especially HMOX1, and to uptake of cystine (through SLC7A11 in the system Xc− cystine-glutamate antiporter) and glutathione production (via glutamate-cysteine ligase; GCLM, which is a known Nrf2 target gene) were highlighted as significantly associated with the DEGs. Thus, after 6 h the response appeared to be epithelial cell-focused, and after 24 h particle-focused.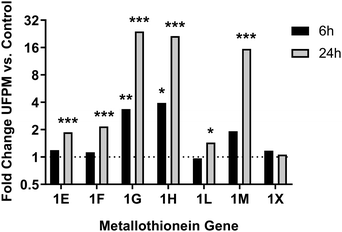 | ||
| Fig. 7 Changes in metallothionein gene expression following exposure to UFPM. RNA-Seq data from cells treated with 5.6 μg cm−2 underground UFPM for 6 h or 24 h, as presented in Fig. 4–6 and Table 1, showed upregulation of multiple metallothionein genes, but that the magnitude and time-specificity of these responses varies between specific metallothioneins. Bars represent mean fold change vs. control; n = 3 individual donors. * – p < 0.05, ** – p < 0.01, *** – p < 0.001 vs. PM-free control by t-test (see Table 1 for full p-values). | ||
Study of time-course of gene expression changes
In view of the discrete responses at 6 h and 24 h, extended time-course experiments from 2–48 h were performed on cultures of cells from the donors used in the above RT-qPCR experiments, to further study the effect of exposure time on gene modulation (Fig. 8, top). CYP1A1 expression was significantly increased at 6 h (see above) and also at 2 h, 4 h, and 8 h (p = 0.031 for each). CYP1A1 expression after 24 h and 48 h exposure failed to reach statistical significance (both p = 0.078). Conversely, MT1G expression increased over time peaking at 24 h (p < 0.05) and 48 h (p < 0.05). These results support observations from the RNA-Seq data and suggest that regulation of gene expression in response to exposure to metal-rich UFPM follows different time courses for individual genes depending on their function.Role of iron and reactive oxygen species in induction of CYP1A1 and MT1G
In order to determine whether particle iron content and/or reactive oxygen species generation were involved in UFPM-induced upregulation of CYP1A1 after 6 h and MT1G after 24 h, cells were exposed to UFPM in the presence of the iron-selective chelator desferrioxamine (DFX; 200 μM) or the ROS scavenger N-acetylcysteine (NAC, 20 mM). Expression of CYP1A1 after 6 h exposure to UFPM was not significantly affected by the presence of DFX or NAC (Fig. 8, bottom). In contrast, induction of MT1G expression by UFPM was significantly reduced in the presence of DFX or NAC, indicating that MT1G expression is at least partially dependent on iron and ROS. This finding supports the involvement of redox-active iron, as identified through the earlier XANES and ARE data.Discussion
Airborne PM10 and PM2.5 in underground railways is more concentrated and more metallic than in ambient air, yet little is known about the detailed biological effects of UFPM from this source. In this paper, we have shown that this type of UFPM is redox-active and exerts temporally-specific effects on PBECs, including rapid responses relating to maintenance of epithelial function, followed by slower induction of processes involved in redox and metal homeostasis, with a predominant role for metallothioneins.Using XANES, the majority of the iron in the UFPM sample was observed as redox-active magnetite (Fe3O4), mirroring PM10 from the Stockholm31 and Seoul undergrounds.32 This also agrees with a study on the Barcelona underground,33 although in another study, haematite (Fe2O3) was shown to dominate in PM10 and PM2.5.34 Our identification of magnetite is consistent with our previous studies showing that underground railway UFPM can generate ROS;14,15 here we demonstrate a functional consequence of this, with increased ARE-regulated transcriptional activation in exposed cells.
In our study, we used mucociliary differentiated PBEC ALI cultures to model the bronchial epithelium. This model is more physiologically relevant than undifferentiated monolayer or cell line cultures, especially as the presence of a mucous barrier may confer on the epithelial surface physical and chemical protection from the particles. Nonetheless, at 6 h genes associated with epithelial maintenance were upregulated, while by 24 h there was a clear upregulation of genes related to redox homeostasis and metal binding. The relatively slow induction of metal binding genes may reflect dissolution of the metals from the particles, but is more likely due to UFPM uptake, which we have observed previously.15 Our observation of “streamlined” responses occurring after a delayed period parallels observations using different PM concentrations.35 Time specificity has also been noted with a different source of dust, with Saudi Arabian PM eliciting antioxidant responses at 24 h exposure (as in the present study), while genes involved in lipid metabolism were affected at 96 h exposure.36 Interestingly, while we have previously shown that underground UFPM induces inflammatory mediator release, we did not find induction of mRNAs encoding interleukins or their receptors. While some studies have found inflammatory changes at the mRNA level, this may depend on the components of the PM involved or the timepoints studied.37
A notable finding of our work is the broad and marked increase in expression of metallothioneins, especially after 24 h exposure. This was initially observed in our RNA-Seq dataset but was validated in a separate set of UFPM-exposed PBEC cultures analysed by RT-qPCR. Metallothioneins are a group of single-chain low molecular weight peptides that are rich in cysteine which allows them to bind heavy metals and also to act as ROS scavengers.38 Metallothionein expression is classically upregulated following liberation of zinc ions from intracellular sources, which then bind to metal regulatory transcription factor 1 (MTF-1) enabling it to interact with the metal response elements (MRE) of metallothioneins and other genes involved in metal homeostasis.39,40 However, recent work has indicated that other metals such as copper or cadmium may be able to tune the specificity of MTF1, with different insults resulting in different DNA binding preferences.41 In addition to heavy metals, other metallothionein inducers include inflammatory cytokines, glucocorticoids, and ROS.42 There is evidence that MT1 and MT2 expression may be regulated through binding of Nrf1 to ARE sites upstream of the MT coding regions, although only a restricted role for Nrf2 is evident.43,44 In studies of nickel-induced lung injury, it has been demonstrated that nickel increases intracellular ROS as well as free intracellular Zn2+ levels to induce MT2A transcript levels.45 However, as only NAC, but not ascorbic acid, prevented nickel-induced MT2A transcription it was suggested that free thiols, rather than ROS per se may regulate the ability of nickel to induce certain MT genes in this system. In our work, suppression of metallothionein expression by DFX or NAC suggests mechanism(s) involving iron and ROS, especially considering the predominance of iron in the UFPM. However, while DFX is often described as an iron-selective chelator,46 it is nonetheless able to bind ions of multiple other metals,47,48 albeit with an affinity several orders of magnitude lower than for the DFX–Fe(III) complex.49 Given that the UFPM that we used also contained other transition metals, these may also be involved, either alone or in concert with iron or other metals. In this regard, given the aforementioned work on MT2A and NAC, it is possible that the action of NAC in our system is at least partly related to provision of free thiols rather than generic antioxidant activity. Furthermore, since the ARE reporter that we used in our luciferase assays can bind either Nrf1 or Nrf2, the mechanism of induction may involve ROS and Nrf1. Further work is required to determine the exact nature of the mechanisms driving increased MT expression by such complex UFPM.
Although metallothioneins are able to bind a range of mono- and divalent metal ions,50,51 including Fe(II) in vitro when the metal-free apoprotein is combined with ferrous iron,52 this is not the case in vivo, where Fe(II) is unable to displace occupation by Zn(II) or Cu(I).50,53 Therefore, it is possible that the metal sequestration arm of the metallothionein defence is relatively ineffective against direct toxicity from iron-rich PM, and it may affect the homeostasis of other metals.54 Indeed, zinc-loaded metallothioneins appear less protective against iron-induced DNA strand breakage compared with copper, which can be bound by such metallothioneins.55 However, the presence of iron may nonetheless have a profound effect on metallothioneins – the metallothionein-zinc complex is able to reduce ferritin-bound Fe(III) to Fe(II), which results in release of redox-active Fe(II) from complex with ferritin, and concomitantly oxidation of the metallothionein thiolate groups, resulting in release of Zn(II). This free Fe(II) is then able to participate in other ROS-generating reactions, thus increasing oxidative stress, while there may also be dysregulated zinc homeostasis.56 Although the predominance of iron in our samples does not necessarily mean this is occurring, it is notable that the only downregulated gene across both 6 h and 24 h timepoints was TFRC, coding for the transferrin receptor, while at 24 h the 51st most upregulated gene was FTH1 (p = 0.022 but not crossing 1.5 FC cut-off), suggesting that the cell may be attempting to sequester intracellular iron and limit further uptake. While MT1 and MT2 are generally associated with zinc homeostasis, there is greater evidence for copper homeostasis being performed by MT3 and MT4, generally localised to the brain, and stratified and cornified epithelium, respectively.57 Beyond MT3 and MT4, MT1 has been suggested to be more adaptable to non-zinc metal ions, perhaps as a result of its greater conformational flexibility compared to MT2, which appears to be a relatively rather poor copper chelator.58 Nonetheless, studies have found that both MT1 and MT2 are, at least under certain conditions, capable of binding copper, along with the aforementioned amelioration of copper-induced DNA damage.58,59 What is less known is whether this is of relevance in the lung, given that there may be isoform-, and thus tissue-specific, differences in metal binding. The above suggests that is it not impossible, however, that oxidative stress and/or the presence of ferritin–Fe(III) may have an impact on the sequestration of copper by metallothioneins in the same way. If this were the case, and given the greater affinity of MT for copper compared to zinc, this may result in displacement of MT-bound zinc, and thus further dysregulation of zinc metabolism.
Previous studies have found increased expression of MT1 in primary airway macrophages after exposure to fine PM for 4 h,60 of metallothionein isoforms at 24 h in human nasal epithelial cells exposed to SRM1648a urban PM,61 and of metallothionein isoforms at 24 h but not 6 h in primary airway epithelial cells in response to urban fine and UFPM which contain higher concentrations of nickel compared with coarse PM.62 These time-specific observations agree with our findings, and the earlier upregulation in phagocytes suggests that PM uptake may be an important factor. Expression of metallothioneins in lung tissue and lymphocytes of diesel exhaust PM-exposed mice has been reported to maintain dose-dependence at higher exposure concentrations than for CYPs.63 Given that systemic responses to PM involving several genes noted in our work have been observed in distal organs of PM-exposed rats,64 and that upregulation of some metallothioneins has been found in ambient UFPM-exposed primary human neurons,65 work on the systemic translocation of metal-rich UFPM and their effects is warranted.
In such studies, there frequently arises the question of the suitability of the PM concentration used for cell exposures. A recent study modelling deposition of traffic-related UFPM in the terminal bronchiolar region following exposure at an airborne concentration of 64.6 μg m−3 (1.5 times that of the present study) suggested a deposition of 11–32 pg min−1 cm−2, depending on whether the subject is at rest or performing light exercise.66 Correcting for the UFPM concentration of 44 μg m−3 on the PM collection day in the present study and a 60 minute average commute time67 gives a deposition concentration of 0.45–1.31 ng cm−2, 3–4 orders of magnitude lower than the concentrations applied to cells in the present study. However, it is known that particle deposition may be strongly concentrated at “hotspots” of airflow turbulence such as bifurcations and loss of laminar airflow such as caused by airway disease, and also increased by oral breathing and increased physical exercise, with these factors combining to increase local deposition by over 4 orders of magnitude.68 Moreover, it is likely that there will not be complete clearance of particles each day, with a fraction of the particles penetrating the mucus and attaching to, or entering, epithelial cells, meaning that cumulative dose may be higher. Furthermore, some people may spend longer in the underground, such as in the case of railway employees or because of increased use of the underground railway. Therefore, and in agreement with other authors using PM with similar airborne and in vitro exposure concentrations,69 our use of 5.6 μg cm−2 in the majority of exposures in this study represents a concentration which could feasibly and realistically be attained in parts of the airways.
Conclusion
This study demonstrates that the ultrafine fraction of PM from a mainline European underground station is rich in iron, existing predominantly as redox-active magnetite Fe3O4, and is able to activate ARE-controlled transcription. The differing gene expression profiles after 6 h and 24 h suggest that responses are organised in such a way as to respond to the risk posed, beginning first with maintenance of the epithelium, and then followed by responses that are especially sensitive to accumulation of metal particles within the cells. The associated upregulation of metallothioneins warrants further functional investigation and may serve as a useful biomarker of exposure to inhaled metal-rich UFPM.Authors contribution
Conceptualisation – ML, DED, methodology – ML, DED, software – JW, AS, CHW, validation – ML, NPS, AY, formal analysis – ML, JW, AS, CHW, investigation – ML, NPS, AMC, resources – ML, GP, RBC, FRC, DED, data curation – JW, AS, CHW, writing – original draft – ML, DED, writing – review and editing – all authors, visualisation – ML, JW, AS, AMC, RBC, CHW, supervision – CHW, DED, project administration – DED, funding acquisition – ML, DED.Conflicts of interest
DED has received personal consultancy fees from, and is a shareholder in, Synairgen PLC. All other authors report no conflicts.Acknowledgements
The authors thank A. John F. Boere, Daan L. A. C. Leseman, and Paul H. B. Fokkens from the Centre for Sustainability, Environment and Health, National Institute for Public Health and the Environment (RIVM), Netherlands, for valuable assistance with collecting ultrafine particulate matter from the underground railway station, Dr Robert A. Ridley and Graham A. Berreen (Brooke Laboratory, Faculty of Medicine, University of Southampton, UK) for help with the culture of PBECs, Dr Kamran Tariq (Faculty of Medicine, University of Southampton) for provision of bronchial brushings, Dr Timothy Hinks (Faculty of Medicine, University of Southampton (now at Nuffield Department of Medicine, University of Oxford)) for providing PBEC donor information, and Professor Roland Wolf (University of Dundee, UK) for the kind gift of the ARE reporter construct. ML is funded by a BBSRC Future Leader Fellowship (BB/P011365/1) and a National Institute for Health Research Southampton Biomedical Research Centre Senior Research Fellowship, and also a research grant from the Asthma, Allergy, and Inflammation Research (AAIR) Charity.References
- R. T. Burnett, C. A. Pope, 3rd, M. Ezzati, C. Olives, S. S. Lim, S. Mehta, H. H. Shin, G. Singh, B. Hubbell, M. Brauer, H. R. Anderson, K. R. Smith, J. R. Balmes, N. G. Bruce, H. Kan, F. Laden, A. Pruss-Ustun, M. C. Turner, S. M. Gapstur, W. R. Diver and A. Cohen, An integrated risk function for estimating the global burden of disease attributable to ambient fine particulate matter exposure, Environ. Health Perspect., 2014, 122, 397–403 CrossRef PubMed.
- R. Burnett, H. Chen, M. Szyszkowicz, N. Fann, B. Hubbell, C. A. Pope, J. S. Apte, M. Brauer, A. Cohen, S. Weichenthal, J. Coggins, Q. Di, B. Brunekreef, J. Frostad, S. S. Lim, H. D. Kan, K. D. Walker, G. D. Thurston, R. B. Hayes, C. C. Lim, M. C. Turner, M. Jerrett, D. Krewski, S. M. Gapstur, W. R. Diver, B. Ostro, D. Goldberg, D. L. Crouse, R. V. Martin, P. Peters, L. Pinault, M. Tjepkema, A. Donkelaar, P. J. Villeneuve, A. B. Miller, P. Yin, M. G. Zhou, L. J. Wang, N. A. H. Janssen, M. Marra, R. W. Atkinson, H. Tsang, Q. Thach, J. B. Cannon, R. T. Allen, J. E. Hart, F. Laden, G. Cesaroni, F. Forastiere, G. Weinmayr, A. Jaensch, G. Nagel, H. Concin and J. V. Spadaro, Global estimates of mortality associated with long-term exposure to outdoor fine particulate matter, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 9592–9597 CrossRef CAS PubMed.
- J. Lelieveld, K. Klingmuller, A. Pozzer, U. Poschl, M. Fnais, A. Daiber and T. Munzel, Cardiovascular disease burden from ambient air pollution in Europe reassessed using novel hazard ratio functions, Eur. Heart J., 2019, 40, 1590–1596 CrossRef PubMed.
- O. K. Kurt, J. J. Zhang and K. E. Pinkerton, Pulmonary health effects of air pollution, Curr. Opin. Pulm. Med., 2016, 22, 138–143 CrossRef CAS PubMed.
- J. F. Pearson, C. Bachireddy, S. Shyamprasad, A. B. Goldfine and J. S. Brownstein, Association Between Fine Particulate Matter and Diabetes Prevalence in the U.S., Diabetes Care, 2010, 33, 2196–2201 CrossRef PubMed.
- X. Zhang, X. Chen and X. B. Zhang, The impact of exposure to air pollution on cognitive performance, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 9193–9197 CrossRef CAS PubMed.
- C. C. Jose, L. Jagannathan, V. S. Tanwar, X. Zhang, C. Zang and S. Cuddapah, Nickel exposure induces persistent mesenchymal phenotype in human lung epithelial cells through epigenetic activation of ZEB1, Mol. Carcinog., 2018, 57, 794–806 CrossRef CAS PubMed.
- F. R. Cassee, M. E. Heroux, M. E. Gerlofs-Nijland and F. J. Kelly, Particulate matter beyond mass: recent health evidence on the role of fractions, chemical constituents and sources of emission, Inhalation Toxicol., 2013, 25, 802–812 CrossRef CAS PubMed.
- A. Valavanidis, T. Vlachogianni, K. Fiotakis and S. Loridas, Pulmonary Oxidative Stress, Inflammation and Cancer: Respirable Particulate Matter, Fibrous Dusts and Ozone as Major Causes of Lung Carcinogenesis through Reactive Oxygen Species Mechanisms, Int. J. Environ. Res. Public Health, 2013, 10, 3886–3907 CrossRef PubMed.
- K. C. De Grove, S. Provoost, G. G. Brusselle, G. F. Joos and T. Maes, Insights in particulate matter-induced allergic airway inflammation: Focus on the epithelium, Clin. Exp. Allergy, 2018, 48, 773–786 CrossRef CAS PubMed.
- M. Steenhof, I. Gosens, M. Strak, K. J. Godri, G. Hoek, F. R. Cassee, I. S. Mudway, F. J. Kelly, R. M. Harrison, E. Lebret, B. Brunekreef, N. A. Janssen and R. H. Pieters, In vitro toxicity of particulate matter (PM) collected at different sites in the Netherlands is associated with PM composition, size fraction and oxidative potential – the RAPTES project, Part. Fibre Toxicol., 2011, 8, 26 CrossRef CAS PubMed.
- M. Loxham, Harmful effects of particulate air pollution: identifying the culprits, Respirology, 2015, 20, 7–8 CrossRef PubMed.
- M. J. Nieuwenhuijsen, J. E. Gomez-Perales and R. N. Colvile, Levels of particulate air pollution, its elemental composition, determinants and health effects in metro systems, Atmos. Environ., 2007, 41, 7995–8006 CrossRef CAS.
- M. Loxham, M. J. Cooper, M. E. Gerlofs-Nijland, F. R. Cassee, D. E. Davies, M. R. Palmer and D. A. Teagle, Physicochemical characterization of airborne particulate matter at a mainline underground railway station, Environ. Sci. Technol., 2013, 47, 3614–3622 CrossRef CAS PubMed.
- M. Loxham, R. J. Morgan-Walsh, M. J. Cooper, C. Blume, E. J. Swindle, P. W. Dennison, P. H. Howarth, F. R. Cassee, D. A. Teagle, M. R. Palmer and D. E. Davies, The effects on bronchial epithelial mucociliary cultures of coarse, fine, and ultrafine particulate matter from an underground railway station, Toxicol. Sci, 2015, 145, 98–107 CrossRef CAS PubMed.
- M. Loxham and M. J. Nieuwenhuijsen, Health effects of particulate matter air pollution in underground railway systems - a critical review of the evidence, Part. Fibre Toxicol., 2019, 16, 12 CrossRef PubMed.
- Y. C. T. Huang, The Role of in Vitro Gene Expression Profiling in Particulate Matter Health Research, J. Toxicol. Environ. Health, Part B, 2013, 16, 381–394 CAS.
- D. M. Cooper and M. Loxham, Particulate matter and the airway epithelium: the special case of the underground?, Eur. Respir. Rev., 2019, 28, 190066 CrossRef PubMed.
- A. Yeomans, E. Lemm, S. Wilmore, B. E. Cavell, B. Valle-Argos, S. Krysov, M. S. Hidalgo, E. Leonard, A. E. Willis, F. Forconi, F. K. Stevenson, A. J. Steele, M. J. Coldwell and G. Packham, PEITC-mediated inhibition of mRNA translation is associated with both inhibition of mTORC1 and increased eIF2 alpha phosphorylation in established cell lines and primary human leukemia cells, Oncotarget, 2016, 7, 74807–74819 CrossRef PubMed.
- S. E. Purdom-Dickinson, Y. Lin, M. Dedek, S. Morrissy, J. Johnson and Q. M. Chen, Induction of antioxidant and detoxification response by oxidants in cardiomyocytes: Evidence from gene expression profiling and activation of Nrf2 transcription factor, J. Mol. Cell. Cardiol., 2007, 42, 159–176 CrossRef CAS PubMed.
- M. S. Leino, M. Loxham, C. Blume, E. J. Swindle, N. P. Jayasekera, P. W. Dennison, B. W. Shamji, M. J. Edwards, S. T. Holgate, P. H. Howarth and D. E. Davies, Barrier disrupting effects of alternaria alternata extract on bronchial epithelium from asthmatic donors, PLoS One, 2013, 8, e71278 CrossRef CAS PubMed.
- J. L. Lordan, F. Bucchieri, A. Richter, A. Konstantinidis, J. W. Holloway, M. Thornber, S. M. Puddicombe, D. Buchanan, S. J. Wilson, R. Djukanovic, S. T. Holgate and D. E. Davies, Cooperative effects of Th2 cytokines and allergen on normal and asthmatic bronchial epithelial cells, J. Immunol., 2002, 169, 407–414 CrossRef CAS PubMed.
- C. Trapnell, L. Pachter and S. L. Salzberg, TopHat: discovering splice junctions with RNA-Seq, Bioinformatics, 2009, 25, 1105–1111 CrossRef CAS PubMed.
- S. Anders, P. T. Pyl and W. Huber, HTSeq-a Python framework to work with high-throughput sequencing data, Bioinformatics, 2015, 31, 166–169 CrossRef CAS PubMed.
- O. Nikolayeva and M. D. Robinson, edgeR for Differential RNA-seq and ChIP-seq Analysis: An Application to Stem Cell Biology, Stem Cell Transcriptional Networks, Methods Protoc., 2014, 1150, 45–79 CAS.
- M. D. Robinson and A. Oshlack, A scaling normalization method for differential expression analysis of RNA-seq data, Genome Biol., 2010, 11, R25 CrossRef PubMed.
- J. Chen, E. E. Bardes, B. J. Aronow and A. G. Jegga, ToppGene Suite for gene list enrichment analysis and candidate gene prioritization, Nucleic Acids Res., 2009, 37, W305–W311 CrossRef CAS PubMed.
- Y. Benjamini and Y. Hochberg, Controlling the False Discovery Rate – a Practical and Powerful Approach to Multiple Testing, J. Roy. Stat. Soc., B Met., 1995, 57, 289–300 Search PubMed.
- F. Supek, M. Bosnjak, N. Skunca and T. Smuc, REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms, PLoS One, 2011, 6, e21800 CrossRef CAS PubMed.
- J. D. Hayes and A. T. Dinkova-Kostova, The Nrf2 regulatory network provides an interface between redox and intermediary metabolism, Trends Biochem. Sci., 2014, 39, 199–218 CrossRef CAS PubMed.
- H. L. Karlsson, L. Nilsson and L. Moller, Subway particles are more genotoxic than street particles and induce oxidative stress in cultured human lung cells, Chem. Res. Toxicol., 2005, 18, 19–23 Search PubMed.
- H. J. Eom, H. J. Jung, S. Sobanska, S. G. Chung, Y. S. Son, J. C. Kim, Y. Sunwoo and C. U. Ro, Iron speciation of airborne subway particles by the combined use of energy dispersive electron probe X-ray microanalysis and Raman microspectrometry, Anal. Chem., 2013, 85, 10424–10431 CrossRef CAS PubMed.
- T. Moreno, V. Martins, X. Querol, T. Jones, K. BéruBé, M. C. Minguillón, F. Amato, M. Capdevila, E. de Miguel, S. Centelles and W. Gibbons, A new look at inhalable metalliferous airborne particles on rail subway platforms, Sci. Total Environ, 2015, 505, 367–375 CrossRef CAS PubMed.
- X. Querol, T. Moreno, A. Karanasiou, C. Reche, A. Alastuey, M. Viana, O. Font, J. Gil, E. de Miguel and M. Capdevila, Variability of levels and composition of PM10 and PM2.5 in the Barcelona metro system, Atmos. Chem. Phys., 2012, 12, 5055–5076 CrossRef CAS.
- X. Ding, M. L. Wang, H. Y. Chu, M. J. Chu, T. Na, Y. Wen, D. M. Wu, B. Han, Z. P. Bai, W. H. Chen, J. Yuan, T. C. Wu, Z. B. Hu, Z. D. Zhang and H. B. Shen, Global gene expression profiling of human bronchial epithelial cells exposed to airborne fine particulate matter collected from Wuhan, China, Toxicol. Lett., 2014, 228, 25–33 CrossRef CAS PubMed.
- H. Sun, M. Shamy, T. Kluz, A. B. Munoz, M. H. Zhong, F. Laulicht, M. A. Alghamdi, M. I. Khoder, L. C. Chen and M. Costa, Gene expression profiling and pathway analysis of human bronchial epithelial cells exposed to airborne particulate matter collected from Saudi Arabia, Toxicol. Appl. Pharmacol., 2012, 265, 147–157 CrossRef CAS PubMed.
- S. C. Jeong, C. Y. Shin, M. K. Song, Y. Cho and J. C. Ryu, Gene expression profiling of human alveolar epithelial cells (A549 cells) exposed to atmospheric particulate matter 2.5 (PM2.5) collected from Seoul, Korea, Mol. Cell. Toxicol., 2014, 10, 361–368 CrossRef CAS.
- B. Ruttkay-Nedecky, L. Nejdl, J. Gumulec, O. Zitka, M. Masarik, T. Eckschlager, M. Stiborova, V. Adam and R. Kizek, The Role of Metallothionein in Oxidative Stress, Int. J. Mol. Sci., 2013, 14, 6044–6066 CrossRef CAS PubMed.
- C. D. Klaassen, J. Liu and S. Choudhuri, Metallothionein: An intracellular protein to protect against cadmium toxicity, Annu. Rev. Pharmacol. Toxicol., 1999, 39, 267–294 CrossRef CAS PubMed.
- K. J. Waldron, J. C. Rutherford, D. Ford and N. J. Robinson, Metalloproteins and metal sensing, Nature, 2009, 460, 823–830 CrossRef CAS PubMed.
- H. I. Sims, G. W. Chirn and M. T. Marr, Single nucleotide in the MTF-1 binding site can determine metal-specific transcription activation, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16516–16521 CrossRef CAS PubMed.
- S. R. Davis and R. J. Cousins, Metallothionein expression in animals: A physiological perspective on function, J. Nutr., 2000, 130, 1085–1088 CrossRef CAS PubMed.
- M. Ohtsuji, F. Katsuoka, A. Kobayashi, H. Aburatani, J. D. Hayes and M. Yamamoto, Nrf1 and Nrf2 Play Distinct Roles in Activation of Antioxidant Response Element-dependent Genes, J. Biol. Chem., 2008, 283, 33554–33562 CrossRef CAS PubMed.
- M. O. Song, M. D. Mattie, C. H. Lee and J. H. Freedman, The role of Nrf1 and Nrf2 in the regulation of copper-responsive transcription, Exp. Cell Res., 2014, 322, 39–50 CrossRef CAS PubMed.
- A. A. Nemec, G. D. Leikauf, B. R. Pitt, K. J. Wasserloos and A. Barchowsky, Nickel Mobilizes Intracellular Zinc to Induce Metallothionein in Human Airway Epithelial Cells, Am. J. Respir. Cell Mol. Biol., 2009, 41, 69–75 CrossRef CAS PubMed.
- D. M. Van Reyk and R. T. Dean, The iron-selective chelator desferal can reduce chelated copper, Free Radical Res., 1996, 24, 55–60 CrossRef CAS PubMed.
- S. J. Stohs and D. Bagchi, Oxidative Mechanisms in the Toxicity of Metal Ions, Free Radical Biol. Med., 1995, 18, 321–336 CrossRef CAS PubMed.
- L. Selley, L. Schuster, H. Marbach, T. Forsthuber, B. Forbes, T. W. Gant, T. Sandstrom, N. Camina, T. J. Athersuch, I. Mudway and A. Kumar, Brake dust exposure exacerbates inflammation and transiently compromises phagocytosis in macrophages, Metallomics, 2020, 12, 371–386 RSC.
- H. Keberle, The Biochemistry of Desferrioxamine and Its Relation to Iron Metabolism, Ann. N. Y. Acad. Sci., 1964, 119, 758–768 CrossRef CAS PubMed.
- M. J. Stillman, Metallothioneins, Coord. Chem. Rev., 1995, 144, 461–511 CrossRef CAS.
- T. T. Ngu and M. J. Stillman, Metal-binding mechanisms in metallothioneins, Dalton Trans., 2009, 5425–5433 RSC.
- Y. Sano, A. Onoda, R. Sakurai, H. Kitagishi and T. Hayashi, Preparation and reactivity of a tetranuclear Fe(II) core in the metallothionein alpha-domain, J. Inorg. Biochem., 2011, 105, 702–708 CrossRef CAS PubMed.
- N. Kojima, C. R. Young and G. W. Bates, Failure of Metallothionein to Bind Iron or Act as an Iron Mobilizing Agent, Biochim. Biophys. Acta, 1982, 716, 273–275 CrossRef CAS.
- W. J. Qiang, Y. P. Huang, Z. H. Wan and B. Zhou, Metal-metal interaction mediates the iron induction of Drosophila MtnB, Biochem. Biophys. Res. Commun., 2017, 487, 646–652 CrossRef CAS PubMed.
- L. Cai, J. Koropatnick and M. G. Cherian, Metallothionein Protects DNA from Copper-Induced but Not Iron-Induced Cleavage in Vitro, Chem. – Biol. Interact., 1995, 96, 143–155 CrossRef CAS PubMed.
- R. Orihuela, B. Fernandez, O. Palacios, E. Valero, S. Atrian, R. K. Watt, J. M. Dominguez-Vera and M. Capdevila, Ferritin and metallothionein: dangerous liaisons, Chem. Commun., 2011, 47, 12155–12157 RSC.
- J. Calvo, H. M. Jung and G. Meloni, Copper metallothioneins, IUBMB Life, 2017, 69, 236–245 CrossRef CAS PubMed.
- E. Artells, O. Palacios, M. Capdevila and S. Atrian, Mammalian MT1 and MT2 metallothioneins differ in their metal binding abilities, Metallomics, 2013, 5, 1397–1410 RSC.
- A. Krezel and W. Maret, The Functions of Metamorphic Metallothioneins in Zinc and Copper Metabolism, Int. J. Mol. Sci., 2017, 18, 1237 CrossRef PubMed.
- Y. C. T. Huang, Z. W. Li, J. D. Carter, J. M. Soukup, D. A. Schwartz and I. V. Yang, Fine Ambient Particles Induce Oxidative Stress and Metal Binding Genes in Human Alveolar Macrophages, Am. J. Respir. Cell Mol. Biol., 2009, 41, 544–552 CrossRef CAS PubMed.
- J. Byun, B. Song, K. Lee, B. Kim, H. W. Hwang, M. R. Ok, H. Jeon, K. Lee, S. K. Baek, S. H. Kim, S. J. Oh and T. H. Kim, Identification of urban particulate matter-induced disruption of human respiratory mucosa integrity using whole transcriptome analysis and organ-on-a chip, J. Biol. Eng., 2019, 13, 88 CrossRef PubMed.
- Y. C. T. Huang, E. D. Karoly, L. A. Dailey, M. T. Schmitt, R. Silbajoris, D. W. Graff and R. B. Devlin, Comparison of Gene Expression Profiles Induced by Coarse, Fine, and Ultrafine Particulate Matter, J. Toxicol. Environ. Health, Part A, 2011, 74, 296–312 CrossRef CAS PubMed.
- A. Srivastava, A. Sharma, S. Yadav, S. J. S. Flora, U. N. Dwivedi and D. Parmar, Gene expression profiling of candidate genes in peripheral blood mononuclear cells for predicting toxicity of diesel exhaust particles, Free Radical Biol. Med., 2014, 67, 188–194 CrossRef CAS PubMed.
- E. M. Thomson, D. Vladisavljevic, S. Mohottalage, P. Kumarathasan and R. Vincent, Mapping Acute Systemic Effects of Inhaled Particulate Matter and Ozone: Multiorgan Gene Expression and Glucocorticoid Activity, Toxicol. Sci, 2013, 135, 169–181 CrossRef CAS PubMed.
- P. Solaimani, A. Saffari, C. Sioutas, S. C. Bondy and A. Campbell, Exposure to ambient ultrafine particulate matter alters the expression of genes in primary human neurons, Neurotoxicology, 2017, 58, 50–57 CrossRef CAS PubMed.
- E. M. Wong, W. F. Walby, D. W. Wilson, F. Tablin and E. S. Schelegle, Ultrafine Particulate Matter Combined With Ozone Exacerbates Lung Injury in Mature Adult Rats With Cardiovascular Disease, Toxicol. Sci, 2018, 163, 140–151 CrossRef CAS PubMed.
- Committee on the Medical Effects of Air Pollution (COMEAP). Statement on the evidence for health effects in the travelling public associated with exposure to particulate matter in the London Underground, 2019.
- R. F. Phalen, M. J. Oldham and A. E. Nel, Tracheobronchial particle dose considerations for in vitro toxicology studies, Toxicol. Sci, 2006, 92, 126–132 CrossRef CAS PubMed.
- L. A. Jimenez, J. Thompson, D. A. Brown, I. Rahman, F. Antonicelli, R. Duffin, E. M. Drost, R. T. Hay, K. Donaldson and W. MacNee, Activation of NF-kappa B by PM10 occurs via an iron-mediated mechanism in the absence of I kappa B degradation, Toxicol. Appl. Pharmacol., 2000, 166, 101–110 CrossRef CAS PubMed.
Footnotes |
| † Current address: La Jolla Institute for Allergy and Immunology, La Jolla, CA, USA. |
| ‡ Current address: Merck Exploratory Science Center, Merck Research Laboratories, Cambridge, MA, USA. |
| This journal is © The Royal Society of Chemistry 2020 |