Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein–inorganic nano hybrid sheets of Pd embedded BSA as a robust catalyst in water for oxidase mimic activity and C–C coupling reactions, and as a sustainable material for micromolar sensing of dopamine†
Sirilata
Polepalli
a,
Bhawna
Uttam
a and
Chebrolu Pulla
Rao‡
 *b
*b
aBioinorganic Laboratory, Department of Chemistry, Indian Institute of Technology Bombay, Powai, Mumbai-400 076, India
bDepartment of Chemistry, Indian Institute of Technology Tirupati, Settipalli post, Tirupati-517 506, Andhra Pradesh, India. E-mail: cprao@iittp.ac.in; cprao@iitb.ac.in; Fax: +91 0877 250 3004; Tel: +91 0877 2503036
First published on 17th August 2020
Abstract
Protein–inorganic hybrids with tunable morphology offer excellent applications in several fields due to their versatile characteristics. Herein, we report a new hybrid material prepared using commercially available protein bovine serum albumin (BSA) and a palladium salt. Employing a green process, distinctive colonies of Pd-nanoparticles (Pd_NPs) (2–5 nm) embedded in BSA hybrid sheets (Pd_NP@BSAsheet) were synthesized and characterized using different microscopy techniques. While naked Pd_NPs show larger particles of ∼52 nm in size, the size of the Pd_NPs embedded in BSA sheets is as small as ∼4 nm and they form well separated colonies by providing a much greater number of distinct reaction centers. Further characterization was performed using thermal, spectral and diffraction techniques. Pd_NP@BSAsheet shows excellent oxidase enzyme mimic activity as demonstrated using 3,3′,5,5′-tetramethylbenzidine (TMB) and o-phenylenediamine (OPD) as substrates. The selective detection of the neurotransmitter dopamine by the in situ generated oxidized species of TMB has been demonstrated in buffer, human serum and urine samples to a micromolar detection limit. The hybrid Pd_NP@BSAsheet was also shown to have superior catalytic activity in Suzuki coupling reactions performed in water as compared to several other literature reported Pd-based catalysts and its reusability has been demonstrated. Thus, the hybrid Pd_NP@BSAsheet material is promising and sustainable as an oxidase enzyme mimic, as a biosensor for sensing crucial neurotransmitter dopamine and as a catalyst for C–C coupling in aqueous medium.
Introduction
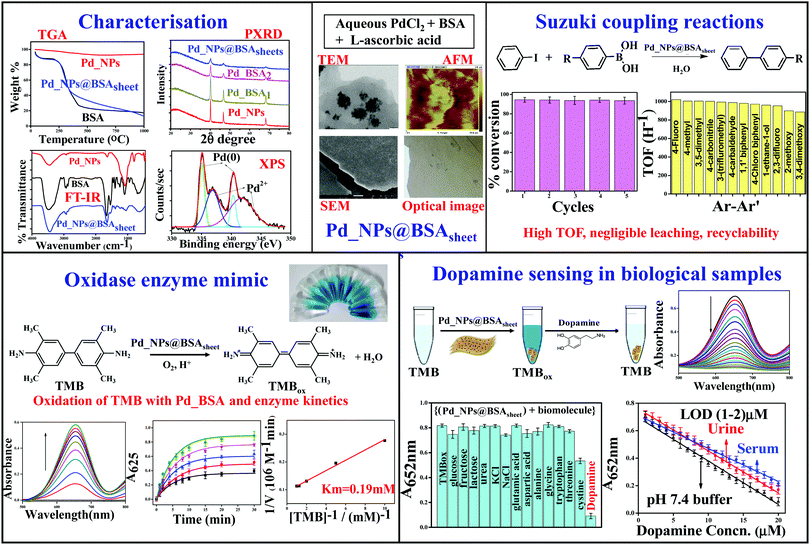
Protein–inorganic hybrid materials have gained immense interest in recent times, due to many desirable properties that they exhibit based on both the inorganic and the protein components present in them. Proteins offer a template for the formation of novel morphological features as the side chains of the amino acid residues provide functional groups suitable for coordination with metal ions. In addition to this, the biocompatibility is an added advantage of naturally occurring proteins. Artificial nanoscale enzymes, viz., nanozymes, have high stability and possesess better catalytic activity as compared to some natural enzymes.1 In spite of several literature reports about inorganic nanozymes,2,3 protein based inorganic materials as artificial enzyme mimics are scarce. The advantages offered by protein–inorganic nanomaterials in different applications like catalysis, sensing and drug delivery have been reviewed recently in the literature.4,5 As protein based materials are water soluble and biocompatible, the corresponding inorganic–protein hybrid materials are in dire need to demonstrate their unusual properties.Bovine serum albumin (BSA) is the most commonly used protein for the preparation of novel inorganic nanohybrid materials. There are a few literature reports on BSA based metal and metaloxide nanoparticles6–10 as artificial enzyme mimics. However, to our knowledge, there is no report on BSA based palladium hybrids with nanosheet morphology. The sheet morphology with Pd_NPs embedded in it provides an increased surface to volume ratio, which results in higher activity. The sheet morphology is important in the controlled synthesis of Pd_NPs in order to result in a greater surface area. Such features are necessary for better performance of the material. Palladium based nanomaterials and gels are versatile as catalysts in organic coupling reactions,11–14 and as enzyme mimics.15–18 In this work, Pd-nanoparticles embedded in hybrid sheets of bovine serum albumin (BSA) were synthesized from aqueous solution in a greener approach with distinct morphological features. Pd_NP@BSAsheet is a new and novel hybrid material of a protein–inorganic system. Such a material is expected to have the characteristics of both the protein and the inorganic components and hence would exhibit unexpected and unprecedented properties. Hence it is important to explore its application potential. To our knowledge, such a sheet morphology material based on BSA and palladium has not been reported earlier. Therefore, such a protein–inorganic hybrid material deserves to be studied for its applications. In the present work, this material has been studied for its enzyme mimic activity, as a sensor and also as a catalyst for C–C coupling. All these aspects are explored in detail in this paper.
Experimental
All the chemicals used in this work were procured from either Sigma Aldrich or Spectrochem Pvt. Ltd, and were used without further purification.Preparation of Pd_NP@BSAsheet
In a typical reaction, 1 mg of PdCl2 was added to 1 mL of aqueous BSA solution having different concentrations (1, 2, and 5 mg), herein referred to as Pd_BSAn (n = 1, 2, 5). Each of these solutions was subjected to vortexing for 8 hours, which resulted in an orange colored suspension, and then was heated at 60 °C in a dry bath for 10 minutes followed by addition of 100 μL of 0.2 M ascorbic acid. The resulting solutions were further heated in a dry bath for 60 minutes to obtain a black suspension, which settles on centrifugation, and washed thoroughly with Milli-Q water 6–7 times in order to remove excess reagents and dried in a desiccator under a vacuum for 3 days.Characterization of the protein–inorganic hybrid Pd_NP@BSAsheet
The protein–inorganic hybrid samples were characterized by spectral, thermal and diffraction methods. The FT-IR spectra were measured on a PerkinElmer infrared spectrometer. X-ray photoelectron spectroscopy (XPS) measurements were performed on a Kraytos Analytical Axis Supra by drop casting the samples on aluminium foil. The thermal analysis of the BSA protein and Pd_NP@BSAsheet was performed on a PerkinElmer Diamond TG/DTA under a nitrogen atmosphere. The samples were heated from 25 to 1000 °C at a rate of 10 °C min−1. For powder X-ray diffraction, Cu Kα radiation of wavelength 1.54 Å was used and scanned over a 2θ range of 5 to 100° on a Panalytical Empyrean instrument.The morphology of the samples was studied by different microscopy techniques. Transmission electron microscopy (TEM) was carried out on a JEOL JEM-2100F, 200 kV. The black suspension (Pd_BSAn) was drop cast on a copper grid and dried in a desiccator under a vacuum. For mapping the elements, scanning transmission electron microscopy (STEM) mode was used. Scanning electron microscopy was performed on a JSM-7600F field emission gun scanning electron microscope (FEG-SEM). The samples for SEM were prepared by drop-casting the Pd_NP@BSAsheet suspension on aluminium foil and dried in a desiccator under a vacuum. The samples were sputtered for 60 s at 10 mA prior to imaging. Atomic force microscopy (AFM) was performed for Pd_NP@BSAsheet using a Veeco Nanoscope IV. Optical microscopy images were taken on a Nikon Eclipse fluorescence microscope.
Oxidase enzyme mimic activity and steady state kinetics of Pd_NP@BSAsheet
The oxidase activity of Pd_NP@BSAsheet was studied using 3,3′,5,5′-tetramethylbenzidine (TMB) and o-phenylenediamine (OPD) as substrates in acetate buffer at pH 4.5. The enzyme kinetics of Pd_NP@BSAsheet was evaluated using varying concentrations of TMB solutions (0.1, 0.2, 0.5, 0.8 and 1 mM) with 100 μg mL−1 of Pd_NP@BSAsheet. The formation of the oxidized TMB (TMBox) was observed by taking the absorption at 652 nm at varying time intervals starting from 1 to 30 minutes. The Michaelis–Menten constant was derived using the Lineweaver–Burk plot: 1/ν = (Km/Vmax) (1/[S] + 1/Km), where v is the initial velocity, Vmax is the maximal reaction velocity, [S] is the substrate concentration and Km is the Michaelis constant (the reciprocal of the reaction rate (1/ν) on the Y-axis and the reciprocal of the substrate concentration (1/[S]) on the X-axis). Similar studies were also carried out at pH = 7.4 using TMB as the substrate.Detection of dopamine using the Pd_NP@BSAsheet_TMBox system
Detection of the neurotransmitter dopamine was performed in different media, i.e., PBS buffer at pH 7.4, human serum and urine samples using the oxidase activity of the Pd_NP@BSAsheet-TMBox system. Firstly, a 1 mM stock solution of dopamine was prepared in buffer at pH 7.4. Then 1 mM TMB solution was oxidized with 100 μg mL−1 Pd_NP@BSAsheet to form TMBox. Dopamine solution was added with increasing concentrations to this reaction mixture containing Pd_NP@BSAsheet-TMBox to observe the decrease in the absorbance of the band at 652 nm. A similar procedure was followed for the experiments performed with human serum and urine samples spiked with dopamine. The selectivity to dopamine among other biologically important molecules was tested by a similar method, using several other interfering bio-relevant molecules, such as glucose, fructose, lactose, urea, K+, Na+, glutamic acid, aspartic acid, alanine, glycine, tryptophan, threonine, and cysteine.C–C coupling reactions with Pd_NP@BSAsheet
To a well-stirred mixture of iodobenzene (55 μL, 0.49 mM), phenylboronic acid (90 mg, 0.74 mM) and K2CO3 (136 mg, 0.98 mM) in water (1.5 mL), 5 mg of the catalyst, Pd_NP@BSAsheet, was added. The reaction mixture was stirred at 80 °C for 8 h. The reaction mixture was extracted with diethyl ether (15 mL) two times. The organic phase was collected and dried over Na2SO4 and concentrated under a vacuum. The crude product obtained was purified by silica-gel column chromatography using ethyl acetate/pet-ether in a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 v/v ratio.
95 v/v ratio.
Results and discussion
Preparation and characterization of Pd_NP@BSAsheet
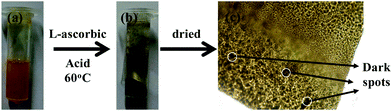
The protein–inorganic Pd_NP@BSAsheet was prepared as given in the experimental section. Aqueous BSA solutions of different concentrations, i.e., 1, 2 and 5 mg mL−1, were used to optimize the sheet formation and the corresponding products are referred to as Pd_BSA1, Pd_BSA2 and Pd_BSA5, respectively. An orange colored suspension was obtained when the reaction mixture was vortexed as seen in Fig. 1a. Treating this with L-ascorbic acid reduces Pd2+ into Pd(0) and the metal is embedded into the BSA sheets. Overall, this is observed as a black suspension with the naked eye as can be noticed from Fig. 1b. Macro sheet formation, visible with the naked eye, was observed only in the case of Pd_BSA5 (Pd_NP@BSAsheet). At lower concentrations of BSA (Pd_BSA1 and Pd_BSA2) the reduction yields a powder material without forming any sheets. Pd_BSA5 acquires a sheet morphology (Pd_NP@BSAsheet) as observed in Fig. 1c and hence all the studies were carried out using Pd_NP@BSAsheet. The hybrid sheets can be observed under a bright field microscope to exhibit a brown uneven surface with darker spots. This catalyst is being referred to as Pd_NP@BSAsheet in this paper.Characterization of Pd_NP@BSAsheet
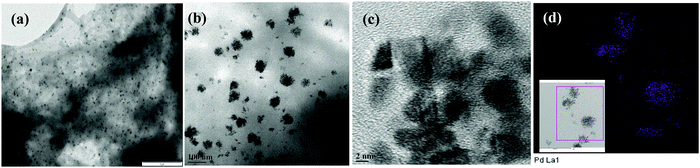
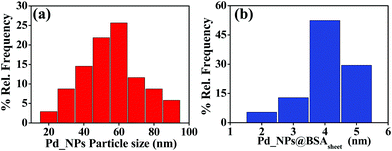
Pd_NP@BSAsheet was characterized by several microscopy techniques. The TEM images of naked Pd nanoparticles (Pd_NPs) without BSA exhibited polydispersity where the size of the particles ranges from 20 to 100 nm due to the lack of any capping agent (Fig. S01a, ESI†). Interestingly, in the presence of BSA, a change in the morphology of Pd_NP formation was observed. While Pd_BSA1 exhibited the formation of spherical aggregates (Fig. S01b, ESI†), Pd_BSA2 exhibited the initial formation of smaller sheets as observed in Fig. S01c (ESI†). When the BSA concentration is 5 mg mL−1 (Pd_NP@BSAsheet), well-formed sheets can be observed in Fig. 2a with small colonies of Pd_NPs embedded in the BSA sheets (Fig. 2b and Fig. S02a–c, ESI†). Another distinctive feature seen in Pd_NP@BSAsheet is the substantial decrease in the size of palladium nanoparticles embedded in the sheets and the size is ∼4 nm (Fig. 2d and Fig. S02d–g, ESI†). Literature reports suggest that the presence of a high percent of tyrosine and histidine residues will result in the nucleation and the growth of the palladium nanoparticles because of their coordination to Pd2+.12,19 The amino acid composition of BSA has approximately 3.4% tyrosine and 2.9% histidine residues. The side chains of these amino acids are responsible for the binding of Pd2+ followed by the formation of characteristic colonies where the protein also acts as a template. Proteins and peptides form different types of nanostructures, such as fibrils, tubes and sheets, under the influence of metal ions.20,21 Amyloidogenic proteins like BSA and lysozyme form self-assembled β-sheet rich regions in one dimension to form fibrils, which in turn assemble further in the second dimension to give nanosheets.22 From EDS (Fig. S03a and b, ESI†) and elemental mapping, the presence of palladium concentrated in the aggregate can be clearly noticed in Fig. 2d, while the other elements (C, N, O, and S) are seen distributed all over the sheet (Fig. S03c–h, ESI†).The particle size distribution of naked Pd_NPs and that of Pd_NPs present in the hybrid Pd_NP@BSAsheet are given in Fig. 3 in order to appreciate the differences observed between these two types of Pd_NPs. Most of the naked Pd_NPs are in the size range of 20–90 nm (Fig. 3a) with a weighted average of ∼52 nm. However, the size of the Pd_NPs present in Pd_NP@BSAsheet exhibits a narrow size distribution of 2–5 nm (Fig. 3b) and these form dense colonies. Thus, the Pd_NPs present in Pd_NP@BSAsheet are one tenth the size of the naked Pd_NPs and are expected to show better catalytic efficiency.
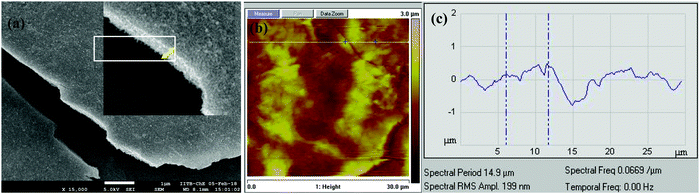
The hybrid Pd_NP@BSAsheet was characterized using other microscopy techniques, such as AFM and SEM. The SEM image and the cross-sectional features given in Fig. 4a and Fig. S04d (ESI†) further confirm that the thickness is ∼250 nm. The AFM images given in Fig. 4b and Fig. S04b (ESI†) show a distinct sheet morphology with a height of 250 nm as noticed from the profile given in Fig. 4c. The AFM and SEM images of the naked Pd_NPs reveal varying sizes (Fig. S04a and c, ESI†) as already noticed even from the TEM images.
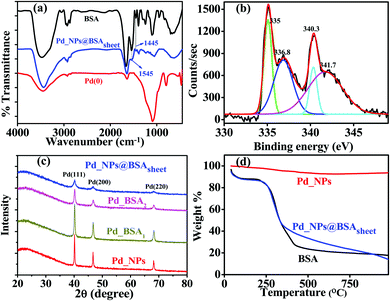
The nano hybrid Pd_NP@BSAsheet was characterized using several techniques. The presence of the protein in Pd_NP@BSAsheet was confirmed by FTIR analysis, where the amide vibrational peaks are observed at 1650, 1545 and 1445 cm−1 (Fig. 5a). The high resolution XPS spectrum of Pd_NP@BSAsheet of Pd 3d shows 3d5/2 and 3d3/2 peaks for Pd(0) at 335.0 and 340.3 eV, respectively, with the partial presence of Pd2+, resulting from the peaks observed at 336.8 (3d5/2) and 341.7 eV (3d5/2) with an area ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 of Pd(0)
3 of Pd(0)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd2+ (Fig. 5b). The PXRD pattern for Pd_NP@BSAsheet seen from Fig. 5c shows peaks arising from (111), (200) and (220) of Pd, with a decrease in crystallinity as the concentration of BSA increases from 1 to 5 mg mL−1. Thermogravimetric analysis of Pd_NP@BSAsheet suggests that these sheets are thermally stable till about 250 °C, with a weight loss of 70% due to the degradation of the organic component at 450 °C. Comparison of this with the control systems (BSA and Pd_NPs) showed that the Pd_NP@BSAsheet sheets are loaded to an extent of ∼11% Pd (Fig. 5d).
Pd2+ (Fig. 5b). The PXRD pattern for Pd_NP@BSAsheet seen from Fig. 5c shows peaks arising from (111), (200) and (220) of Pd, with a decrease in crystallinity as the concentration of BSA increases from 1 to 5 mg mL−1. Thermogravimetric analysis of Pd_NP@BSAsheet suggests that these sheets are thermally stable till about 250 °C, with a weight loss of 70% due to the degradation of the organic component at 450 °C. Comparison of this with the control systems (BSA and Pd_NPs) showed that the Pd_NP@BSAsheet sheets are loaded to an extent of ∼11% Pd (Fig. 5d).
Oxidase mimic activity of Pd_NP@BSAsheet
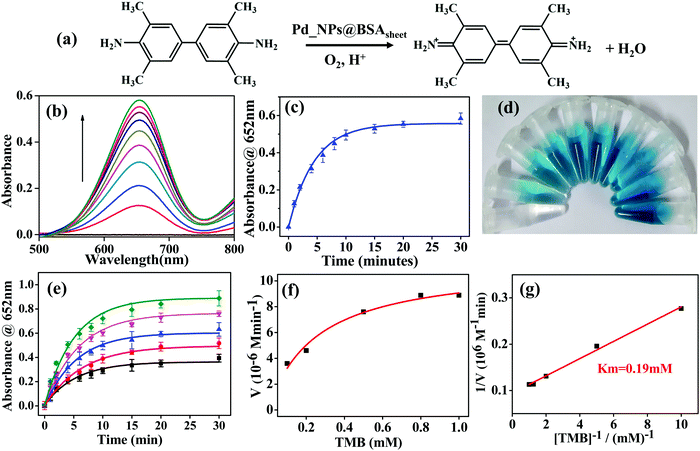
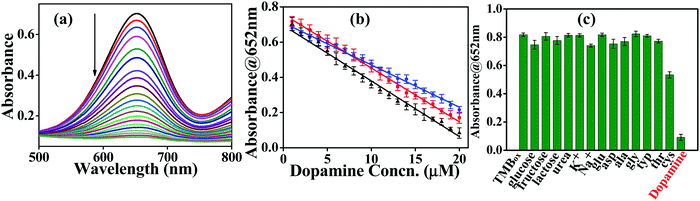
The oxidase activity of Pd_NP@BSAsheet was investigated using TMB as a substrate (Fig. 6a). Pd_NP@BSAsheet catalyzes the oxidation of TMB with no addition of H2O2 to give a blue colored product (TMBox) which shows an absorption maximum at 652 nm (Fig. 6b), and thus acts as an oxidase enzyme mimic. The oxidase activity at different time intervals in the range of 1 to 30 min can be seen from Fig. 6c. A gradual increase in the blue color with increasing time of the reaction can be noticed from Fig. 6d. Time dependent oxidation of the TMB substrate using Pd_NP@BSAsheet as a catalyst was performed to evaluate the enzyme kinetics using the Michaelis–Menten equation and Lineweaver–Burk plot. From the absorbance vs. time plot (Fig. 6e), the initial velocity was estimated by taking ε of TMBox as 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 for the band at 652 nm as given in the literature.23 Comparing the Km value of Pd_NPs@BSAsheet with several literature reports shows the superior oxidase enzyme mimic activity of the system (Table 2). The plot of the reaction velocity vs. substrate concentration showed a typical Michaelis–Menten saturation curve (Fig. 6f). From the double-reciprocal Lineweaver–Burk plot, the Michaelis–Menten parameters were estimated to be Vmax = 10.5 × 10−6 M min−1 and Km = 0.19 mM (Fig. 6g). The oxidase mimic activity of Pd_NP@BSAsheet was also studied at pH 7.4 and the data are given in Fig. S05a–c (ESI†). The specific activity of Pd_NP@BSAsheet is 179.2 U mL−1, which is higher as compared to the natural enzyme laccase (106.38 U mL−1),24 suggesting better activity for this material. Thus, the data suggest that the Pd_NP@BSAsheet nano hybrid acts as a better oxidase enzyme mimic.
000 M−1 cm−1 for the band at 652 nm as given in the literature.23 Comparing the Km value of Pd_NPs@BSAsheet with several literature reports shows the superior oxidase enzyme mimic activity of the system (Table 2). The plot of the reaction velocity vs. substrate concentration showed a typical Michaelis–Menten saturation curve (Fig. 6f). From the double-reciprocal Lineweaver–Burk plot, the Michaelis–Menten parameters were estimated to be Vmax = 10.5 × 10−6 M min−1 and Km = 0.19 mM (Fig. 6g). The oxidase mimic activity of Pd_NP@BSAsheet was also studied at pH 7.4 and the data are given in Fig. S05a–c (ESI†). The specific activity of Pd_NP@BSAsheet is 179.2 U mL−1, which is higher as compared to the natural enzyme laccase (106.38 U mL−1),24 suggesting better activity for this material. Thus, the data suggest that the Pd_NP@BSAsheet nano hybrid acts as a better oxidase enzyme mimic.
| Substrate used | Product obtained (number) |
|---|---|
| Phenyl boronic acid | 1,1′-Biphenyl (1a) |
| 4-Fluorophenyl boronic acid | 4-Fluoro-1,1′-biphenyl (1b) |
| 4-Chlorophenyl boronic acid | 4-Chloro-1,1-biphenyl (1c) |
| 4-Methylphenyl boronic acid | 4-Methyl-1,1′-biphenyl (1d) |
| 4-Formylphenyl boronic acid | [[1,1′-Biphenyl]-4-carbaldehyde] (1e) |
| 4-Acetylphenyl boronic acid | 1-{[1-1′Biphenyl]-4-yl}ethane-1-one (1f) |
| 4-Cyanophenyl boronic acid | [1,1′-Biphenyl]-4-carbonitrile (1g) |
| 2-Methoxyphenyl boronic acid | 2-Methoxy-1,1′-biphenyl (1h) |
| 3-(Trifluoromethyl)phenyl boronic acid | 3-(Trifluoromethyl)-1,1′-biphenyl (1i) |
| 3,5-Dimethylphenyl boronic acid | 3,5-Dimethyl-1,1′-biphenyl (1j) |
| 3,4-dimethoxyphenyl boronic acid | 3,4-Dimethoxy-1,1′-biphenyl (1k) |
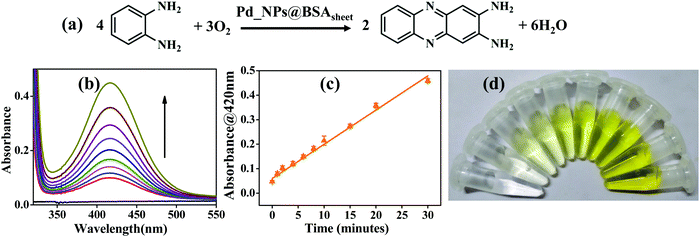
In addition to the study carried out with TMB, the oxidase activity was also evaluated using o-phenylenediamine (OPD) as a substrate with Pd_NP@BSAsheet as a catalyst (Fig. 7a). With an increase in time, the absorbance of the oxidised OPD (OPDox) observed at 420 nm increases as noticed from Fig. 7b. The absorption peak for OPDox has been reported in the literature.18 A plot of absorbance vs. time shows a linear fit as given in Fig. 7c. The gradual colour change of OPDox can also be seen from the photograph of the vials given in Fig. 7d.
Detection of dopamine using the Pd_NP@BSAsheet_TMBox system
Dopamine is an important neurotransmitter in the human body with a critical role in neural networks and motor functions. Abnormal levels of dopamine are responsible for life threatening disorders like Parkinson's disease,29 dementia and clinical depression.30 Although there are several literature reports for efficient detection of dopamine,31–33 a facile colorimetric method for its sensing in bodily fluids with no necessity of H2O2 is always in demand. Utilizing the oxidase activity of Pd_NP@BSAsheet, a colorimetric method has been developed using the Pd_NP@BSAsheet_TMBox system for easy detection of dopamine in biological samples such as human serum and urine. The absorption peak at 652 nm corresponding to TMBox decreases with increasing concentration of dopamine (Fig. 8a) and the blue color gradually diminishes. A plausible mechanism for the detection of dopamine by Pd_NP@BSAsheet is the reduction of TMBox to TMB with concurrent oxidation of dopamine to aminochrome, which can be monitored spectrophotometrically.34 A linear relationship has been observed between the concentration of dopamine (in the range of 1–20 μM) and the absorbance of TMBox at 652 nm in all three cases, viz., pH 7.4 buffer, urine, and serum samples (Fig. 8b), and the corresponding limit of detection (LOD) comes out to be 1.01, 1.14 and 1.77 μM, respectively. The LOD was calculated based on the literature reported procedure of 3σ/s, where ‘s’ is the slope of the calibration curve and ‘σ’ is the SD of the response.35 The efficiency of the Pd_NP@BSAsheet_TMBox system used for sensing dopamine in the present case is better than many of the reported ones given in Table 3. Ours is the only catalyst which functions without the use of H2O2. In order to test the greater selectivity for dopamine by Pd_NP@BSAsheet_TMBox, sensing studies were performed with several biologically relevant molecules, including sugars and amino acids as noticed from the histogram given in Fig. 8c. Thus, Pd_NP@BSAsheet_TMBox can be used as a simple and efficient colorimetric method for the detection of dopamine in biologically relevant media.| Material | Method of sensing | Linear range (μM) | Limit of detection (μM) | Ref. |
|---|---|---|---|---|
| Polydimethylsiloxane (PDSM)/glass microfluidic chip | Electrochemical | 10–500 | 3.92 | 36 |
| Reduced graphene oxide–Au nanocage composite | Electrochemical | 5–250 | 3 | 37 |
| Reduced graphene oxide–Ag nanoparticles | Electrochemical | 10–800 | 5.4 | 38 |
| Gold nanoparticles | Colorimetric | 2.5–20 | 2.5 | 39 |
| Silver nanoparticles | Colorimetric | 3.2–20 | 1.2 | 40 |
| Pd_NP@BSAsheet | Colorimetric | 1–20 | 1.01 | This work |
Pd_NP@BSAsheet as a heterogeneous catalyst for C–C coupling in water
Organic C–C coupling reactions are generally performed with palladium-based catalysts under an inert atmosphere using organic solvents to a large extent. However, when palladium is converted into a nano-form in the presence of BSA to result in Pd_NP@BSAsheet, this material turned out to be an alternate, heterogenous catalyst to perform Suzuki coupling reactions in aqueous solution with very high efficiency. This is a green, protein-based unique catalyst suitable for C–C coupling reactions in water. The reaction conditions were optimized for the formation of product 1a (Table 1) by varying the catalyst amount, temperature and time (Fig. S06a–c, ESI†) and it was found that 5 mg of catalyst (0.059 mol% and 6.3 ppm of Pd) at 80 °C and 8 hours of reaction time are optimum to obtain a high product yield. The possibility of leaching was tested by using the left-over mother liquor of the reaction mixture in place of the catalyst. The yield of the biphenyl product when the mother liquor was used as the catalyst was as low as 2%, suggesting that almost no Pd is leached from Pd_NP@BSAsheet. To further explore the substrate scope, a library of substituted phenylboronic acid derivatives with different functionalizations was used in the cross-coupling reactions with Pd_NP@BSAsheet as a catalyst. Both the electron donating and electron withdrawing groups showed high conversion rates for the C–C coupled products and this is evident from Fig. SI07 and Table S01 (ESI†) and the characterisation data of the C–C coupled products, including GC-MS data for all the coupled products, are given in Fig. SI08 and SI09 (ESI†).Recyclability of Pd_NP@BSAsheet
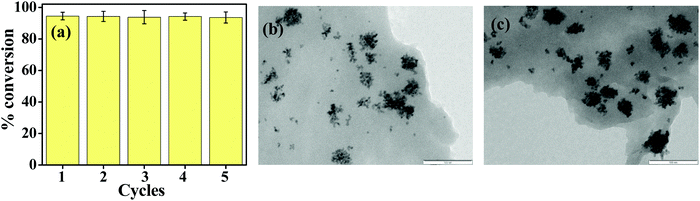
Pd_NP@BSAsheet was regenerated by separating it from the reaction mixture and washing several times with water followed by drying it in a desiccator. The regenerated Pd_NP@BSAsheet was reused and demonstrated for five consecutive cycles of C–C coupling and from the histogram no significant loss of catalytic activity could be seen (Fig. 9a). The morphology of Pd_NP@BSAsheet after 5 cycles was studied by TEM microscopy. Comparison of this (Fig. 9b) with that of the fresh Pd_NP@BSAsheet (Fig. 9c) shows that the sheet morphology is intact and the Pd_NPs are distributed on it with the same characteristics in both cases. Thus, the nano hybrid Pd_NP@BSAsheet is facile to use and separate from the reaction mixture, and is recyclable with high TOF values and negligible leaching.The efficiency of Pd_NP@BSAsheet as a green catalyst in Suzuki coupling reactions is compared with several literature reports as given in Table 4. In order to make this table, only those materials synthesized based on bio-molecules or biological extracts are considered for comparison. To our knowledge there is no previous report on a complete protein based Pd catalyst with a hybrid sheet morphology. As observed from the table, these sheets show better activity even at a low catalyst mol% in water, while most of the other reported reactions use large concentrations of the catalyst. Thus, Pd_NP@BSAsheet acts as a sustainable, recyclable and green catalyst for Suzuki coupling reactions to take place in water.
| Material | Catalyst mol% | Solvent | Temperature | % yield | Ref. |
|---|---|---|---|---|---|
| Pd NPs with Pulicaria glutinosa extract | 5 | H2O/SDS | 100 °C | ∼100 | 41 |
| Au/Pd bimetallic NPs with Euphorbia condylocarpa root extract | 0.2 | H2O | 80 °C | 92 | 42 |
| Pd NPs with hearth wood extract of Artocarpus lakoocha | 0.5 | H2O | 80 °C | 94 | 43 |
| Pd/TiO2 NPs with Myrtus communis leaf extract | 0.7 | NMP: H2O | 120 °C | 98 | 44 |
| Pd NPs with cationic cellulose nanofibers | 0.02 | DMF | 110 °C | 98 | 45 |
| Pd-DNA-Fe3O4 | 9.4 | Deep eutectic solvent | 85 ± 5 °C | 92.2 | 46 |
| Pd_NP@BSAsheet | 0.059 | H2O | 80 °C | 98.5 | This work |
Conclusions and comparisons
An unprecedented protein–inorganic hybrid material, Pd_NP@BSAsheet, has been developed and we demonstrated successfully three of its applications spanning across different areas, as an artificial enzyme mimic, and in sensing and C–C coupling catalysis. The reaction conditions were optimized in order to obtain the sheet morphology for the protein, BSA. The templated formation of such a sheet morphology controlled the size as well as the distribution of the embedded Pd_NPs. The distribution of Pd_NPs in the sheet resembles the formation of colonies filled with NPs of ∼4 nm size with monodispersity, while these are aggregated to give NPs of ∼60 nm in the absence of such protein templation, suggesting the role of BSA in the formation of this protein–inorganic hybrid nanosheet material. All this was clearly demonstrated through extensive characterization performed using TEM, SEM, AFM, PXRD, FT-IR, XPS and TGA analysis. The hybrid nanosheets showed excellent oxidase enzyme mimic activity as demonstrated using TMB and OPD substrates. The kinetics study gave Vmax and Km values of 10.5 × 10−6 M min−1 and 0.19 mM, respectively. The oxidized TMB present in the reaction mixture (Pd_NP@BSAsheet_TMBox) exhibits selective detection of neurotransmitter dopamine present in pH 7.4 buffer, human serum and urine samples. The LOD was 1–2 μM in all the three media, suggesting that the protein–inorganic hybrid catalyst would work well in biological systems. Compared to various literature reported sensors, the present one, viz., Pd_NP@BSAsheet_TMBox, showed superior sensing ability without the use of any H2O2 as can be noticed from the data given in Table 3. Pd_NP@BSAsheet also acts as a sustainable green catalyst for Suzuki coupling reactions in water. For all the 11 different substrates, it showed >90% catalytic activity with high TOF values. Recyclability of the C–C coupling was demonstrated for 5 cycles and the sheet morphology of the catalyst was intact even after 5 cycles as observed based on TEM, supporting that the material is robust, recoverable and reusable. Pd_NP@BSAsheet possesses higher catalytic ability even at a low mol% of catalyst in water as compared to several bio-based materials given in Table 4. The green synthesis and the application aspects of the protein inorganic nano hybrid material, Pd_NP@BSAsheet, are summarized in Fig. 10. Thus, Pd_NP@BSAsheet nanosheets find useful applications, in chemical biology as an enzyme mimic, in analytical biochemistry to sense micromolar concentrations of neurotransmitter dopamine and in organic synthesis to give C–C coupled products even in aqueous reactions with recyclability, and hence are a versatile and sustainable hybrid nanobiomaterial.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
CPR acknowledges Department of Science and Technology for the J. C. Bose National Fellowship (SB/S2/JCB/066/2015) and IIT Tirupati for providing an MHRD Professorship. CPR expresses his gratitude to IIT Bombay for fostering his academic and research activities for over three decades. S. P. acknowledges INSPIRE-DST for the fellowship {IF150417} and B. U. acknowledges CSIR for the award of a Senior Research Fellowship {09/087(0835)/2015-EMR-I}. We thank the central facilities of IIT Bombay (SAIF and IRCC) for TEM, SEM, AFM TGA, and XPS and department central facilities. We thank Ms Aekta Upadhyay and Mr Rahul Nag for helping with measuring the SEM and TEM micrographs, respectively.References
- H. Li, T. Wang, Y. Wang, S. Wang, P. Su and Y. Yang, Ind. Eng. Chem. Res., 2018, 57, 2416–2425 CrossRef CAS.
- L. Song, Y. Zhu, Z. Yang, C. Wang and X. Lu, J. Mater. Chem. B, 2018, 6, 5931–5939 RSC.
- A. Tripathi, K. D. Harris and A. L. Elias, ACS Omega, 2020, 5, 9123–9130 CrossRef CAS.
- R. Wang, Y. Zhang, D. Lu, Z. Liu and R. Zare, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 320–328 CAS.
- X. Wu, M. Hou and J. Ge, Catal. Sci. Technol., 2015, 5, 5077 RSC.
- X. Liu, Q. Wang, H. Zhao, L. Zhang, Y. Su and Y. Lv, Analyst, 2012, 137, 4552–4558 RSC.
- X. Liu, Q. Wang, Y. Zhang, L. Zhang, Y. Su and Y. Lv, New J. Chem., 2013, 37, 2174–2178 RSC.
- H. Zang, L. Han and F. Li, Sens. Actuators, B, 2019, 286, 460–467 CrossRef.
- S.-B. He, P. Balasubramanian, Z.-W. Chen, Q. Zhang, Q.-Q. Zhuang, H.-P. Peng, H.-H. Deng, X.-H. Xia and W. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 14876–14883 CrossRef CAS.
- L. Han, H. Zhang, D. Chen and F. Li, Adv. Funct. Mater., 2018, 28, 1800018 CrossRef.
- M. Maity and U. Maitra, J. Mater. Chem. A, 2014, 2, 18952–18958 RSC.
- Y.-O. Kim, H.-S. Jang, Y.-H. Kim, J. M. You, Y.-S. Park, K. Jin, O. Kang, K. T. Nam, J. W. Kim, S.-M. Lee and Y.-S. Lee, RSC Adv., 2015, 5, 78026–78029 RSC.
- B. J. Gallon, R. W. Kojima, R. B. Kaner and P. L. Diaconescu, Angew. Chem., Int. Ed., 2007, 46, 7251–7254 CrossRef CAS.
- N. Narkhede, B. Uttam and C. P. Rao, ACS Omega, 2019, 4, 4908–4917 CrossRef CAS.
- Q. Wang, L. Zhang, C. Shang, Z. Zhang and S. Dong, Chem. Commun., 2016, 52, 5410–5413 RSC.
- X. Xia, J. Zhang, N. Lu, M. J. Kim, K. Ghale, Y. Xu, E. McKenzie, J. Liu and H. Ye, ACS Nano, 2015, 9, 9994–10004 CrossRef CAS.
- L. Rastogi, D. Karunasagar, R. B. Sashidhar and A. Giri, Sens. Actuators, B, 2017, 240, 1182–1188 CrossRef CAS.
- N. Narkhede, B. Uttam and C. P. Rao, Inorg. Chim. Acta, 2018, 483, 337–342 CrossRef CAS.
- R. Coppage, J. M. Slocik, B. D. Briggs, A. I. Frenkel, R. R. Naik and M. R. Knecht, ACS Nano, 2012, 6, 1625–1636 CrossRef CAS.
- K. Tabbasum and C. P. Rao, RSC Adv., 2015, 5, 16828–16836 RSC.
- K. Tabbasum and C. P. Rao, RSC Adv., 2014, 4, 53044–53054 RSC.
- T. P. J. Knowles, T. W. Oppenheim, A. K. Buell, D. Y. Chirgadze and M. E. Welland, Nat. Nanotechnol., 2010, 5, 204–207 CrossRef CAS.
- M. N. Ivanova, E. D. Grayfer, E. E. Plotnikov, L. S. Kibis, G. Darabdhara, P. K. Boruah, M. R. Das and V. E. Fedorov, ACS Appl. Mater. Interfaces, 2019, 11, 22102–22112 CrossRef CAS.
- N. Gaur, K. Narasimhulua and Y. Pydisetty, RSC Adv., 2018, 8, 15044–15055 RSC.
- H. Song, Z. Li, Y. Peng, X. Li, X. Xu, J. Pan and X. Niu, Analyst, 2019, 144, 2416–2422 RSC.
- J. Wang, Y. Wang, D. Zhang and C. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 29614–29624 CAS.
- L. Jiang, S. Fernandez-Garcia, M. Tinoco, Z. Yan, Q. Xue, G. Blanco, J. J. Calvino, A. B. Hungria and X. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 18595–18608 CrossRef CAS.
- C.-J. Yu, T.-H. Chen, J.-Y. Jiang and W.-L. Tseng, Nanoscale, 2014, 6, 9618–9624 RSC.
- J. Segura-Aguilar, I. Paris, P. Muñoz, E. Ferrari, L. Zecca and F. A. Zucca, J. Neurochem., 2014, 129, 898–915 CrossRef CAS.
- A. Zhang, J. L. Neumeyer and R. J. Baldessarini, Chem. Rev., 2007, 107, 274–302 CrossRef CAS.
- H. B. Wang, Y. Li, G. L. Dong, T. Gan and Y. M. Liu, New J. Chem., 2017, 41, 14364–14369 RSC.
- S. Dutta, C. Ray, S. Mallick, S. Sarkar, Y. Negishi and T. A. Pal, J. Phys. Chem. C, 2015, 119, 23790–23800 CrossRef CAS.
- M. N. Ivanova, E. D. Grayfer, E. E. Plotnikov, L. S. Kibis, G. Darabdhara, P. K. Boruah, M. R. Das and V. E. Fedorov, ACS Appl. Mater. Interfaces, 2019, 11, 22102–22112 CrossRef CAS.
- J. Segura-Aguilar and I. Paris, in Handbook of Neurotoxicity, ed. R. Kostrzewa, Springer, New York, 2014, vol. 2, ch. 9, pp. 865–884 Search PubMed.
- V. Thomsen, D. Schatzlein and D. Mercuro, Spectroscopy, 2008, 18, 112–114 Search PubMed.
- J.-S. Liu, Q.-L. Xiao, D. Ge, Y.-Y. Zhang, W.-Z. Zhang, Z. Xu, C. Liu and L.-D. Wang, Chin. J. Anal. Chem., 2015, 43, 977–982 CAS.
- W. H. Li, L. Ma, B. X. Wu, Y. Zhang and Z. A. Li, Anal. Methods, 2017, 9, 3819–3824 RSC.
- B. Kaur, T. Pandiyan, B. Satpati and R. Srivastava, Colloids Surf., B, 2013, 111, 97–106 CrossRef CAS.
- R. Baron, M. Zayats and I. Willner, Anal. Chem., 2005, 77, 1566–1571 CrossRef CAS.
- M. R. H. Nezhad, J. Tashkhourian, J. Khodaveisic and M. R. Khoshi, Anal. Methods, 2010, 2, 1263–1269 RSC.
- M. Khan, M. Khan, M. Kuniyil, S. F. Adil, A. Al-Warthan, H. Z. Alkhathlan, W. Tremel, M. N. Tahir and M. R. H. Siddiqui, Dalton Trans., 2014, 43, 9026–9031 RSC.
- M. Nasrollahzadeh, M. S. Sajadi, A. Rostami-Vartooni and M. Khalaj, RSC Adv., 2014, 4, 43477–43484 RSC.
- D. Baruah, R. N. Das, S. Hazarika and D. Konwar, Catal. Commun., 2015, 72, 73–80 CrossRef CAS.
- M. Nasrollahzadeh and M. S. Sajadi, J. Colloid Interface Sci., 2016, 465, 121–127 CrossRef CAS.
- Z. Jebali, A. Granados, A. Nabili, S. Boufi, A. M. B. do Rego, H. Majdoub and A. Vallribera, Cellulose, 2018, 25, 6963–6975 CrossRef CAS.
- S. Chakraborty, M. H. Mruthunjayappa, K. Aruchamy, N. Singh, K. Prasad, D. Kalpana, D. Ghosh, N. S. Kotrappanavar and D. Mondal, ACS Sustainable Chem. Eng., 2019, 7, 14225–14235 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: The supporting information contains TEM data, EDS and elemental mapping data, AFM and SEM images, optimization of the Suzuki coupling reaction conditions, the substrate scope of the Pd_NP@BSAsheet catalyst in Suzuki coupling reactions and 1H-NMR spectral data of the cross coupled products. See DOI: 10.1039/d0ma00512f |
| ‡ The corresponding author spent over three decades and held Institute Chair Professorship at IIT Bombay till mid 2019. |
| This journal is © The Royal Society of Chemistry 2020 |