Cuscuta chinensis flavonoids down-regulate the DNA methylation of the H19/Igf2 imprinted control region and estrogen receptor alpha promoter of the testis in bisphenol A exposed mouse offspring
Yuanyuan
Wei†
a,
Chao
Han†
a,
Shuying
Li
a,
Yuqing
Cui
a,
Yongzhan
Bao
ab and
Wanyu
Shi
 *ab
*ab
aInstitute of Traditional Chinese Veterinary Medicine, Agricultural University of Hebei, Baoding 071001, China. E-mail: shiwanyu2010@126.com; Tel: +86-138-0326-9157
bHebei Provincial Engineering Center for Chinese Veterinary Herbal Medicine, Baoding 071000, China
First published on 2nd January 2020
Abstract
Exposure to the emerging contaminant bisphenol A (BPA) is ubiquitous and associated with reproductive disorders. The BPA effect as an endocrine disruptor is widely known but other mechanisms underlying developmental disease, such as epigenetic modifications, still remain unclear. The objective of this study was to investigate whether Cuscuta chinensis flavonoids (CCFs) can be used as a dietary supplement to reverse BPA-induced epigenetic disorders, by analyzing the molecular processes related to BPA impairment of testicular development. BPA and different concentrations of CCFs were administered to the dams at gestation day (GD) 0.5–17.5. The testis and serum of male mice were collected at postnatal day (PND) 21 and PND 56 for the detection of related indicators. Our results showed that compared with the BPA group, CCFs could significantly increase the serum contents of testosterone (T), estradiol (E2) in males at PND 21 and PND 56, as well as the contents and transcription levels of DNA methyltransferase 3A (Dnmt3A), Dnmt3B in males at PND 21 and that of estrogen receptor alpha (ERα) at PND 56. The expressions of Dnmt1 and ERα at PND 21 and ERβ at both PND 21 and PND 56 in males were significantly decreased with the administration of different concentrations of CCFs (P < 0.01 or P < 0.05). CCFs also significantly inhibited the BPA-induced hypermethylated status of the ERα promoter and H19/Igf2 imprinting control region (ICR) in the testis at PND 56. These results indicated that CCFs could decrease the methylation levels of ERα and H19/Igf2 genes by inhibiting the expression of DNA methyltransferases (DNMTs), thereby decreasing the levels of reproductive hormones and receptors in adult males, and ultimately alleviating the negative effect of BPA on testicular development in male mice.
1. Introduction
Early-life dietary nutrition has a significant impact on the developmental fate of individuals and disease susceptibility through an altered epigenome.1 Throughout the developmental period epigenetic marks are laid down to maintain proper expression of imprinted genes.2 Genomic imprinting results in monoallelic, parent-of-origin dependent expression of a small but significant subset of autosomal genes in Therian mammals.3 This unique gene expression regulation is controlled by DNA methylation, histone modification and non-coding RNAs. Abnormal methylation or loss of methylation can shut down these critical genes or lead to overexpression of gene products. Because of their monoallelic expression, imprinted genes are particularly vulnerable to the de-regulation of expression caused by an epigenetic distortion.2 In primordial germ cells, genome-wide demethylation eliminates previous parent-specific methylation marks that regulate imprinted gene expression. Following this erasure, methylation patterns in imprinted genes are re-established in a sex-specific manner, earliest in developing male germ cells. Imprinted genes maintain their primary methylation marks throughout life and during the epigenomic re-establishment that follows fertilization of the next generation.4 Imprinting is vulnerable to deregulation at the time of primary imprint mark erasure and establishment during gametogenesis. Therefore, the environmental factors that alter these gene imprints will lead to serious developmental disorders and enhanced susceptibility to disease.5 The instability of epigenome allows it to respond and adapt to environmental stressors and can profoundly affect phenotypes.2 Maternal diet has been shown consistently to alter the epigenome of the offspring, producing different phenotypes and altered disease susceptibilities.6BPA is a chemical material often added to plastics and inner coatings of food and beverage containers7 and can leach out of these products when exposed to high temperatures.8 As a kind of environmental endocrine disruptor (EED),9 BPA can adversely affect the later development of an individual by affecting the expression of genes in the body. Prenatal exposure to environmental contaminants like BPA has adverse effects on the epigenome of the offspring, and parental epigenetic dietary supplementation prevents environment pollution-induced adverse effects.6 Certain bioactive compounds in “epigenetics diets” may provide clinical benefits in offspring through inhibiting histone deacetylases (HDACs) or DNA methyltransferases (DNMTs).10,11
The flavonoids of Cuscuta chinensis (CCFs) are the main components of the Chinese herbal medicine Cuscuta chinensis. Modern researchers found that CCFs could increase the weights of the testis and epididymis in immature rats and promote the secretion of testosterone in rat Leydig cells.12,13 Studies have found that the flavonoid component of the Cuscuta chinensis—aspartame has the function of improving reproductive endocrine.12 The results of a sperm capillarity test and a hypotonic swelling test conducted for evaluating sperm motility and membrane function showed that CCFs had a promoting effect on sperm motility and membrane function.14 It is suggested that CCFs may be a dietary supplement to alleviate the toxic effects of BPA by reducing the apoptosis of testicular cells.15 However, little is known about the effect of flavonoids on DNA methylation. DNA methylation is one of the most important manifestations of epigenetic regulation, controlled by DNMTs, and affects the expression of imprinted genes. The imprinted gene has the specificity of temporal and spatial expression, and plays an important role in the growth and development of animals in different tissues and cells. There are different imprint genes, and their roles are not the same.16 The function of many imprinted genes is related to the growth and development of individuals and the occurrence of tumours. Abnormal expression of imprinted genes often triggers a variety of diseases with complex mutations and phenotypic defects as well as tumours,17 thereby affecting the epigenetic pattern of the body, genetic imprinting, etc. Tollefsbol's lab created the term “epigenetics diet” in 2011. It refers to a class of biologically active dietary compounds such as isothiocyanates in broccoli, genistein in soybeans, resveratrol in red grapes, and other common foods. They have all been shown to alter the epigenome, resulting in healthy results.18 Epigenetic diets can inhibit the progression of certain developmental diseases by modulating epigenetic modification enzymes such as DNMTs and histone deacetylases, as well as certain non-coding RNAs.18,19
The mammalian epigenome undergoes two major cycles of elimination and remodelling, including gametogenesis and early embryogenesis, during which the epigenome is susceptible to environmental factors.20–22 The absence or abnormality of DNMTs leads to the abnormality of the imprint centre of genomic imprinting (i.e. H19/Igf2), which will hinder the normal embryonic development.23 Moreover, ERα plays a vital role in reproduction.24 Therefore, to investigate the effects of CCFs on the reproductive toxicity induced by BPA, a BPA exposure model of embryonic mice was established and CCF treatment was carried out. The methylation of ERα and H19/Igf2 genes in the testis of F1 female mice at two time points of weanling (PND 21) and sexual maturity (PND 56) was detected, along with the changes of hormones and DNMTs.
2. Materials and methods
2.1. Chemicals and reagents
Cuscuta chinensis flavonoids (purity ≥ 85%, used corn oil to dissolve CCFs) were purchased from Nanjing Zelang Biotechnology Co., Ltd (Nanjing, China); BPA (>99% purity) and corn oil were purchased from Sigma-Aldrich Inc. (Louis, MO, USA). The Eastep® Super Total RNA Extraction Kit, Taq® PCR Master Mix, and GoScript™ Reverse Transcription System were obtained from Promega (Beijing) Biotech Co. Ltd (Beijing, China). A plasmid extraction kit (B518161) was purchased from Sangon Biotech (Shanghai) Co., Ltd (Shanghai, China). The EZ DNA Methylation-Gold™ Kit (D5005) and Competent Cell Preparation Kit (B529307) were purchased from Zymo Research (Orange County, CA, USA).2.2. Experimental animals
Nine-week-old Kunming mice, SPF grade, were purchased from SPF (Beijing) Biotechnology Co., Ltd with the license of SCXK (Beijing) 2016-0002. The mice were housed in cages and maintained under standard laboratory conditions (12 h/12 h light/dark cycle; 23 ± 0.5 °C; free access to food and water) for one week before the start of the experiment. Four female mice were placed overnight with 1 male. Detection of a vaginal plug in the next morning was designated as gestation day (GD) 0.5.25 The protocols using animals in our study were approved by the Council for Animal Care in Hebei province (approval number 2019-006).2.3. Groups and exposure
One hundred pregnant mice were randomly divided into 5 groups of twenty in each group. Group A was the control group, which was given 0.2 ml of corn oil per mouse at GD 0.5–17.5 via gavage. Group B was a model group of BPA, which was given 0.2 ml of corn oil containing BPA at 5 mg kg−1 day−1 (no observed adverse effect levels, NOAEL, https://comptox.epa.gov/dashboard/dsstoxdb/results?search=Bisphenol+A) via gavage, which was typically used in previous research studies.26–28 Group C was BPA + CCFs and was given 0.2 ml of corn oil containing BPA at 5 mg kg−1 and CCFs at 20 mg kg−1; group D received BPA at 5 mg kg−1 + CCFs at 30 mg kg−1 and Group E received BPA at 5 mg kg−1 + CCFs at 40 mg kg−1. The selected dosage of CCFs was used in previous studies.15 At postnatal day (PND) 21 and PND 56, ten F1 males of each group were sacrificed under anaesthesia with pentobarbital sodium (Fig. 1). A blood sample was collected, and the testes were collected and stored at −80 °C.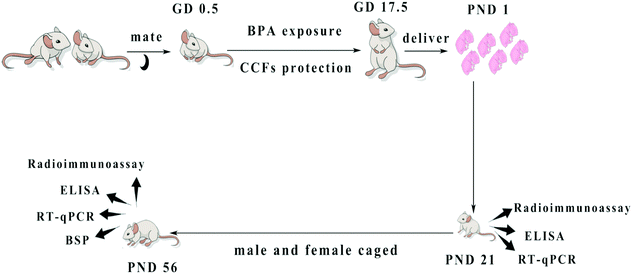 | ||
| Fig. 1 The design and methods of BPA exposure and CCF protection. Pregnant mice exposed to BPA were administered with CCFs from GD 0.5 to GD 17.5. The F1 males were sacrificed at PND 21 and PND 56. | ||
2.4. Radioimmunoassay
Blood was collected from the retro-orbital sinus under anaesthesia. The serum was obtained from the blood by centrifugation at 3200g for 15 min. Radioactivity counting (CPM) was performed on each tube pellet. The serum estradiol (E2) and testosterone (T) contents were analysed using a multi-tube radioimmunoassay counter purchased from Zhongcheng Electromechanical Technology Development Co., Ltd (Hefei, China).2.5. Enzyme-linked immunosorbent assay
One hundred mg testicular tissue was homogenized into 1 ml suspension solution of 0.9% saline. After centrifugation at 3200g for 15 min, the supernatant was collected. Testicular Dnmt1, Dnmt3A, Dnmt3B, ERα and ERβ levels were determined according to the instructions of the Shanghai Horatio Biotechnology Co., Ltd (Shanghai, China) ELISA kit.2.6. Quantitative real-time PCR
The total RNA extracted from the testis using the total RNA extraction kit (Promega, Beijing, China) was reverse transcribed to obtain cDNA. The transcription levels of Dnmt1, Dnmt3A, Dnmt3B and ERα mRNA in the testis were determined by RT-qPCR (Table 1). Primers were designed and synthesized by Takara (Dalian, China). Amplification was performed for 45 cycles consisting of 5 seconds at 95 °C, and 30 seconds at 64 °C, and then 30 seconds at 72 °C. The internal reference gene is GAPDH, and the relative expression level of each gene is calculated according to the 2−ΔΔCt method.| Gene | Primer sequence (5′–3′) | T m (°C) | GC (%) |
|---|---|---|---|
| Dnmt1 | Forward CTTCGGCAACATCCTGGACA | 64.5 | 55 |
| Reversed ACTGGACAGCAGGCAGAGCTTA | 63.7 | 55 | |
| Dnmt3A | Forward TGCGCCAGAAGTGCAGAAAC | 64.9 | 55 |
| Reversed CGTCATACTGGTAAGCACACTCCAA | 64.5 | 48 | |
| Dnmt3B | Forward GAATGCGCTGGGTACAGTGG | 63.9 | 60 |
| Reversed GCCAGATTAAAGTGCTGGCTGAA | 64.8 | 48 | |
| ERα | Forward AATGATGGGCTTATTGACCAACCTA | 63.8 | 40 |
| Reversed AGAATCTCCAGCCAGGCACAC | 64 | 57 | |
| GAPDH | Forward AAATGGTGAAGGTCGGTGTGAAC | 64.4 | 40 |
| Reversed CAACAATCTCCACTTTGCCACTG | 64 | 48 |
2.7. Bisulfite sequencing PCR
After the genomic DNA was extracted from the testis (the testes were collected from F1 mice at PND 56), bisulfite treatment of DNA was performed according to a modified protocol by Mann and Wang et al. Followed by PCR amplification (ABI, Verity 96 well, USA), the methylation status of the promoter of ERα, DNMT3A, DNMT3B and the imprinting control region (ICR) of the H19/Igf2 imprinted gene was analyzed by cloning and sequencing (ABI, 3730XL, USA). Using Meth Primer online software, one CpG island with 9 CpG sites was identified in the promoter region of the ERα gene. A bisulfite sequencing PCR (BSP) primer targeted the CpG islands in the promoter region of ERα, and was designed using the Methyl Primer software. Treating genomic DNA with bisulfite resulted in the conversion of an unmethylated cytosine into uracil. However, the methylated cytosine was found to remain unchanged. The percentage of methylation of the CpG island was calculated as the “CG” sites divided by the sum of the “CG” + “TG” sites. PCR amplification conditions were as follows: 95 °C for 5 min, 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, 35 cycles, and a final extension at 72 °C for 8 min. Primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd and are shown in Table 2.| Name | Primer sequence (5′–3′) | Cycle number | PCR product (bp) |
|---|---|---|---|
| DNMT3A CpG | F: GTGTTYGATGTTTTAGGATGGTTT | 35 | 262 |
| R: TCCCRAACCRATATCAAATACCTA | |||
| DNMT3B CpG | F: AGGAGGTAGGGYGAGGAAGTT | 35 | 278 |
| R: CCAATAAACCACCCRAACTAATC | |||
| ERα CpG | F:TGTAGGAGTTTAGTTGTTTTGTTTG | 35 | 246 |
| R:CRATCCTACCCTACTAATTCAAAAAC | |||
| H19/Igf2 ICR | F: ATAGGGGAGAAAATTTAATTAGTTG | 35 | 382 |
| R: AAAAAACCATACCCTATTCTTAAAC |
2.8. Statistical analysis
The statistical analysis was performed using SPSS 21.0 software. The values were presented as mean ± standard deviation (SD). Multiple comparisons between the groups were performed using the LSD method. One-way ANOVA was performed after the comparison of variance homogeneity tests among multiple groups. When the variance was not uniform, two independent sample T-tests were used. The difference was significant at P < 0.05, and the difference at P < 0.01 was extremely significant.3. Results
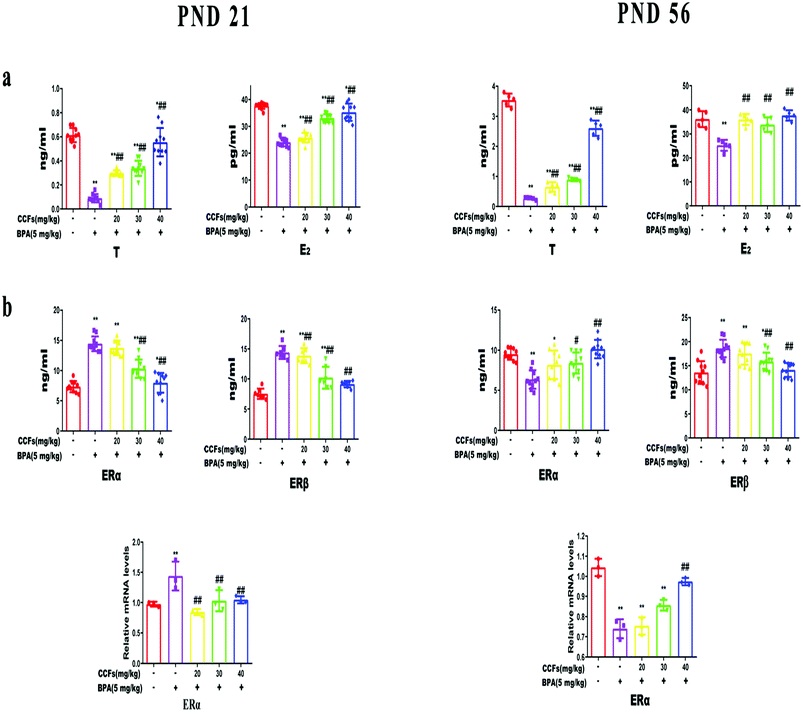
3.1. The effects of CCFs on hormone and estrogen receptor levels
Compared with the control group, the serum T and E2 levels of F1 males in the BPA group were significantly decreased at PND 21 and PND 56 (P < 0.01). Compared with the BPA model group, the serum T and E2 levels were significantly increased (P < 0.01 or P < 0.05) at all concentrations (20 mg kg−1, 30 mg kg−1, 40 mg kg−1) of CCF groups (Fig. 2a). No significant differences were observed at all the concentrations of CCF groups and in the serum E2 levels of F1 males of the control group at PND 56.Compared with the control group, the levels of testicular ERβ of F1 males in the BPA model group were significantly increased at PND 21 and PND 56 (P < 0.01) (Fig. 2b). The expression of ERα was significantly increased at PND 21 and significantly decreased at PND 56 (P < 0.01). After the administration of 30 mg kg−1 and 40 mg kg−1 CCFs, the levels of ERβ decreased significantly, and the levels of ERα decreased significantly at PND 21 while they increased significantly at PND 56 (P < 0.01 or P < 0.05). The contents of ERβ in the 40 mg kg−1 CCF group at PND 21 and PND 56 had no statistical difference compared with the control group.
3.2. The effects of CCFs on the mRNA transcriptional levels of testicular DNMTs
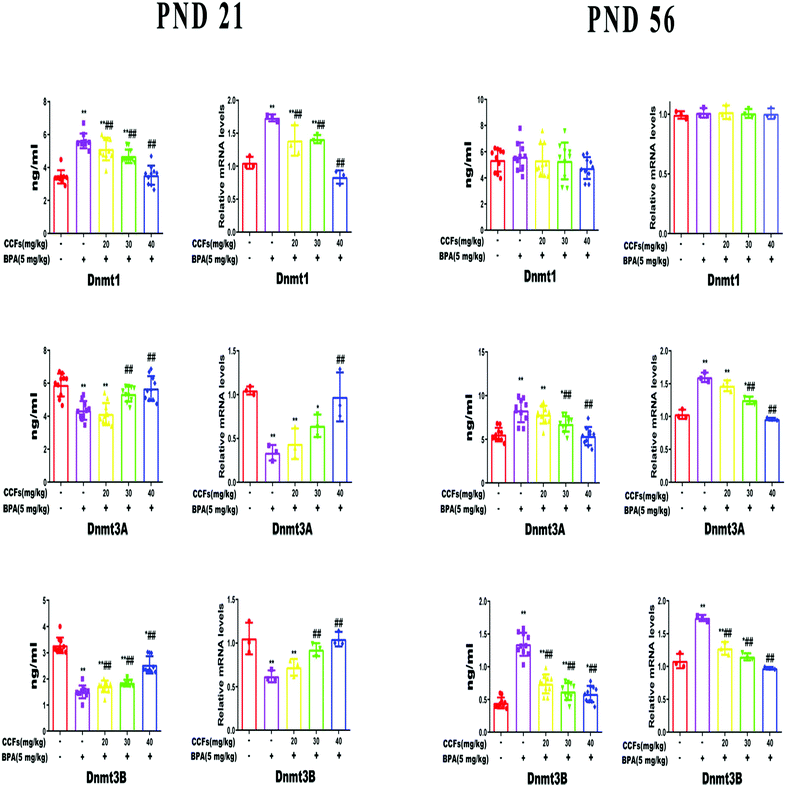
The testicular Dnmt1 content and mRNA transcription level of F1 males were significantly increased in the BPA group at PND 21 (P < 0.01), but had no significant difference at PND 56 compared with the control group (P > 0.05). BPA significantly decreased the expression of Dnmt3A and Dnmt3B of F1 males at PND 21 and increased that at PND 56 (P < 0.01). Compared with the BPA group, 40 mg kg−1 CCFs significantly reduced the expression of Dnmt1 at PND 21 and Dnmt3A at PND 56 as well as Dnmt3B. Besides, the levels of testicular Dnmt3A and Dnmt3B increased with the administration of 30 mg kg−1 and 40 mg kg−1 CCFs at PND 21 (P < 0.01), see Fig. 3.3.3. The effects of CCFs on the methylation status in the promoter region of the ERα gene
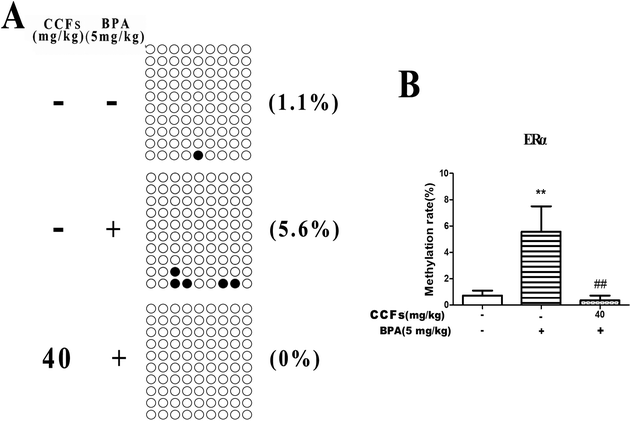
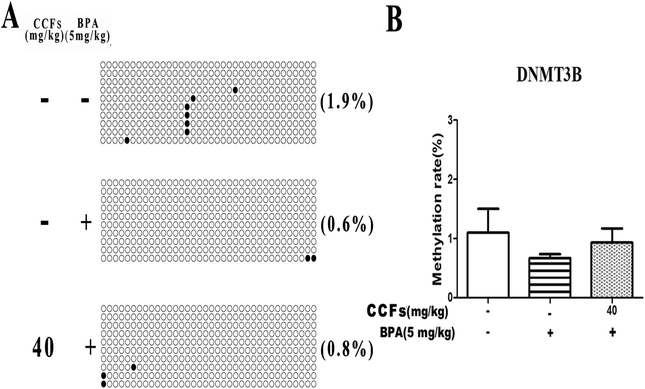
Each row of sequence indicates an individual clone. The black circle represents a methylated CG, and the white circle represents an unmethylated CG (Fig. 4A). As shown in Fig. 4, the BSP results showed that 0.73% ± 0.62% and 5.57% ± 3.35% (mean ± SD) of CpGs in the CpG islands of the ERα promoter region were methylated in the control and BPA groups. After treatment with 40 mg kg−1 CCFs, the CpG methylation rate significantly decreased (0.36% ± 0.60%), compared with the BPA group (P < 0.01; Fig. 4B). There was no significant difference between the 40 mg kg−1 CCF group and control group (P > 0.05).3.4. The effects of CCFs on the methylation status in the promoter region of the DNMT3A gene
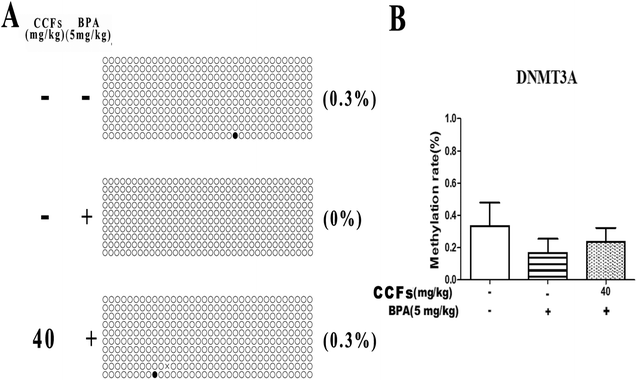
Representative clones of each group are shown in Fig. 5A. Each row represents a unique methylation status of the 16 successfully sequenced clones of the analysed testis. Each circle within the row represents a single site, and white and black circles represent unmethylated and methylated sites, respectively. The methylation sites in the BPA group were with an average of 0.17% ± 0.10% (mean ± SD). The percentages of methylated clones in the control and 40 mg kg−1 CCF groups were with average percentages of 0.33% ± 0.21% and 0.23% ± 0.12%, respectively. Multiple comparisons revealed that the difference between the CCF and BPA groups was not statistically significant (P > 0.05; Fig. 5B).3.5. The effects of CCFs on the methylation status in the promoter region of the DNMT3B gene
We investigated 13 sites in DNMT3B CpG. Representative clones of each group are shown in Fig. 6A. Each row represents a unique methylation status of the 16 successfully sequenced clones of the analysed testis. Each circle within the row represents a single site, and white and black circles represent unmethylated and methylated sites, respectively. As shown in Fig. 6, the methylation of DNMT3B CpG in the BPA group was with an average of 0.67% ± 0.12% (mean ± SD). The percentages of methylated clones in the control and 40 mg kg−1 CCF groups were with average percentages of 1.10% ± 0.70% and 0.93% ± 0.42%, respectively. There was no significant difference between the three groups (P > 0.05; Fig. 6B).3.6. The effects of CCFs on the methylation status in the H19/Igf2 imprinted gene
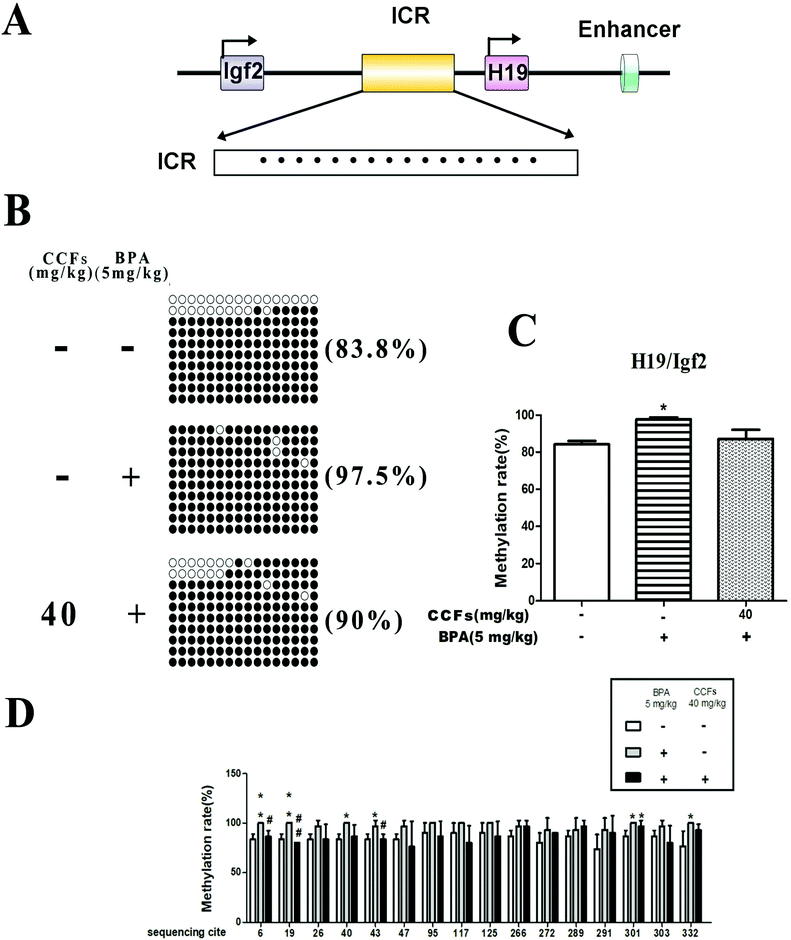
Representative clones of each group are shown in Fig. 7B. Each row represents a unique methylation status of the 16 successfully sequenced clones of the analysed testis. Each circle within the row represents a single site, and white and black circles represent unmethylated and methylated sites, respectively. The methylated clones in the BPA group were with an average of 97.70% ± 1.61% (mean ± SD). Compared with the BPA group, the percentages of methylated clones in the control and 40 mg kg−1 CCF groups were significantly lower, with average percentages of 84.17% ± 3.17% and 87.10% ± 8.53%, respectively. Multiple comparisons revealed that the difference between the CCF and BPA groups was statistically significant (P < 0.05 or P < 0.01; Fig. 7C). Fig. 7D reveals the results of comparing each group of individual methylation sites. There were significant differences between the CCF and BPA groups at the sequencing sites 6, 19, 40, 43, 301, and 332 (P < 0.05 or P < 0.01, Fig. 7D).4. Discussion
Development is a plasticity process that is sensitive to environmental disturbances including nutrients, stress, drugs and environmental pollutants.29,30 In 2002, the fetal origins of disease created the term “Developmental Origins Health and Diseases” (DOHaD), which focuses on the importance of nutritional insults in the development of disease susceptibility in later life.29 One similar concept, the Fetal Basis of Adult Disease (FEBAD) hypothesis assumes that prenatal insults, such as nutritional or environmental stimuli, can interfere with developmental procedures and lead to a higher risk of disease later in life.20Some bioactive compounds may offset or impair the damage caused by environmental pollution to the epigenome.6 For example, BPA is a public health concern hormone-damaging chemical and dietary supplementation of methyl donors (such as vitamin B12, folic acid, choline, etc.) and isoflavone genistein can reverse BPA-induced epigenetic dysregulation.31 It has been found that CCFs can reverse the pathological changes of the placenta and decidua structure following abortion, regulate the balance of Th1/Th2 type cytokines, and thus play a role in maintaining pregnancy, suggesting that CCFs can be the main pharmacodynamics basis of Cuscuta chinensis.32,33 Therefore, we wonder whether the flavonoids can be used as a dietary supplement to reverse the genomic dysregulation induced by BPA.
Studies have found that T and E2 have multiple regulatory mechanisms for male reproduction, including the aromatization hypothesis of testicular metabolism and the role of estrogen receptors.34 Testosterone can be converted to estradiol by aromatase and then affects male reproduction through ERα and ERβ. Estrogen and androgen participate in the complete manifestation of male sexual behavior.35 Our study found that the serum T and E2 levels in the BPA model group were significantly decreased, but the expression of testicular ERα and ERβ showed different trends. BPA has the dual effects of weak estrogen and anti-androgen, so that it can play a role in endocrine disruption by decreasing the serum contents of T and E2 and disrupting the expression of testicular ERα and ERβ.
It may be due to the long-term effects of BPA, which may damage the testis, reduce the activity of hormone-producing cells, and increase the expression of DNMTs in adult male mice, ultimately resulting in the methylation of hormone-producing genes and the inhibition of the transcription of hormones and estrogen. BPA—a weak estrogen—as a stimulant for ERα expression in immature (PND 21) mice does harm the production of that when mice mature (PND 56). The expression of ERβ was increased with mice at PND 56. It should be a compensatory response. CCFs can significantly restrain the BPA-induced decrease in serum E2, T contents in offspring males at PND 21 and PND 56, and regulate the abnormal expression of testicular ERα and ERβ. It may be because CCFs restore the methylation levels of ERα and the H19/Igf2 imprinting gene that regulates reproduction by normalizing the transcription level of DNMTs.
Epigenetics may be one of the most important molecular mechanisms linking environmental stimuli, fetal programming and adult phenotypes.6,36 Epigenetic modification is an attractive therapeutic target due to its reversibility.37 DNA methylation is one of the important ways of epigenetic regulation. In mammals, DNMTs are responsible for genome-wide methylation establishment and maintenance. Alterations in DNMTs can lead to abnormalities in genomic DNA methylation and disorders of epigenetic regulation.38 Several pieces of evidence have suggested that hypermethylation of promoter CpG islands is associated with the repression of transcription. The mechanisms of genomic imprinting are likely complex, but as yet remain unclear. And parental-specific DNA methylation during germ line development is associated with parental-specific closure of certain genes. Proper sperm production is associated with the establishment of appropriate epigenetic marks in developing germ cells. A study on mice showed that abnormal methylation patterns resulting from prenatally BPA exposure were established before germ layer differentiation in the embryonic stem cells.31 Several studies have demonstrated the association between abnormal spermatogenesis and epigenetic disturbances with the major focus on DNA methylation.23 Gene-specific analysis of DNA methylation in humans found that altered methylation patterns of the placenta and the fetal liver and kidney were associated with prenatal exposure to BPA.39–41 DNA methylation is the primary mechanism for generating genomic imprints.42 Notably, perinatal exposure to BPA suggests that male infertility can be inherited across generations through epigenetic deregulation in the male germ line.43–45
In the mammalian genome, approximately 1% is a CpG site, with mCpG accounting for 70%–80%. Methylation of CpG islands in which CpG sites are aggregated interferes with the binding of some transcription factors to gene regulatory regions, or directly inhibits RNA polymerase activity and inhibits gene expression.46 DNA methylation silences gene expression which plays a role in immune disorders.47 In the early stage of gamete formation, the imprints from both the father and the mother will all be eliminated. The parent allele will produce a new methylation pattern when spermatozoa form sperm. The maternal allele methylation pattern is formed during egg production, so alleles from the father and mother before fertilization have different methylation patterns, a gene with this phenomenon is called an imprinted gene. For example, DSCAM is an imprinting gene that is extremely important in neuronal development.48H19/Igf2 is an imprinting gene related to fetal growth and development.49 In a study conducted by Nasri et al. (2011), H19 abnormality in a methylation imprinting locus can cause male infertility.23
Zhou et al. found that aflatoxin B1 (AFB1) can induce an increase in DNMT3A in cells, thereby producing a toxic effect, which is in line with our study.50 Our study found that the expression of Dnmt1 in male offspring at PND 21 and the expression of Dnmt3A and Dnmt3B at PND 56 were significantly increased after exposure to BPA. Dnmt1 is mainly involved in the maintenance of methylation, and a significant increase in the offspring males at PND 21 will silence most of the genes that should be transcribed, hindering development. In order to redress the balance, the body decreases the levels of Dnmt3A and Dnmt3B. Dnmt3A and Dnmt3B are involved in de novo synthetic methylation, and a significant increase in male offspring at PND 56 brings chromatin to a tighter state, increases DNA double-strand binding, inhibits the process of unwinding, generates gene silencing and maintains silencing.51 CCFs can significantly regulate the expression of DNMTs in the testis, and the expression of DNMTs tends to be normal, thus protecting the reproductive function in offspring adult males. Besides, we found that the methylation of Dnmt3A and Dnmt3B CpG in the promoter was not statistically different. The reason may be that Dnmt3A and Dnmt3B are DNA methyltransferases themselves, and the ratio of methylation may be narrow, as these enzymes are required for modification. Their effects should often be reflected in the transcription factor levels.
We also found that the methylation of ERα CpGs and H19/Igf2 imprinting genes in the BPA group was increased. We considered that it is due to an increase in Dnmt3A and Dnmt3B and is responsible for the decrease in ERα expression. What is more, the abnormal imprinting of H19/Igf2 can cause embryonic growth and developmental malformation.42,52,53 This abnormal level of DNA methylation may be transmitted to the next generation through germ cells, affecting future generations. But CCFs can significantly improve these anomalies according to our results.
5. Conclusion
Although early nutrition does not alter DNA, it can significantly affect development by regulating gene expression. In summary, our data suggest that BPA induces a decrease in ERα expression in the testis of mature mice, which may ultimately relate to an increase in the expression of Dnmt3A and Dnmt3B causing a significant decrease in DNA methylation of the CpG islands in the ERα promoter region and H19/Igf2 imprinted locus. CCFs can decrease the expression of Dnmt3A and Dnmt3B as a dietary supplement, reduce the methylation level of related genes, and protect reproduction. These findings provide additional insight into the molecular mechanisms underlying the pathophysiology of EEDs and the protection of CCFs.Conflicts of interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No. 31872506).References
- K. A. Lillycrop and G. C. Burdge, Maternal diet as a modifier of offspring epigenetics, J. Dev. Origins Health Dis., 2015, 6, 88–95 CrossRef CAS PubMed.
- A. J. Bernal and R. L. Jirtle, Epigenomic Disruption: The Effects of Early Developmental Exposures, Birth Defects Res., Part A, 2010, 88, 938–944 CrossRef CAS PubMed.
- S. K. Murphy and R. L. Jirtle, Imprinting evolution and the price of silence, Bioessays, 2003, 25, 577–588 CrossRef CAS PubMed.
- M. K. Skinner, Environmental epigenomics and disease susceptibility, EMBO Rep., 2011, 12, 620–622 CrossRef CAS PubMed.
- R. Das, D. D. Hampton and R. L. Jirtle, Imprinting evolution and human health, Mamm. Genome, 2009, 20, 563–572 CrossRef PubMed.
- S. Z. Li, M. Chen, Y. Y. Li and T. O. Tollefsbol, Prenatal epigenetics diets play protective roles against environmental pollution, Clin. Epigenet., 2019, 11, 82 CrossRef PubMed.
- J. Abbasi, Scientists Call FDA Statement on Bisphenol A Safety Premature, JAMA, J. Am. Med. Assoc., 2018, 319, 1644–1646 CrossRef PubMed.
- S. Patel, E. Brehm, L. Y. Gao, S. Rattan, A. Ziv-Gal and J. A. Flaws, Bisphenol A Exposure, Ovarian Follicle Numbers, and Female Sex Steroid Hormone Levels: Results From a CLARITY-BPA Study, Endocrinology, 2017, 158, 1727–1738 CrossRef PubMed.
- E. M. Martin and R. C. Fry, Environmental Influences on the Epigenome: Exposure-Associated DNA Methylation in Human Populations, Annu. Rev. Public Health, 2018, 39, 309–333 CrossRef PubMed.
- A. Vaiserman, Epidemiologic evidence for association between adverse environmental exposures in early life and epigenetic variation: a potential link to disease susceptibility?, Clin. Epigenet., 2015, 7, 96 CrossRef PubMed.
- T. M. Hardy and T. O. Tollefsbol, Epigenetic diet: impact on the epigenome and cancer, Epigenomics, 2011, 3, 503–518 CrossRef CAS PubMed.
- D. N. Qin, B. R. She, Y. C. She and J. H. Wang, Effects of flavonoids from Semen Cuscutae on the reproductive system in male rats, Asian J. Androl., 2000, 2, 99–102 CAS.
- S. J. Kim, M. R. Kim, S. Y. Hwang, W. J. Bae, S. Kim, S. H. Hong, J. Y. Lee, T. K. Hwang, Z. Wang and S. W. Kim, Preliminary report on the safety of a new herbal formula and its effect on sperm quality, World J. Mens Health, 2013, 31, 254–261 CrossRef PubMed.
- J. O. Omirinde, P. C. Ozegbe and M. O. Oyeyemi, Comparative evaluation of the sperm characteristics and morphology of adult Wistar rats fed either low or normal protein-energy diets and orally dosed with aqueous Cuscuta australis extracts, Niger. J. Physiol. Sci., 2014, 29, 55–61 CAS.
- Y. Wei, S. Li, C. Han, Y. Bao and W. Shi, Cuscuta chinensis flavonoids alleviate bisphenol A-induced apoptosis of testicular cells in male mice offspring, Andrologia, 2019, 51, e13427 CrossRef PubMed.
- A. P. Feinberg, Methylation meets genomics, Nat. Genet., 2001, 27, 9–10 CrossRef CAS PubMed.
- T. M. Holm, L. Jackson-Grusby, T. Brambrink, Y. Yamada and W. M. Rideout, 3rd and R. Jaenisch, Global loss of imprinting leads to widespread tumorigenesis in adult mice, Cancer Cell, 2005, 8, 275–285 CrossRef CAS PubMed.
- T. M. Hardy and T. O. Tollefsbol, Epigenetic diet: impact on the epigenome and cancer, Epigenomics, 2011, 3, 503–518 CrossRef CAS PubMed.
- J. H. Park, S. H. Kim, M. S. Lee and M. S. Kim, Epigenetic modification by dietary factors: Implications in metabolic syndrome, Mol. Aspects Med., 2017, 54, 58–70 CrossRef CAS PubMed.
- C. M. Law, D. J. Barker, C. Osmond, C. H. Fall and S. J. Simmonds, Early growth and abdominal fatness in adult life, J. Epidemiol. Community Health, 1992, 46, 184–186 CrossRef CAS PubMed.
- M. A. Hanson and P. D. Gluckman, Developmental origins of health and disease - Global public health implications, Best Pract. Res. Clin. Obstet. Gynaecol., 2015, 29, 24–31 CrossRef CAS PubMed.
- M. A. Hanson and P. D. Gluckman, Developmental origins of health and disease: New insights, Basic Clin. Pharmacol. Toxicol., 2008, 102, 90–93 CrossRef CAS PubMed.
- F. Nasri, B. Gharesi-Fard, B. Namavar Jahromi, M. A. Farazi-Fard, M. Banaei, M. Davari, S. Ebrahimi and Z. Anvar, Sperm DNA methylation of H19 imprinted gene and male infertility, Andrologia, 2017, 49, e12766 CrossRef PubMed.
- L. Sun, Z. Gao, L. Luo, H. Tan and G. Zhang, Estrogen affects cell growth and IGF-1 receptor expression in renal cell carcinoma, OncoTargets Ther., 2018, 11, 5873–5878 CrossRef CAS PubMed.
- Y. Wei, C. Han, S. Li, Y. Cui, Y. Bao and W. Shi, Maternal exposure to bisphenol A during pregnancy interferes ovaries development of F1 female mice, Theriogenology, 2019, 142, 138–148 CrossRef PubMed.
- M. Susiarjo, I. Sasson, C. Mesaros and M. S. Bartolomei, Bisphenol a exposure disrupts genomic imprinting in the mouse, PLoS Genet., 2013, 9, e1003401 CrossRef CAS PubMed.
- J. G. Bromer, Y. Zhou, M. B. Taylor, L. Doherty and H. S. Taylor, Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response, FASEB J., 2010, 24, 2273–2280 CrossRef CAS PubMed.
- Y. Y. Wei, C. Han, Y. M. Geng, Y. Q. Cui, Y. Z. Bao, W. Y. Shi and X. H. Zhong, Maternal exposure to bisphenol A during pregnancy interferes testis development of F1 male mice, Environ. Sci. Pollut. R, 2019, 26, 23491–23504 CrossRef CAS PubMed.
- J. J. Heindel, J. Balbus, L. Birnbaum, M. N. Brune-Drisse, P. Grandjean, K. Gray, P. J. Landrigan, P. D. Sly, W. Suk, D. C. Slechta, C. Thompson and M. Hanson, Developmental Origins of Health and Disease: Integrating Environmental Influences, Endocrinology, 2015, 156, 3416–3421 CrossRef CAS PubMed.
- D. J. Hoffman, R. M. Reynolds and D. B. Hardy, Developmental origins of health and disease: current knowledge and potential mechanisms, Nutr. Rev., 2017, 75, 951–970 CrossRef PubMed.
- D. C. Dolinoy, D. Huang and R. L. Jirtle, Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13056–13061 CrossRef CAS PubMed.
- H. X. Ma, Z. L. You and R. G. Wang, Effect of total flavones from Cuscuta chinensis on expression of Th type-1/Th type-2 cytokines, serum P and PR in abortion rats model, Zhong Yao Cai, 2008, 31, 1201–1204 CAS.
- H. X. Ma, Z. L. You and X. Y. Wang, Effect of total flavones from Cuscuta chinensis on expression of Fas/FasL, PCNA and HB-EGF in SD rats model with bromocriptine-induced abortion, Zhong Yao Cai, 2008, 31, 1706–1709 CAS.
- D. G. Zuloaga, D. A. Puts, C. L. Jordan and S. M. Breedlove, The role of androgen receptors in the masculinization of brain and behavior: what we've learned from the testicular feminization mutation, Horm. Behav., 2008, 53, 613–626 CrossRef CAS PubMed.
- D. Wu and A. C. Gore, Changes in androgen receptor, estrogen receptor alpha, and sexual behavior with aging and testosterone in male rats, Horm. Behav., 2010, 58, 306–316 CrossRef CAS PubMed.
- C. A. Hamm and F. F. Costa, Epigenomes as therapeutic targets, Pharmacol Ther., 2015, 151, 72–86 CrossRef CAS PubMed.
- E. Li, C. Beard and R. Jaenisch, Role for DNA methylation in genomic imprinting, Nature, 1993, 366, 362–365 CrossRef CAS PubMed.
- C. Potter, J. McKay, A. Groom, D. Ford, L. Coneyworth, J. C. Mathers and C. L. Relton, Influence of DNMT genotype on global and site specific DNA methylation patterns in neonates and pregnant women, PLoS One, 2013, 8, e76506 CrossRef CAS PubMed.
- M. S. Nahar, C. Liao, K. Kannan, C. Harris and D. C. Dolinoy, In utero bisphenol A concentration, metabolism, and global DNA methylation across matched placenta, kidney, and liver in the human fetus, Chemosphere, 2015, 124, 54–60 CrossRef CAS PubMed.
- E. M. Martin and R. C. Fry, Environmental Influences on the Epigenome: Exposure- Associated DNA Methylation in Human Populations, Annu. Rev. Public Health, 2018, 39, 309–333 CrossRef PubMed.
- C. Faulk, J. H. Kim, T. R. Jones, R. C. McEachin, M. S. Nahar, D. C. Dolinoy and M. A. Sartor, Bisphenol A-associated alterations in genome-wide DNA methylation and gene expression patterns reveal sequence-dependent and non-monotonic effects in human fetal liver, Environ. Epigenet., 2015, 1 Search PubMed , pii: dvv006.
- Z. Wang, L. Xu and F. He, Embryo vitrification affects the methylation of the H19/Igf2 differentially methylated domain and the expression of H19 and Igf2, Fertil. Steril., 2010, 93, 2729–2733 CrossRef CAS PubMed.
- S. Salian, T. Doshi and G. Vanage, Perinatal exposure of rats to Bisphenol A affects fertility of male offspring–an overview, Reprod. Toxicol., 2011, 31, 359–362 CrossRef CAS PubMed.
- S. Salian, T. Doshi and G. Vanage, Perinatal exposure of rats to Bisphenol A affects the fertility of male offspring, Life Sci., 2009, 85, 742–752 CrossRef CAS PubMed.
- M. Manikkam, C. Guerrero-Bosagna, R. Tracey, M. M. Haque and M. K. Skinner, Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures, PLoS One, 2012, 7, e31901 CrossRef CAS PubMed.
- M. Zhou, N. J. Sng, C. E. LeFrois, A. L. Paul and R. J. Ferl, Epigenomics in an extraterrestrial environment: organ-specific alteration of DNA methylation and gene expression elicited by spaceflight in Arabidopsis thaliana, BMC Genomics, 2019, 20, 205 CrossRef PubMed.
- J. Wang, D. Van Den Berg, A. E. Hwang, D. Weisenberger, T. Triche, B. N. Nathwani, D. V. Conti, K. Siegmund, T. M. Mack, S. Horvath and W. Cozen, DNA methylation patterns of adult survivors of adolescent/young adult Hodgkin lymphoma compared to their unaffected monozygotic twin, Leuk. Lymphoma, 2019, 60, 1429–1437 CrossRef CAS PubMed.
- L. Allach El Khattabi, S. Backer, A. Pinard, M. N. Dieudonne, V. Tsatsaris, D. Vaiman, L. Dandolo, E. Bloch-Gallego, H. Jammes and S. Barbaux, A genome-wide search for new imprinted genes in the human placenta identifies DSCAM as the first imprinted gene on chromosome 21, Eur. J. Hum. Genet., 2019, 27, 49–60 CAS.
- C. Martinet, P. Monnier, Y. Louault, M. Benard, A. Gabory and L. Dandolo, H19 controls reactivation of the imprinted gene network during muscle regeneration, Development, 2016, 143, 962–971 CrossRef CAS PubMed.
- X. Zhou, F. Gan, L. Hou, Z. Liu, J. Su, Z. Lin, G. Le and K. Huang, Aflatoxin B1 Induces Immunotoxicity through the DNA Methyltransferase-Mediated JAK2/STAT3 Pathway in 3D4/21 Cells, J. Agric. Food Chem., 2019, 67, 3772–3780 CrossRef CAS PubMed.
- Z. Li, H. Dai, S. N. Martos, B. Xu, Y. Gao, T. Li, G. Zhu, D. E. Schones and Z. Wang, Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells, Genome Biol., 2015, 16, 115 CrossRef PubMed.
- H. Lou, F. Le, M. Hu, X. Yang, L. Li, L. Wang, N. Wang, H. Gao and F. Jin, Aberrant DNA Methylation of IGF2-H19 Locus in Human Fetus and in Spermatozoa From Assisted Reproductive Technologies, Reprod. Sci., 2018, 26, 997–1004 CrossRef PubMed.
- Y. J. Wan, M. T. Deng, G. M. Zhang, C. F. Ren, Z. F. Liu and F. Wang, Analysis of H19/Igf2 Methylation Status in the Sperm of Cloned Goats and Their Offspring, Cell. Reprogram., 2018, 20, 66–75 CrossRef CAS PubMed.
Footnote |
| † Equal contributors. |
| This journal is © The Royal Society of Chemistry 2020 |