Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biological responses to physicochemical properties of biomaterial surface
Maryam
Rahmati
a,
Eduardo A.
Silva
 b,
Janne E.
Reseland
a,
Catherine
A. Heyward
c and
Håvard J.
Haugen
b,
Janne E.
Reseland
a,
Catherine
A. Heyward
c and
Håvard J.
Haugen
 *a
*a
aDepartment of Biomaterials, Institute of Clinical Dentistry, University of Oslo, 0317 Oslo, Norway. E-mail: h.j.haugen.odont.uio.no
bDepartment of Biomedical Engineering, University of California, One Shields Avenue, Davis, CA 95616, USA
cOral Research Laboratory, Institute of Clinical Dentistry, University of Oslo, 0317 Oslo, Norway
First published on 9th July 2020
Abstract
Biomedical scientists use chemistry-driven processes found in nature as an inspiration to design biomaterials as promising diagnostic tools, therapeutic solutions, or tissue substitutes. While substantial consideration is devoted to the design and validation of biomaterials, the nature of their interactions with the surrounding biological microenvironment is commonly neglected. This gap of knowledge could be owing to our poor understanding of biochemical signaling pathways, lack of reliable techniques for designing biomaterials with optimal physicochemical properties, and/or poor stability of biomaterial properties after implantation. The success of host responses to biomaterials, known as biocompatibility, depends on chemical principles as the root of both cell signaling pathways in the body and how the biomaterial surface is designed. Most of the current review papers have discussed chemical engineering and biological principles of designing biomaterials as separate topics, which has resulted in neglecting the main role of chemistry in this field. In this review, we discuss biocompatibility in the context of chemistry, what it is and how to assess it, while describing contributions from both biochemical cues and biomaterials as well as the means of harmonizing them. We address both biochemical signal-transduction pathways and engineering principles of designing a biomaterial with an emphasis on its surface physicochemistry. As we aim to show the role of chemistry in the crosstalk between the surface physicochemical properties and body responses, we concisely highlight the main biochemical signal-transduction pathways involved in the biocompatibility complex. Finally, we discuss the progress and challenges associated with the current strategies used for improving the chemical and physical interactions between cells and biomaterial surface.
1. Introduction
In the case of severe tissue injuries, the body is not able to successfully repair the injured tissues.1 Owing to the emergence of tissue engineering and regenerative medicine fields, many promising treatment strategies are currently available for repairing and/or replacing damaged tissues.2,3 However, there is no doubt that, if not all, most of these approaches are dependent on using chemical principles for designing biomaterials as biological substitutes that mimic and/or stimulate tissue functions. The American National Institutes of Health defines a biomaterial as “any substance or combination of substances, other than drugs, synthetic or natural in origin, which can be used for any period of time, which augments or replaces partially or totally any tissue, organ or function of the body, to maintain or improve the quality of life of the individual”.4,5 Biomaterials engineering is a highly interdisciplinary research field in which scientists (mostly with a background in chemistry) introduce biological alternatives for replacing or enhancing tissue and/or organ functions.1,6 Over the past few decades, chemical scientists and engineers have achieved substantial progress in designing promising biomaterials as key diagnostic or therapeutic solutions for several disorders.7–9Although considerable effort is devoted to developing biomaterials as successful tissue replacements for clinical applications, most of the suggested strategies fail to match the functional properties of targeted tissues in vivo, due to their poor biocompatibility.1 The nature of the interaction of biomaterials with the surrounding biological microenvironment defines their biocompatibility. There is still a critical gap in successfully matching the biomaterial surface physicochemical characteristics to biochemical signal-transduction pathways in vivo. This gap could be owing to our poor understanding of biochemical signaling pathways, lack of reliable techniques for designing biomaterials with optimal physicochemical properties, and/or poor stability of biomaterial properties after implantation.10–13 The designed biomaterials for tissue engineering applications should have a strong affinity to targeted cells by sending chemical and physical signals to stimulate neo-tissue formation. Establishing strong positive interactions between the biomaterial surface and cells is entirely dependent on both the materials and targeted biological system chemistry.14
From the biochemical point of view, the features of cellular niche are highly important. The cellular niche is a highly complex tissue-specific microenvironment within a particular anatomic location providing physicochemical signals for cell communication.15 Different mechanotransduction, macromolecular adsorption and biochemical signaling pathways, which can be dependent on the tissue type, play key roles in determining the material's success after implantation. The main biochemical signaling pathways of local innate immune cells and their receptors, the other neighboring tissues or factors around the targeted tissue, and the biological systems a material might face are very diverse in different tissues.16,17
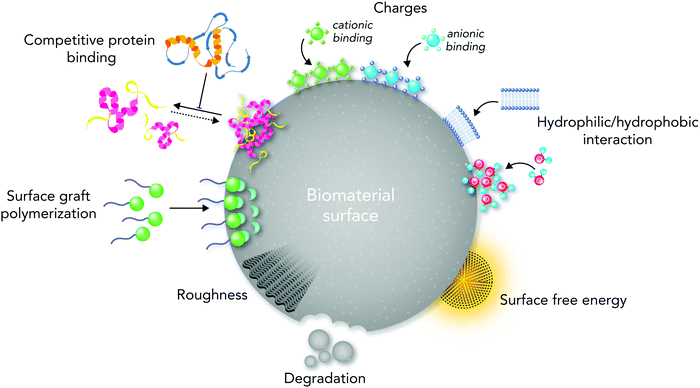
From the materials engineering point of view, each physicochemical property of the biomaterial surface (such as topographical features, stiffness, functional groups, and interfacial free energy) can profoundly affect biochemical mechanisms (Fig. 1). In addition, the commonly applied techniques and chemical strategies for modifying the surface properties can influence biomaterial–cell interactions.
Despite several reviews in the literature that address the importance of surface properties in regulating cell responses,10,11,13,18–20 none of the recently published reviews have comprehensively discussed the vital roles of chemistry in regulating biological pathways, manipulating biomaterial surface properties, and directing molecular and cellular responses after biomaterial implantation. In addition, it is time to provide an updated state-of-the-art and future perspective for researchers in this field based on the recent chemical, physical and biological research findings. This review aims at emphasizing the key roles of chemistry in determining the biocompatibility of biomaterials by presenting an overview of both biochemical and chemical engineering principles and challenges in designing biocompatible systems. As biochemical signaling pathways of the immune system are critical factors in determining the success of biomaterials, we briefly highlight the main biochemical signaling mechanisms and concepts of biocompatibility. Then, we address the current progress and challenges in biological responses to biomaterial surface physicochemical properties such as topographical features, functional groups, interfacial free energy, ion enrichment, and biological moieties. Although we use biomaterials in different tissue engineering applications, reliable evaluation of biological responses is still a big challenge. Altogether, this review provides an overview of the progress and challenges of each part to the readers; however, due to the complex nature of biological responses to biomaterials, not all related issues are possible to discuss here.
2. Using biomaterials for tissue regeneration applications
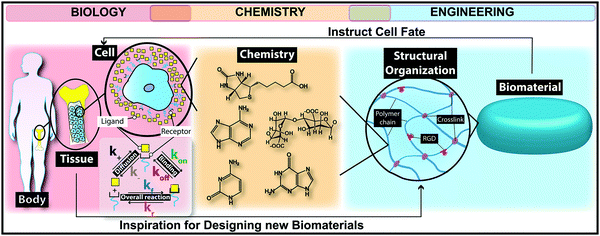
The self-renewal potential of tissues decreases or completely disappears over time due to several reasons such as increasing age, reducing the amount and capability of host stem cell/progenitor populations, naturally poor repair potential of tissues, or undesirable inflammatory responses in damaged tissues and/or organs.21,22 Tissue engineering and regenerative medicine approaches represent a clinically appealing and promising strategy to repair biological processes associated with injured tissues.22 In the past few decades, scientists have used various cell types as key elements in different tissue regeneration therapies.23–25 However, if cells are transplanted freely into the body, only a small proportion might reach the targeted tissue.26Biomedical scientists use naturally occurring chemical processes as an inspiration to design new biomaterials. Different classes of biomaterials are designed to offer suitable microenvironments for enhancing cell engraftment, including both naturally occurring and synthetic polymers, ceramics, metals and composites (Fig. 2).26,27 Implanted biomaterials in tissue engineering are categorized generally into two main groups: (i) auto-, allo- or xeno-based cellularized or decellularized scaffolds known as natural/physiological polymers (e.g. proteins, polysaccharides and decellularized tissue matrices) and (ii) other materials such as synthetic polymers, implants, ceramics and composites.5 Chemical strategies can be employed for designing a wide range of naturally occurring and synthetic biomaterials while stimulating cells to secrete and deposit the native extracellular matrix (ECM) locally.28–30 The substantial progress in chemical and tissue engineering fields has led to the existence of smart biomaterials as promising therapeutic solutions for several devastating disorders. Nowadays, we clinically use biomaterials as valid therapeutic candidates for various tissue regeneration applications such as musculoskeletal system,31 cardiovascular system,32 neural system,33 and skin.34 In addition, biomaterials can affect the results of regenerative medicine strategies such as cell-based therapies, and engineered living tissues or organs.35
For successfully using biomaterials in the medicine world, designed biomaterials should be able to enhance the cell survival and functions after transplantation as well as stimulate autologous tissue growth.36,37 The designed biomaterials for tissue regeneration applications should provide provisional mechanical support and mass transport to stimulate biochemical signaling pathway functions toward tissue healing.38 Additionally, biomaterials could increase the success of tissue regeneration by sending physicochemical signals with spatiotemporal precision toward cells.39 With this concept, a biomaterial is dynamically involved in providing some physicochemical cues to targeted cells resulting in neo-tissue formation.40 For initiating biochemical signaling pathways, considering the presence of soluble signaling molecules such as growth factors and cytokines is also important.41
On the other hand, scaffolds designed from one material type would not be able to meet the requirements for tissue regeneration applications, which is owing to the absence of a controlled degradation rate, optimal physicochemical properties, and stimulating ideal biochemical signaling pathways.42,43 Thus, composite biomaterials designed by combining the chemistries of different materials tend to exhibit greater success in stimulating tissue regeneration after implantation.44,45 Manipulating the biomaterial surface physico-chemistry based on the targeted site is essential for achieving optimal biological performance. Indeed, selecting biomaterials for tissue engineering applications is reliant on their physicochemical surface properties such as surface roughness,46 architecture,47 charge,48 energy,49 and functional groups.50 Hence, the effects of each physicochemical surface property on the biological performance of biomaterials should be precisely investigated in vitro and in vivo.37
3. The evolution of the definition of host responses
In the early 20th century, a prodigious revolution took place in both therapeutic and diagnosis strategies through designing synthetic biomaterials by manipulating the chemistry of materials.51 Since naturally occurring chemistry was used for designing biomaterials, modifying and/or proving their biological safety were among the most challenging issues in this field. Some decades ago, James Anderson defined foreign body reactions to biomaterials by demonstrating short- and long-term inflammatory responses to biomaterials and the substantial roles of macrophages in each step.16,52–54 Owing to Anderson group's work, the biomedical scientists’ understanding of molecular and cellular responses to biomaterials increased so that these days at the time of designing each biomaterial its effects on the foreign body responses determine its biocompatibility.Although this definition favors non-degradable inert biomaterials, it cannot thoroughly define the body responses to the recently developed biomaterials with bioactive degradable surfaces suitable for tissue regeneration.10,11,13,55 Owing to the advances in chemistry, the recently developed biomaterials are designed and formulated to stimulate different biochemical signaling pathways. In these cases, we could not define biocompatibility as only not having any adverse effects.56 The designed biomaterials should have a strong affinity for targeted cells to stimulate biochemical signaling pathways toward the neo-tissue formation. This ability is entirely dependent on the specific chemical characteristics of both the material system and the biological environment of targeted tissue.14
The biomaterial surface physicochemical properties such as charges, functional groups, biological moieties, and ion enrichment play key roles in directing biological responses to biomaterials.14,56 From the biochemical perspective, different mechanotransduction, physiological, macromolecular adsorption and biochemical signaling pathways are crucial, which can be different from tissue to tissue. Because different tissues and cells have different chemical signals and physical characteristics, it is hard to say whether a material compatible with one tissue will make positive interactions with other cell types and tissues.10,13 In addition, although both innate and adaptive immune systems respond to biomaterial implantation, their biochemical cues are different from each other, which requires evaluating their responses individually.57
4. The classical perspective of biological responses to biomaterials
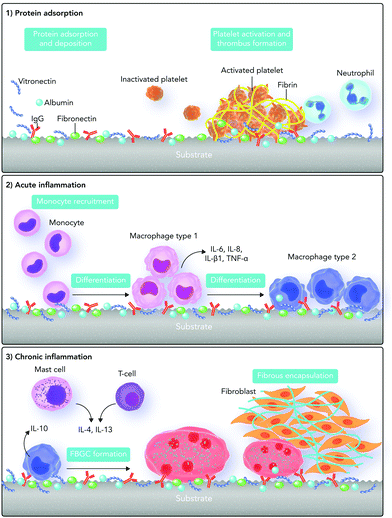
The host responses to biomaterials mainly originate from biochemical signals and cues. As our knowledge in the biochemistry field has tremendously grown since the first definition of biocompatibility, we should update our definitions regarding host responses to biomaterials. Here we briefly discuss the out-of-date concept of cellular responses and biochemical signaling pathways involved in foreign body responses to biomaterials. Then, we provide an overview of the recently updated biochemical signaling pathways in the next sections.Foreign body responses to implanted biomaterials are generally defined as a sequence of body reactions, which start instantly after biomaterial implantation.14,56 With this concept, after biomaterial implantation, the tissue injury stimulates several chemical signaling cascades, which result in a sequence of acute and chronic inflammatory as well as wound healing responses.58,59 Protein adsorption, neutrophils, and type 1 macrophages direct the acute inflammatory phase. This phase is essentially responsible for provisional matrix formation and wound site cleaning, which can take from some hours to days.60
After the release of some biochemical cues, blood vessels start expanding and consequently more blood flows into the injured area. Some blood and tissue proteins (such as fibronectin, fibrinogen, vitronectin, complement C3, albumin and growth factors) as well as leukocytes are released, which adhere to the blood vessel endothelium.59–61
Proteins are made from 20 natural amino acids. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered as proteins and are commonly called peptides, or sometimes oligopeptides. Each amino acid has a general backbone network of {–NH–CαHR–CO–}, where R describes a specific side group structure that gives the amino acid its specific functional properties. Based on the R structure, the amino acids are divided into three main types: nonpolar, polar, and charged amino acids, in which each class has an affinity to surfaces with unique physicochemical properties.14,62 Furthermore, the size of proteins affects their adsorption to the biomaterial surface. Small proteins move faster and are responsible for the primary adsorption on surfaces. However, the larger proteins have higher affinity to the surface, which is owing to their greater surface area.63
Moreover, the protein conformation defines its structure, bioactivity and communication with other biomolecules on the surface.64 Most proteins have at least one active region to adsorb on the biomaterial surface, ligands, and receptors. The receptor domain of extracellular molecules accepts a signal from the upstream part and as a result changes its conformation, leading to stimulating the formation of a ligand binding domain.14 These receptor binding proteins are connected into a chain, which transmit the biochemical signals across the cell membrane.14 Researchers can achieve different protein–surface interactions through modifying the physicochemical properties of proteins.14
Monocytes released into the area differentiate into type 1 and type 2 macrophages. Type 1 macrophages are responsible for the acute inflammatory phase and release pro-inflammatory factors. On the other hand, type 2 macrophages are responsible for the chronic inflammatory phase and release anti-inflammatory factors.59,60,65,66
Monocytes, type 2 macrophages, and lymphocytes control the chronic inflammatory phase. During this phase, tissue granulation, fibroblast infiltration, and neovascularization occur, which can subsequently lead to the formation of blood vessels and connective tissue to allow wound healing to proceed.66,67 In the wound healing phase, the proliferation of fibroblasts and vascular endothelial cells changes the fibrin clot into an extremely vascularized granulation tissue. The presence of several growth factors is important in this phase including platelet-derived growth factor, fibroblast growth factor, transforming growth factor-β, transforming growth factor-α/epidermal growth factor, interleukin-1 (IL-1), and tumor necrosis factor.68–70 Fibroblasts are also active in synthesizing collagen and proteoglycans, which lead to replacement of the granulation tissue with ECM (Fig. 3). Depending on the severity of injury at the implanted site, tissue type, and biomaterial properties, the acute phase takes less than one week and the chronic phase about two weeks.71,72
Based on this traditional definition of foreign body responses to biomaterials, the ability of a biomaterial to stimulate minimal inflammatory responses defines its success. Hence, the focus in designing biomaterials is on reducing foreign body responses through directing macrophage responses. However, because allowing natural body responses to occur is more useful for both biomaterial integration and function, this definition started to be redefined over the past few years.73–75 The synchronization between inflammation and its resolution is essential for wound healing, which is dependent on the biochemical signaling pathways and cues.76 To enhance the healing process, biomaterials are currently designed with a focus on improving their chemical interactions with immune system components.77–79
5. The role of innate and adaptive immune systems and biochemical cues in biological responses to biomaterials
The immune system is the main biological network, which releases biochemical cues responsible for protecting the body against foreign materials and keeping homeostasis. It consists of two main parts: innate and adaptive immune systems. Just after the instant recognition of foreign materials, the innate immune system causes a non-specific inflammatory response through a chain of biochemical reactions.16,77,80,81 Responsible cells in the innate immune system consist of phagocyte cells (including dendritic cells, monocytes, and macrophages) and lymphocytes (natural killer cells, gamma delta T-cells, and innate lymphoid cells).16,81,82 However, the adaptive immune system is responsible for showing particular antigen responses and making a long-term memory through B and T lymphocytes.16,81,83A suitable immune system response requires organized crosstalk between these two systems, where chemical cues are intrinsically present and play pivotal roles. After biomaterial implantation, the degradation products and subsequent chemical surface changes of biomaterials can stimulate the immune system.71 The interactions between the surface and the immune system are reliant on the targeted tissue nearby the biomaterial causing tissue-specific biochemical responses.81 After biomaterial implantation, the native vasculature is likely to be disrupted, which could induce interactions between blood and the implanted biomaterial.56
Depending on the biomaterial surface physicochemistry, the plasma constituents including proteins, lipids, sugars, and ions can be adsorbed on it.77 Platelets, which through aggregation and coagulation form a fibrin-rich clot, are also a part of the blood exudate. The formed clot is a temporary provisional matrix for supporting cellular and molecular functions.84 The adsorbed proteins elicit biochemical signaling pathways and make interactions with the innate immune system cells such as neutrophils, monocytes, fibroblasts and endothelial cells through their particular recognition sites including C-termini, N-termini, proline–histidine–serine–arginine–asparagine (PHSRN) and arginine–glycine–aspartic acid (RGD).85–87
Neutrophils are commonly the first responders to foreign materials. These cells are stimulated when the adsorbed proteins (RGD, PHSRN), microbes (pathogen associated molecular patterns or PAMPs), and/or dead cell residues (damage-associated molecular patterns or DAMPs) bind to their ligands through biochemical reactions.87–90 The adsorbed proteins bind to macrophage type 1 antigen, lymphocyte function-associated antigen 1, and integrin alphaXbeta2. However, DAMPs and PAMPs bind to toll-like receptors (TLRs) and some specific pattern recognition receptors (PRRs), which also exist on the surface of macrophages and dendritic cells.87,91
Neutrophils stimulate the expression of cytokines as pro-inflammatory chemical mediators through sending biochemical signals.92,93 These chemical mediators stimulate directed chemotaxis of other innate inflammatory cells and dendritic cells, which leads to the stimulation of adaptive immunity responses through B and T lymphocytes.77,94
DAMPs are endogenous molecules that under normal physiological conditions are sequestered intracellularly and cannot be recognized by the innate immune system.95 Nevertheless, under cellular stress or injury conditions, they are released into the extracellular environment leading to the transmission of biochemical signals to cells for initiating inflammatory responses under sterile conditions.95 The DAMP release from cells depends on the type of cell injury or death. Chromatin-associated protein, high-mobility group box 1, heat shock proteins, and purine metabolites are prototypical DAMPs derived from damaged cells.95–98 Furthermore, ECM degradation can send biochemical signals for releasing DAMPs. DAMPs can initiate inflammatory responses, and the lack of DAMPs in the environment leads to a decrease of inflammatory biochemical cues.99
There are different types of PRRs in the innate immune system that stimulate the expression of various types of pro-inflammatory cytokines and biochemical markers. According to the subcellular location of PRRs, they are classified into two main groups: (i) TLRs and C-type lectin receptors, which are transmembrane proteins, and (ii) RIG-I-like receptors (retinoic acid-inducible gene-I-like receptors, RLRs) and NOD-like receptors (NLR), which exist in the intracellular compartments. PAMPs and DAMPs activate these receptors and subsequently inflammasome complexes.100,101
The inflammasome complex contains a cytosolic sensor that can be a PRR of the NLR, absent in melanoma 2 (AIM2) receptors, and an effector protein.102 There are various types of PRRs, which can form inflammasomes such as NLRP1, NLRP3, NLRC4 (the NLR family of intracellular proteins) and AIM2.103–105
In response to PAMPs and DAMPs, the pentameric or heptameric assembly of PRRs can oligomerize the caspase recruitment domain in filaments. This might cause the inflammasome formation through stimulating caspase-1.106 The NLRP3 inflammasome is the most known inflammasome, which contains the NLRP3 scaffold, caspase recruitment domain adaptor protein, caspase-1, and accessory protein serine/threonine-protein kinase.107,108 Monocytes, macrophages, granulocytes, dendritic cells, epithelial cells and osteoblasts mainly express this inflammasome.109
After cellular injury through biomaterial implantation, DAMPs and PAMPs activate the NLRP3 inflammasome through sending biochemical signals.110,111 Examples of such stimuli from the DAMP group are crystalline matter such as asbestos, calcium influx, mitochondrial reactive oxygen species (ROS), and extracellular neurotransmitter adenosine triphosphate (ATP).112 However, this process is not yet fully understood and needs further detailed studies.73 The inflammasome can through subsequent control over the rest of immune response processes either cause the resolution of inflammation and tissue regeneration, or lead to chronic inflammation and fibrosis.113 After inflammasome expression, the migrated monocytes/macrophages adhere to the temporary provisional matrix formed on the biomaterial surface.114
After 24 to 48 hours, the activated neutrophils die through apoptosis and release some vesicles and lipids through biochemical signals (e.g. lipoxins and resolvins), which have anti-inflammatory influences.87,115,116 Hence, neutrophils through binding to PAMPs and DAMPs and initiating inflammasome responses are vital for activating type 1 macrophages and the acute inflammatory phase. Apoptotic neutrophils are also crucial for stimulating macrophage polarization from type 1 to type 2 and the following inflammation resolution. Therefore, if their lifespan is extended and/or if they increase in number at the biomaterial surface, chronic inflammation can occur at the site.117
After the polarization of type 1 macrophages to type 2, they locally release several growth factors (such as transforming growth factor beta and vascular endothelial growth factor) while stimulating fibroblast and endothelial cell migration and proliferation by sending biochemical signals. Fibroblasts produce collagen to form the ECM, whereas endothelial cells nourish the formation of new blood vessels to offer essential nutrients for neo-tissue formation as well as for waste removal.118 In the chronic inflammatory phase, T lymphocytes, mainly helper T cells, play key roles in controlling the expression of pro- and anti-inflammatory mediators.119 In this system, B lymphocytes are responsible for making antibodies (Fig. 4).120 Immune-modulatory biomaterials should direct biochemical signaling pathways and cues, which are responsible for the functions of neutrophils, PAMPs, DAMPs, inflammasomes, endothelial cells, and mesenchymal stem cells (MSCs).121 As describing the details of innate and adaptive immune system mechanisms and the responsible biochemical cues is out of the scope of this review, we refer the readers to the following seminal review papers.16,73,77,80,95
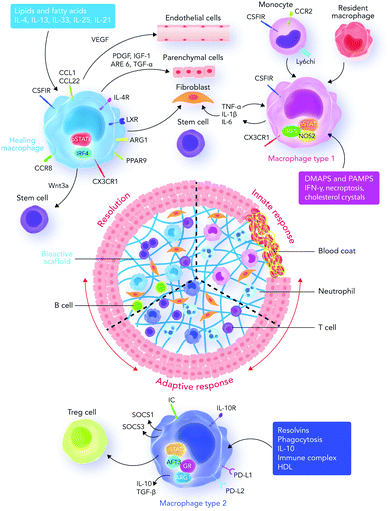 | ||
| Fig. 4 Innate and adaptive immune system responses. Recruited and resident macrophages start experiencing marked phenotypic and functional changes in response to damage-associated molecular patterns (DAMPs), pathogen associated molecular patterns (PAMPs), growth factors, cytokines, and other mediators released into the interface area. The main phenotypic changes regulate inflammation, tissue repair, regeneration, and resolution. Macrophages then express different types of factors which can direct different functions in fibroblasts, epithelial cells, endothelial cells, and stem and progenitor cells to promote tissue repair. During the final stages of the healing process, a regulatory pro-resolving phenotype, which confirms the suppression of the tissue-damaging inflammatory response, is expected. If the process does not successfully proceed, persistent inflammation and/or maladaptive repair processes can cause tissue-destructive fibrosis. Sometimes, the recruited monocytes lead to the formation of a resident macrophage phenotype in tissues.122 Abbreviations: DAMP, damage-associated molecular pattern; PAMP, pathogen-associated molecular pattern; Treg cell, regulatory T cell; IRF5, interferon regulatory factor 5; NOS2, nitric oxide synthase 2; LXR, liver X receptor; AREG, amphiregulin; Arg1, arginase-1; IRF4, interferon regulatory factor 4; PPARg, peroxisome proliferator-activated receptor g; FGF, fibroblast growth factor; GAL-3, galectin-3; TGF, transforming growth factor; GR, glucocorticoid receptor; ATF3, activating transcription factor 3; SOCS, silencer of cytokine signaling. | ||
6. The roles of ion channels in regulating immune system responses
Cell surface receptors play key roles in receiving biochemical signals (from chemical substances such as hormones, growth factors, cell adhesion molecules, nutrients, and neurotransmitters) and initiating biochemical and/or biophysical signaling in the cells.123–126 Ion channels are a class of surface receptors, which control many cellular signaling events in cells.127–129 Ion exchanges between the intra and extracellular environments create the mechanisms essential for controlling the cell metabolism and activation state.130 In addition, ion channels are important regulators of cell–cell communication. As a result, genes encoding proteins responsible for regulating membrane permeability to ions are also vital in most of the complex intra and extracellular signaling events.130 Because of the key roles of immune cells in controlling foreign body responses, we discuss some ion channels that regulate innate and adaptive immune system responses here.Ion channels direct immune responses mostly by regulating endosomal pH and intracellular calcium concentrations.131,132 Regulating the intracellular calcium amounts is dependent on the biophysical properties of the ion channels and their ability to control the calcium passage across the membrane.130 The calcium permeability can be changed by activating particular ligands, feedforward responses to the calcium release from intracellular stores, changes in cell polarization, and the strength of sodium driving force.130
In adaptive immune system cells (B- and T-lymphocytes), regulating the intracellular calcium amount is important as releasing calcium from intracellular stores activates the immune response pathways.133 In addition, the CAV1 (caveolin 1) ion channel (as a subfamily of L-type voltage-gated calcium channels) is vital in activating B- and T-lymphocytes.134,135
Increasing ROS activates transient receptor potential melastatin (TRPM) 2 ion channels.136 The TRPM2 activation causes the release of calcium from immune cells. In addition, TRPM2 has a key role in activating the NLRP3 inflammasome causing the expression of cytokines and chemokines from immune cells.136
Some studies have reported the importance of ion channels in regulating microglia functions as the resident macrophage cells of the central nervous system.137,138 In microglia, the P2X and N-methyl-D-aspartate (NMDA) receptor families respond to the neurotransmitters adenosine triphosphate (ATP) and glutamate, respectively.139 By regulating the intracellular calcium concentration, these receptors can affect microglial activation. P2X receptors (P2XRs) are trimeric plasma membrane channels, permeable to small inorganic cations (e.g. Na+, K+, and Ca2+).127,140 However, some P2XR channels are permeable to both cationic and anionic organic ions.141 Ferreira et al.142 studied the Ca2+-activated K+ channel (KCa3.1)-dependent responses in microglia under ROS.142 They concluded that increasing the cyclic guanosine monophosphate (cGMP) concentration leads to protein kinase activation and, subsequently, ROS formation in mitochondria. The ROS formation causes endoplasmic reticulum calcium release, which subsequently binds to calmodulin to activate the KCa3.1 channel.142
Connexin and pannexin cell–cell channels, unopposed hemichannels as well as P2 receptors are essential in initiating and regulating the inflammatory responses.143 For instance, the activation of connexin and pannexin channels leads to the release of ATP and other metabolites to the extracellular media. Extracellular ATP can stimulate intracellular signaling pathways by acting on P2 receptors, which leads to inflammation.143
Overall, the activation of ion channels can be “danger” signals propagating the inflammatory responses of immune systems.143 Their biochemical effects on cell homeostasis affect the immune system functions.133 Therefore, the activation of ion channels can be vital in directing the host responses to biomaterials. However, more research on understanding the ion channel roles in regulating signaling pathways and directing cell–biomaterial interactions is vital.
7. The role of mesenchymal stem cells in biological responses to biomaterials
MSCs have many roles in modulating the immune system responses to implanted biomaterials, particularly in biochemical signaling pathways responsible for stimulating the innate immune system.144,145 These cells can have immunosuppressive roles by releasing several soluble biochemical factors responsible for controlling the functions of lymphoid and myeloid cells.145–147 For example, prostaglandin E2 (PGE2) synthesized by MSCs can stimulate macrophages to have an adapted directing phenotype by increasing IL-10 and decreasing tumor necrosis factor-α and IL-12 expression.148 The biochemical soluble factors released by MSCs (e.g. IL-10, PGE2 and IL-1b) can play vital roles in the crosstalk between MSCs and macrophages, mainly in macrophage type 1 to 2 polarization.149 In addition, interferon gamma and tumor necrosis factor-alpha cytokines expressed from T cells can stimulate macrophage polarization by stimulating MSCs to release cyclooxygenase-2 and indoleamine 2,3-dioxygenase.150MSCs can also control T regulatory lymphocytes (Tregs) and T helper-based immunosuppressive activities by releasing heme oxygenase-1 and its metabolic by-product carbon monoxide.145,151 Because type 2 macrophage polarization is linked with Tregs stimulation, MSCs are vital in controlling the crosstalk between innate and adaptive immune systems.145,151 These cells can down-regulate the expression of some lymphocyte growth factors, differentiation of antigen presenting cells and effector T cells as well as epithelial cell proliferation by releasing PGE2.145 The readers can find a good level of details concerning the role of MSCs in regulating immune system responses to foreign materials in these review papers.145,148,152
8. The role of mechanotransduction pathways in biological responses to biomaterials
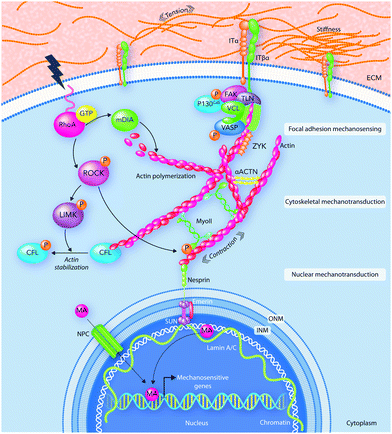
The local microenvironment and physical forces surrounding the cells can play a crucial role in several physiological mechanisms including embryonic development, in adult physiology, and in a wide variety of different disorders and diseases. For example, in tissue development, the local physical forces can control dorsal closure, epithelial morphogenesis and skeletal growth, ECM remodeling, vascular inflammation as well as tissue regeneration processes.153–155 In addition, at the cellular scale, cell-generated contractile forces can dictate both the cytoskeleton assembly and cellular architecture formation.155,156Conversely, cells translate these mechanical stimuli into biochemical responses in a process that is typically referred to as mechanotransduction.157,158 Therefore, the cell ability to sense the mechanical properties surrounding them is a key decision-making factor influencing cellular responses to biomaterials.159,160Fig. 5 shows how myosin motors and mechanosensors play a role in mechanotransduction pathways.11,161 The cellular membrane is the main location of force transmission from the ECM to cells. When cells encounter a stiff substrate, several multiprotein complexes known as focal adhesions are activated and become the central hub of cell–ECM interactions.
The mechanosensing activity of focal adhesions includes recognizing and transporting mechanical signals from the ECM to the cellular cytoskeleton. Many of the focal adhesion complexes have both transmembrane and intracellular components. The intracellular layer is an interface between the transmembrane components and the actin cytoskeleton.157,158,162 The molecular composition of the focal adhesion core can be very diverse and is mainly sensitive to the ECM composition and mechanics.163,164 Focal adhesions are created after the assembly of transmembrane proteins for physical interactions with ECM components.
Chen W. et al.165 reported that the “inside-outside signaling” mechanism inside cells or extracellular mechanical stimuli control integrin affinity for its ECM ligand.165 The activated integrins assemble and strengthen the molecular links at the cell–matrix interface.166 The ECM structure can elicit the expression of certain integrin subsets, which in combination with other biochemical signaling pathways can lead to particular cellular responses to physical forces.167 Artola et al.168 revealed that cells can adjust their force production to be ideal at tissues with different physiological conditions by controlling the expression of various integrin types.168
In addition, cells can control their own mechanical properties through changing their cytoskeletal architecture, which is a dynamic network of filamentous and cross-linking proteins.169 Cytoskeleton networks contain three main components including actin fibers, microtubules and intermediate filaments.170 The F-actin sliding on the motor protein myosin II provides the cytoskeleton contractility.171
In summary, the mechanical properties of the cell microenvironment display a direct effect on several cellular functions after their translation to biochemical signals. Therefore, it is undeniable that the mechanotransduction pathways and their following biochemical signals play critical roles in directing host responses to biomaterials.11,161 We will discuss the effects of biomaterial surface physicochemistry on the biochemical signals caused by mechanotransduction pathways in more detail in the subsequent sections.
9. Biomaterial strategies for directing biological responses
9.1. Impact of biomaterial surface physical properties on biological responses
The physical properties of the biomaterial surface can direct biophysical and biochemical signaling pathways involved in cellular responses to the surface. However, the mechanisms involved in cell responses to these surface properties are not yet fully understood. In the following sections, we provide a brief overview of the current progress and challenges in this field.9.1.1.1. Biological responses to feature size and geometry of biomaterial surface. In the natural processes of tissue healing and/or regeneration, the curvature or topographical features of other surrounding cells and ECM guide the injured cell functions.174,175 Hence, the surface topography of scaffolds can affect the cell fate determination, adhesion, polarization, and migration through manipulating physicochemical signaling pathways.176 Topographical features including shape, size, and geometric structures can direct cellular functions through influencing either the cytoskeleton organization and protein orientation or protein unfolding.
Actin filaments can spread out on the 2D structure of flat surfaces; however, the curved surfaces offer a 3D network for cells to grow inside the material.177 Rianna et al.178 studied the mechanotransduction pathways and biochemical factors behind cell responses to topographical patterns through investigating the mechanical properties of peripheral and nuclear regions of cultured NIH-3T3 cells on azopolymer scaffolds with various topographical patterns. They designed micrometer scale patterns in either parallel ridge or square lattice geometry and then studied the mechanical cell responses by atomic force microscopy (AFM). Their results indicated that surface topographical features stimulate the cytoskeleton network to generate some forces, which affect nucleus functions.178 Scientists and engineers used a wide range of strategies for improving the topographical features of biomaterials and therefore controlling cell functions in a non-invasive manner.179–181 The topographical patterns can be represented either anisotropically by grooves and ridges or isotropically through random spreading of protrusions and pits.182
Regarding anisotropic patterns, studies investigate the alignment of cells alongside the anisotropic direction regardless of the topography scale. However, in isotropic topographies, which have more effects on cell signaling pathways, the topography size plays a key role in controlling cell responses to patterns.183,184 Among the topographic parameters, the size of designed patterns, in both micro- and nano-scales, can play a vital role in controlling cell functions. The micro-scale patterns can profoundly influence the overall cell morphology; however, the nano-scale topographies are more critical in controlling the molecular and subcellular physicochemical sensing pathways.182,185
Padmanabhan et al.186 investigated the role of surface topography size and stiffness of metallic glass nanorod arrays on cell–cell fusion. They revealed that the topographic features in nano-scale size can dominate biochemical signals in decreasing fusion through controlling cytoskeletal remodeling-associated signaling pathways.186 Researchers focus on manipulating protein adsorption mechanisms and signaling pathways via designing substrates with nano-scale surface topography.187,188 In addition, the nano-scale substrates can be used to answer basic questions concerning protein adsorption/desorption at the nano-scale. Wang et al.189 developed a patterned poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) brush with sub-100 nm structures over large areas by combining block copolymer micelle lithography and surface-initiated atom transfer radical polymerization (ATRP).189 The PDMAEMA brushes were neutralized and collapsed at pH 9, while positively charged and swollen at pH 4. The authors studied bovine serum albumin (BSA) adsorption on PDMAEMA brushes using laser scanning confocal microscopy, AFM, and quartz crystal microbalance with dissipation (QCM-D). Because of the steady sub-100 nm topography of the patterned brushes, the authors could observe the protein adsorption mechanisms inside and outside of brushes using the AFM technique (Fig. 6).189
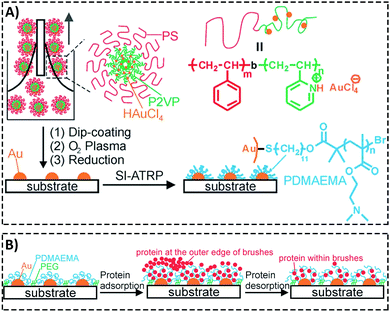 | ||
| Fig. 6 (A) A representative graphic of fabricating patterned poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) brushes with sub-100 nm structures over large areas using a combination of block copolymer micelle lithography and surface-initiated atom transfer radical polymerization (ATRP). (B) A schematic illustration of protein adsorption mechanisms inside and outside of brushes with a nanostructured surface. Reproduced from ref. 189 published by The Royal Society of Chemistry. | ||
However, there are still some contradictions between the research results, which could be owing to the following reasons:
(i) Considering one individual defined scale for topographical features while biological environments are rich in physicochemical gradients.
(ii) Mostly, researchers focus on cell responses to one individual physical or chemical property. However, we recommend considering the synergistic effects of topographical and chemical properties of the surface on each other. Surface nanofunctionalization has attracted much attention as a promising strategy for enhancing cell responses to biomaterials, which we will address later in this paper.
Liu et al.190 designed some surface nanotopography gradients through surface immobilization of gold nanoparticles in a density-dependent manner. They modified the surface chemistry of scaffolds via coating a thin plasma polymerized film with allylamine (AA) or acrylic acid (AC) chemical composition on the surface (Fig. 7A). They revealed that surface nanotopography plays the main role in stimulating the initial cell adhesion and spreading. However, both topographical and chemical properties of the surface govern cell differentiation.190 After culturing osteoblast-like SaOS-2 cells on surfaces, surface nanotopography could enhance the stimulating effects of allylamine chemical treatment on osteogenic differentiation (Fig. 7B). Furthermore, in the natural in vivo conditions, the biological interactions with surface topographical features occur in an inhomogeneous and dynamic environment. These inhomogeneous dynamic interactions between micro/nano topographical patterns and molecules are complicated because local changes in other surface features, mainly chemistry, control attractive and repulsive forces on the surface.191 Some strategies are available to design dynamic topographical features without disturbing environmental conditions or affecting the surface chemistry of scaffolds.
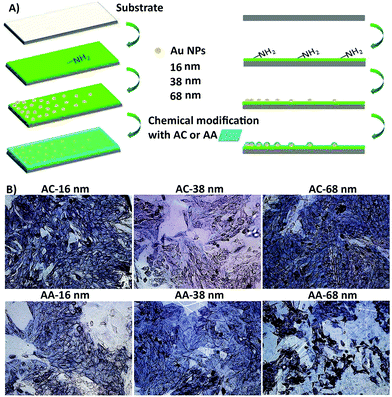 | ||
| Fig. 7 (A) An illustration of the process to design the biomaterial with different surface topography gradient designed from three different sized gold nanoparticles (16, 38, 68 nm) coated with acrylic acid (AC) and allylamine as uppermost surface chemistry modification (AA) (i.e. AC 16, AC 38, and AC 68). (B) They cultured osteoblast-like SaOS-2 cells for seven days. The surface nanotopography could improve the influence of AA chemical treatment on osteogenic differentiation, especially on AA 68 surfaces. The ALP expression of cells was highest at position 8 mm of AA. Scale bar = 100 μm. Reprinted from ref. 190. Copyright © 2018, Elsevier. | ||
Hernandez et al.181 suggested an in vitro approach for stimulating cells with dynamic topographical features of protein-based hydrogel surfaces. They modified scaffolds in situ in real time by positioning a pulsed near-infrared laser focus inside a hydrogel, which leads to enhancement of the chemical cross-linking and consequently local contraction of the protein matrix. Fig. 8 shows that exposing hydrogels to a series of scan patterns can generate, remove, or retreat topographical patterns without having destructive effects on other surface properties or cell functions.181 By using a laser-based confocal microscope technique, researchers can also design a synthetic scaffold capable of controlling cell orientation and migration in time and space without affecting surface chemistry.192 However, stimulating spatiotemporal dynamic topographic parameters without affecting surface chemistry is still in its infancy and needs more research.
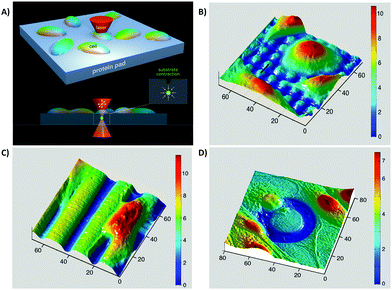 | ||
| Fig. 8 In situ imprinting of topographical properties on protein-based hydrogel scaffolds. (A) Multiphoton excitation of the photosensitizer in the scaffold through a pulsed near-IR laser beam enhanced hydrogel contraction. Through limiting the excitation to 3D defined volumes, the authors could achieve imprinting without exposing cell or material surfaces to optical destruction. (B–D) In situ imprinting under cultured cells. The authors imprinted different topographies in the presence of cells such as micropost arrays (B), grooves (C), and annular depressions (D). All numbers are in units of μm. Reprinted with permission from ref. 181. Copyright © 2018, American Chemical Society. | ||
9.1.1.2. Biological responses to biomaterial surface roughness. Surface roughness relates to the texture of the biomaterial surface and is commonly represented with the roughness value (Ra/Sa), which quantitatively represents the roughness grade.176,193 The broadly used techniques for producing and controlling surface protrusions and/or depressions are blasting, electropolishing, nanoparticle/fiber formation, and nanofabrication technologies such as photolithography.176,194,195 Using chemical surface treatments such as acid etching can further increase the surface roughness when compared with traditional machining techniques.196 The biomaterial surface roughness can directly dictate specific cellular responses. For example, Barr et al.197 studied the biocompatibility of 13 commercially available breast implants by focusing on macrophage responses to their roughness feature. They showed that macrophage responses to surface roughness determine the biocompatibility of implants.197
By considering the current assumptions about the effects of surface roughness on biological functions, one may identify several challenges:
(i) The first challenge relates to the other suggested parameters, which besides Ra can play key roles in determining surface roughness. Anselme and co-workers198–200 showed that the fractal dimension (Δ) and the developed surface can also be key parameters in determining surface roughness. The fractal dimension parameter can be useful in measuring surface disorder; however, the developed surface factor is related to the surface detachment index.198 The mean distance between peaks (RSm), the sum of the average height of the five highest profile peaks and the average depth of the 5 deepest profile valleys calculated from the parallel line to the mean line (Sz), kurtosis (Sku), skewness (Ssk) and fluid core index (Sc) are important roughness parameters.201–205
(ii) We cannot decide about host responses to biomaterials based on improving only one physicochemical property or (iii) testing with one or two cell types in vitro. However, several research groups reported designing biocompatible materials by only improving their roughness through mainly focusing on Ra measurement and in vitro testing with one or two cell types. Table 1 shows that increasing the surface roughness of one biomaterial can have positive effects on one cell type or protein (a phenomenon called rugophilia); however, it can have negative influences on other types. Increasing the surface roughness can decrease or not affect the proliferation and/or differentiation of leukocytes, keratinocytes, and monocytes; however, it can improve osteoblast proliferation on the surface.206–208
| Materials | R a | Other modified physicochemical properties | Cell responses | Ref. |
|---|---|---|---|---|
| Abbreviations: smooth surface (SS), rough surface (RS), poly(ε-caprolactone) (PCL), not available (NA), a human osteosarcoma cell line (MG-63 cells), mean distance between peaks (RSm), CFRPEEK–nanohydroxyapatite ternary composites (PEEK/n-HA/CF), root mean square average roughness (Rq), a mouse osteoblast-like cell line (MC3T3-E1), N-isopropylacrylamide (NIPAM), polydimethylsiloxane (PDMS), a human liver cancer cell line (HepG2), tricalcium (TCP), dicalcium silicate (C2S), poly(L-lactide) (PLLA), mouse embryonic fibroblasts (NIH 3T3), human bone osteosarcoma cell (U-2 OS), polyhydroxybutyrate (PHB), two melanoma cell lines (VGP WM115 cells) and (WM266-4 cells), polyethylene oxide (PEO), 1,4-polyisoprene (PI), calcium phosphates (CaP), hydroxyapatite (HA). | ||||
| Silicon | 1.07–80.03 μm | NA | Surface roughness in this range induces poor macrophage polarization and an innate potential to increase the pro-inflammatory response. Depending on the amount, Ra has variable influences on inflammatory factors. | 197 |
| Polystyrene | ∼121, 505, 867 nm | NA | Rough surfaces with nano-meter dimensions make E-cadherin junctions and human gingival keratinocytes to grow gradually or imperfectly. | 208 |
| PCL | 654 ± 91 nm | Hydrophilic surface | Increasing the initial cell attachment, proliferation, and differentiation of MG-63 cells. | 223 |
| PCL | ∼0.5–4.7 μm & RSm ∼ 214–33 μm | NA | Depending on the surface roughness gradient, faster, slower, or similar osteogenic commitment and gene expression of MSCs can be observed on rough surfaces in comparison with smooth surfaces. | 201 |
| PCL | 1, 1.3, 2 μm | HA coating | Increasing the surface roughness increases osteoblast attachment and differentiation. | 213 |
| Increasing the activity of the osteoclast marker, tartrate-resistant acid phosphatase, by decreasing roughness. | ||||
| PDMS & PNIPAM | 2.6 ± 0.7–163.6 ± 11.7 nm | NA | Increasing the roughness causes a decrease in both amount and regions of MSC and HepG-2 attachment. | 216 |
| PLLA | 1 × 1–20 × 20 μm | NA | Increasing the surface roughness increases fibroblast proliferation; however, it decreases osteoblast proliferation. | 224 |
| PHB | Pristine PHB = 32.9 nm | Laser surface treatment | Surface roughness can play a leading role in determining NIH 3T3 responses. | 225 |
| Treated PHB = 270 nm | ||||
| PEO & PI | R a = NA | Hydrophobicity | VGP WM115 cell adhesion forces are higher on surfaces with various hydrophobicity and roughness in comparison with WM266-4 cells. | 226 |
| R q = 1–5 nm | ||||
| Zirconia | 1.7 & 3 μm | NA | 1.7 μm Ra shows better osteoblast responses both in vitro and in vivo compared to 3 μm Ra. | 227 |
| PEEK/n-HA/CF | ∼0.09–2.95 μm. | Surface treatment | Moderate surface roughness increases MG-63 cell attachment/proliferation. | 228 |
| R q ∼ 0.17–3.64 μm | Suitable surface roughness increases bioactivity and osseointegration in vivo. | |||
| α-Tricalcium phosphate and αTCP | R a = 0.46–2.29 μm, Rq = 0.67–2.78 μm, Rz = 4.68 | 1.5 wt% or 3.0 wt% of C2S | Increasing MSC adhesion and proliferation by increasing surface roughness. | 202 |
| CaP | 12.52 μm & 0.9–1.7 μm. | Chemical treatment with HA and β-TCP | The influence of surface roughness depends on surface chemistry. | 229 |
| HA causes higher mineralizing activity of MSCs at Ra = 1.5 μm. | ||||
| β-TCP increases the osteogenic differentiation of MSCs when Ra = 1.7 μm. | ||||
| TiAl6V4 | ∼0.30–1.80 μm | NA | Surfaces with Ra in the 0.50–1.00 μm range increase MC3T3-E1 cell proliferation. | 230 |
| Ti6Al4V | 0.114, 0.277, 0.316 μm | Surface treatment and melatonin | Increasing the surface roughness increases osteoblast adhesion after 24 h cell culture. | 196 |
| Adding melatonin to the surface increases osteoblast proliferation. | ||||
| Titanium | 100–400 nm | NA | Increasing the surface roughness increases osteoblast differentiation, macrophage tendency to polarize to type 1 macrophages, and osteogenic ability. | 231 |
| Titanium | 0.02–3.63 μm | NA | Surface roughness can have a combinational influence on osteoclast genesis and osteogenic differentiation of macrophages. | 212 |
| Titanium | 100 nm | NA | Increasing the surface roughness inhibits MC3T3-E1 cell functions through decreasing cell attachment, proliferation, and calcification ability. | 232 |
| Stainless steel | SS = 2 nm & RS = 0.9 μm | NA | Mechanotransduction pathways play key roles in determining MSC responses to surface roughness. | 214 |
| TiAl6V4 & 316 L stainless steel | 0.01–0.1 μm | NA | Surface roughness in this range has no effect on osteoblast adhesion mechanisms when roughness is isotropic and groove width is lower than a critical amount. | 215 |
| It can only affect osteoblast orientation on wider grooves. | ||||
(iv) Even in one cell type, the surface roughness can affect different cell functions including cell adhesion, migration, proliferation, and differentiation in various ways. Increasing the surface roughness reduces the proliferation and increases the differentiation of osteoblasts.209–211 When we evaluate host responses to biomaterials in tissues containing different cell types, the differences between responses of various cells to surface roughness can pose many challenges.212,213
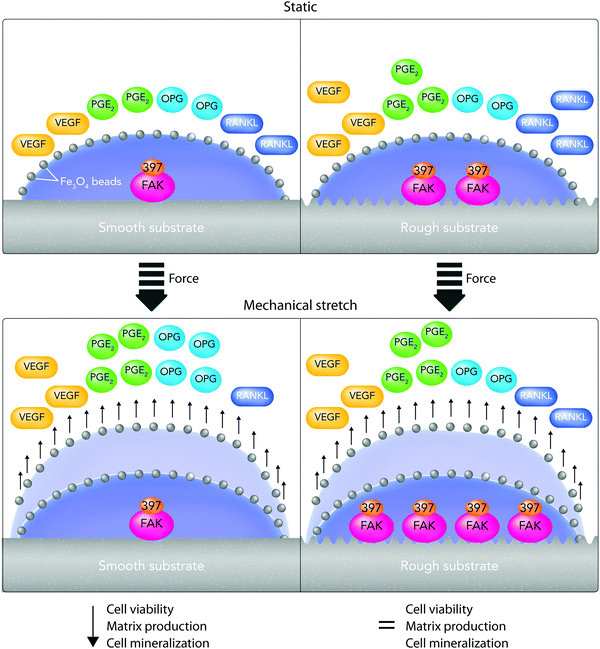
(v) On the other hand, there is a considerable contradiction between the literature outcomes when it comes to one particular cell type response to surface roughness. These contradictions could arise from the current misuse of biocompatibility definition by ignoring critical biochemical signal transduction pathways, which indeed might play vital roles in determining cell responses.11,13 For example, Saldana et al.214 studied the role of mechanotransduction pathways in controlling human-MSC (hMSCs) responses to stainless steel surface roughness. In this study, the surface Ra was manipulated with 2 nm or 0.9 μm for smooth and rough samples, respectively. Fig. 9A provides an overall perspective of the multitude of receptors and proteins that were involved in this study. It shows that under static conditions, improving stainless steel roughness increases the expression of biochemical markers including PGE2, vascular endothelial growth factor (VEGF), and receptor activator of nuclear factor kappa-B ligand (RANKL) as well as the phosphorylation of focal adhesion kinase (FAK) to its active form (Tyr-397).214 Moreover, Fig. 9B indicates that applying tensile forces to the plasma membrane of hMSCs improves VEGF secretion on smooth surfaces as well as PGE2 amounts and osteoprotegerin/RANKL proportion on both smooth and rough surfaces. Although mechanical stretch does not affect smooth surfaces, it stimulates FAK phosphorylation at Tyr397 on rough surfaces. Overall, showing the influence of stretch in enhancing FAK phosphorylation at Tyr397 on rough surfaces (Ra = 0.9 μm) is evidence for the current hypothesis that mechanotransduction pathways can be substantial factors in directing cell responses to surface roughness.214
(vi) Although surface roughness can have positive or negative effects on cell responses in short-term periods (less than 48 h), its influence can change over time. Therefore, it is recommended to consider both short-and long-term cell responses to surface roughness. For example, Lee et al.209 suggested a new strategy for controlling the sensitivity of different cell functions to surface roughness through using shape memory materials. They designed a shape memory (meth) acrylate copolymer with thermomechanical properties, which had a time-dependent dynamic surface change from smooth to rough under cell culture conditions. They used soft lithography techniques for making rough surfaces and then by applying compression decreased the surface roughness to generate smooth areas. Their results showed that under static conditions, surface roughness does not affect osteoblast amount, alkaline phosphatase specific activity (ALP), as well as osteoprotegerin and VEGF expression; however, it enhances osteocalcin expression. After three days of culture of cells on rough surfaces under dynamic conditions, surface roughness caused a decrease in DNA content and an increase in osteocalcin and osteoprotegerin expression.209
(vii) Another challenge involves the determination of the critical roughness value for each specific biomaterial type. The reported roughness value is different from study to study. Although a few statistical studies have revealed the critical roughness value on different surfaces,198,215 it is still difficult to define one critical roughness value, which can properly guide cell functions. Therefore, researchers suggest decreasing dissimilarities between cell responses by considering an average roughness gradient, rather than individual numbers.201,216 For instance, Zhou et al.216 designed some polydimethylsiloxane (PDMS) substrates with surface roughness gradients by using a combination of microfluidics and photopolymerization techniques. They grafted N-isopropylacrylamide (NIPAM) with concentration gradients onto PDMS substrates, which produced a gradient of roughness ranging from 2.6 ± 0.7 nm to 163.6 ± 11.7 nm on the surface (Fig. 10A). The applied gradient improves cell attachment on the surface in both MSCs and hepatocellular carcinoma cell lines (HepG-2) (Fig. 10B).216
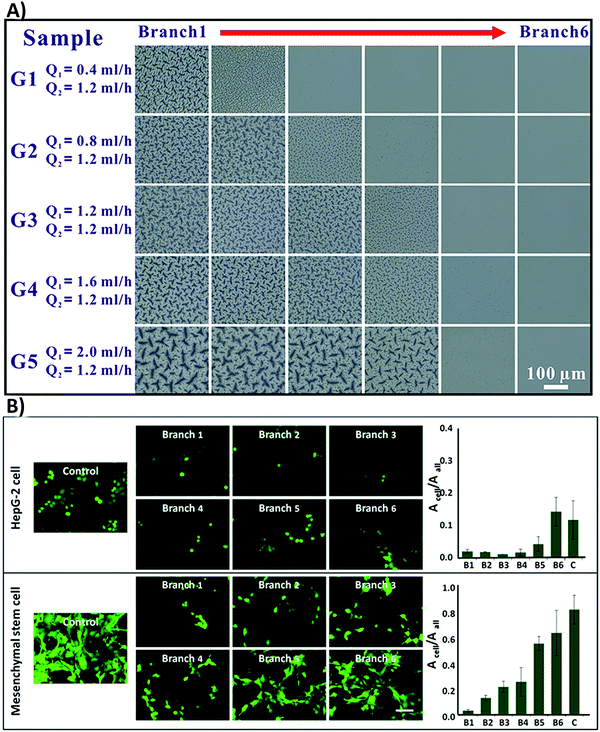 | ||
| Fig. 10 (A) The optical images of the polydimethylsiloxane (PDMS) surface grafted with N-isopropylacrylamide (NIPAM) with concentration gradients. A total of five samples were represented with flexible Q1 and fixed Q2 (microchannels were injected with red (Q1) and green (Q2) dye solutions to improve the contrast). Branch one contained the highest NIPAM concentration and branch six had the lowest. (B) Fluorescence images of MSC and HepG-2 cell attachment on the surface with various roughness grades. The authors stained HepG-2 cells with cell tracker dye (green) and MSCs were transfected with GFP (green). Acell was applied to determine the total cell adhesion region; however, Aall was the total area of the vision (Acell/Aall is the proportion between the total area of cell adhesion and the captured vision). The scale bar is 50 μm. Reprinted with permission from ref. 216. Copyright © 2015, American Chemical Society. | ||
(viii) Designing biomaterials with multiscale surface roughness in both micro- and nano-scale can also improve cell responses. A roughness gradient in both micro- and nano-scale can improve interactions between various cell or protein types on the surface.217
(ix) The synergistic effects of roughness with other physical and chemical properties, which are different depending on the situation, can also cause contradictions. Rough surfaces with different topographies or stiffness have various influences on cell functions.218 The same roughness value in two different types of materials can affect cell responses differently, which can be owing to the differences in their chemistry.219–222 Fukuda et al.220 investigated the osseointegration ability of a poly(ether ether ketone) implant by enhancing its surface roughness and/or surface chemistry (Fig. 11A). Fig. 11B shows that phosphorylation of the surface enhances cell responses on both smooth and rough surfaces, with more improvement on rough surfaces. However, only modifying the surface roughness cannot improve MSC responses. Fig. 11C shows the substantial effects of the combined surface modification strategies on bone regeneration in the rabbit tibia.220 Both chemistry and roughness properties of the surface play roles in neo-tissue formation.
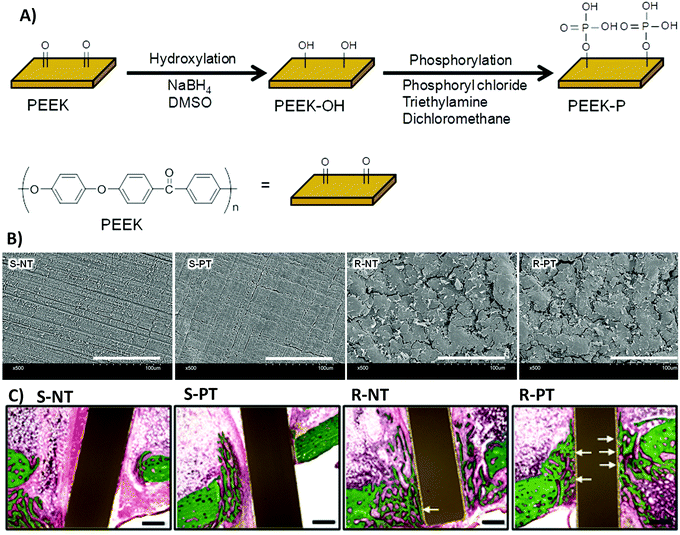 | ||
| Fig. 11 (A) A schematic of poly(ether ether ketone) (PEEK) surface phosphorylation. (B) Scanning electron microscopy (SEM) images of the biomaterial surface. S-NT: unmodified PEEK, S-PT: phosphorylated PEEK with a smooth surface, R-NT: sandblasted PEEK, R-PT: phosphorylated PEEK with a sandblasted surface. (C) Biomaterial implantation in the rabbit tibia. Illustrative histological images of neo-tissue formation and osseointegration of samples four weeks after surgery. The white arrows show neo-tissue formation at the interface with substrates. Scale bars, 500 μm. Reprinted from ref. 220. Copyright © 2018, Springer Nature in accordance to Creative Commons Attribution License. | ||
| Materials | Stiffness values | Stiffness modification approach | Cell responses | Ref. |
|---|---|---|---|---|
| Abbreviations: mouse fibroblasts (NIH3T3), thiolated heparin (Hep-SH), diacrylated poly(ethylene glycol) (PEG-DA), polyacrylamide (PAAm), adipose-derived stem cells (ADSCs), poly(ethylene glycol)-diacrylate (PEGDA), molecular weight (MW), human dermal fibroblasts (HDFs), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), polyelectrolyte multilayer (PEM), hepatic stellate cells (HSCs), methacrylated hyaluronic acid (MeHA), gelatin-hydroxyphenylpropionic acid (Gtn-HPA), storage modulus (G′), glycosaminoglycans (GAGs), poly-L-lysine (PLL), thiol group modified hyaluronan (HA-SH), poly(glycerol sebacate) (PGS), alkaline phosphates (ALP), n-octyl methacrylate–diethylene glycol dimethacrylate (DEGDMA/nOM), valvular interstitial cell (VIC), alpha smooth muscle actin (aSMA), polycaprolactone (PCL), polylactide (PLA), polyglycolide (PGA), umbilical cord mesenchymal stem cells (UC-MSCs), methyl acrylate/methyl methacrylate (MA/MMA), methacrylated hyaluronic acid (MeHA), human adipose derived stem cell (hASC), major quantitative measurements (MQMs), sulfated glycosaminoglycans (sGAG). | ||||
| PDMS | ∼2–85 kPa | Adding cross-linkers with different concentrations | The NIH3T3 adhesion is related to the exposure of cell-binding motif of fibronectin. | 247 |
| Motif exposure depends on the surface stiffness. | ||||
| MQMs: NA or difficult to summarize. | ||||
| PDMS | 0.6–2.7 MPa | Changing Sylgard 527 and 184 concentration | Surface stiffness does not affect MC3T3-E1 cell spreading; however, it can influence MC3T3-E1 osteogenic differentiation, but there is no rule like “the stiffer, the better”. | 249 |
| MQMs: (I) the vinculin expression on surfaces with 2.7 MPa stiffness is almost 1.5 times higher than that on surfaces with the lowest stiffness. (II) After three weeks of cell culture, the expression of osteogenesis markers on surfaces with 0.6 MPa stiffness is almost 1.5 times higher than that on surfaces with 2.7 MPa stiffness. | ||||
| PDMS or PAAm | 0.1–40 kPa | UV cross-linking | Increasing the surface stiffness increases the spreading speed of A549 cells. | 239 |
| In PAAm, the bulk stiffness affects cell behaviors; however, in PDMS, the surface stiffness plays a key role in regulating cell responses. | ||||
| MQMs: the cell spreading speed on stiff surfaces is almost 4-fold higher than that on soft surfaces. | ||||
| Hep-SH & PEG-DA | 10–110 kPa | Changing the concentration of the precursor solution | MQMs: (I) softer surfaces support the expression of heparin and maintenance of hepatocyte phenotype 5 times better than rough or stiff surfaces. | 250 |
| (II) Cell density on soft and stiff heparin gels is 690 cells per mm2 and 540 cells per mm2, respectively. | ||||
| PAAm | 6.1 or 46.7 kPa | Changing the concentration of bisacrylamide | Compared to topography and dimension, surface stiffness and/or dimension are predominant in controlling MSC proliferation. | 251 |
| Stiffness supports the osteogenic or neuronal differentiation of rBMSCs on a stiff or soft surface, respectively. | ||||
| MQMs: cell proliferation on stiff and soft surfaces is 3.49 ± 0.96 and 2.50 ± 0.42, respectively. | ||||
| PAAm | 100 Pa, 10 or 30 kPa | NA | Increasing the surface stiffness values increases foreign body responses (primary rat microglial cells and astrocytes) to surfaces. | 235 |
| MQMs: compared to soft surfaces, stiff surfaces show a significant increase in the inflammatory response (P = 1.9 × 10−6), immune cell trafficking (P = 9.6 × 10−5), cellular growth and proliferation (P = 7.5 × 10−5), cell-mediated immune response (P = 2.2 × 10−4), and antigen presentation (P = 1.1 × 10−4) of microglia. | ||||
| PAAm | 0.51, 3.7, and 22 kPa | Changing monomer concentration | Nuclear localization of osteogenesis transcription factors is dependent on surface stiffness. | 252 |
| ALP expression is improved only in MSCs, which are not only adhered to stiffer surfaces, but also have a direct contact with other cells. Both cell–cell contact and stiffness are important in guiding the cell fate. | ||||
| MQMs: NA or difficult to summarize. | ||||
| PAAm | 13–16, 35–38, 48–53, & 62–68 kPa | Changing bisacrylamide concentration | Increasing surface stiffness increases UC-MSC adhesion. | 253 |
| Tendency for adipogenic differentiation on softer surfaces, for muscle differentiation on surfaces with moderate stiffness, and for osteogenesis on high-stiffness surfaces. | ||||
| MQMs: regarding cell proliferation, after 2 days, the percentages of S phase cells on matrices of 13-16, 35-38, 48-53, and 62–68 kPa are 26.82, 26.64, 24.43, and 22.39%, respectively. | ||||
| PAAm | 1–25 kPa | Changing monomer concentration | ECM proteins can influence NIH 3T3 responses to surface stiffness. | 238 |
| NIH 3T3 exhibits durotaxis on fibronectin coated stiffness gradients but not on the lamin coated surfaces. | ||||
| MQMs: NA or difficult to summarize. | ||||
| PAAm | 0.5, 1.7, 2.9, 4.5, 6.8 & 8.2 kPa | Changing monomer and cross-linker concentration | Some stiffness values might be non-durotactic for hASC cells, other values can be durotactic and cause cell migration and differentiation. | 245 |
| MQMs: NA or difficult to summarize. | ||||
| PAAm | 0.04–0.95 kPa | Changing monomer and cross-linker concentration | Schwann cells exhibit durotaxis in response to stiffness gradients in the biological stiffness range of peripheral nerve tissue. | 244 |
| Median cell velocity on even surfaces is 0.67 μm min−1, on surfaces with low gradient stiffness is 1.55 μm min−1, and on surfaces with high gradient stiffness is 1.38 μm min−1. | ||||
| PAAm | 0.167 or 49.6 kPa | Changing monomer and cross-linker concentration | Hypoxia can affect MSC cell responses to surface stiffness. | 254 |
| MQMs: NA or difficult to summarize. | ||||
| PAAm | 5–60 kPa | Changing monomer and cross-linker concentration | Primary breast cancer cells experience significant phenotypic changes after culturing on surfaces with different stiffness. | 255 |
| MQMs: (i) an approximate 4-fold decrease in FBXW7 gene expression in cells seeded onto soft surfaces compared to glass. (II) A 10-fold overexpression of CYP1A1 in MDA-MB-453 cells on soft surfaces compared to glass. (III) A 3-fold decrease of cell apoptosis on soft surfaces. | ||||
| ECM-derived polymers | 15–194 kPa | Using PEGDA with different MW | Surface biochemical and mechanical cues synergize only at particular mixtures to increase bone differentiation of ADSCs. For osteocalcin gene expression, intermediate stiffness (55 kPa) and low concentration of fibronectin are optimal. | 240 |
| MQMs: (I) a 58-fold and 46-fold osteocalcin expression at 20% w/v PEGDA for both MW 3400 and MW 5000, respectively, compared to the control group. (II) Increasing fibronectin from 10 to 25 μg ml−1 decreases osteocalcin to 12–47-fold of the control. | ||||
| Glass slides | 0.5–110 MPa | Using a weak PEM system and EDC cross-linker | Compared to chemistry and wettability, surface stiffness is a stronger driving force in determining HDF cell fate. | 256 |
| MQMs: (I) after 6 days of cell culture, a 5-fold increase in cell number on soft regions compared to the control group. (II) However, a 10-fold increase in cell number on stiff regions compared to the control group. | ||||
| Self-assemble peptide nanofibers | 22.9 ± 5 or 7.3 ± 0.9 kPa | Supramolecular interactions | NIH 3T3 neuronal polarity and maturity are faster on softer nanofiber surfaces compared to stiff surfaces. | 257 |
| Surface stiffness can affect neuronal growth by adjusting its dynamics. | ||||
| MQMs: after 20 h cell culture, 67.1 ± 6.2% neurons on stiff surfaces reach developmental stage 2; however, 60.9 ± 2.6% neurons on soft substrates reach developmental stage 3 at the same time. | ||||
| MeHA | 2.1 or 24 kPa | UV cross-linking | Stiffer surfaces with E ∼24 kPa better support HSC cell adhesion and differentiation compared to softer ones. | 258 |
| MQMs: after 28 days of HSC cell culture, the cell mean area on soft and stiff surfaces is ∼200 and 5500 μm2, respectively. | ||||
| MeHA | 5, 12, and 23 kPa | Changing HA concentration | HMSC cell volume or size has a potential influence on foreign responses to surface stiffness. | 248 |
| MQMs: greater cell volume leads to more focal adhesion by increasing stiffness (62%, 38%, and 10% focal adhesion formation on surfaces with 2800, 3600, and 6000 μm3 cell volumes, respectively). | ||||
| Gtn-HPA | G′ = 570–2750 Pa | Changing H2O2 and Gtn-HPA concentration | Cartilage regeneration is dependent on surface stiffness values of scaffolds. | 259 |
| Scaffolds with medium stiffness (1000 Pa) better guide chondrocyte cell responses toward cartilage repair compared to softer or stiffer surfaces. | ||||
| MQMs: (I) after 10 days of cell culture, a hydrogel with medium stiffness produces the highest level of sGAG, which is twice that of low stiffness hydrogels. (II) The collagen expression increases by increasing stiffness, with 1.5-fold difference. | ||||
| PEG | 130 & 3170 kPa | Changing PEG-DA MW and concentration | Surface stiffness plays a key role in regulating MSC differentiation. | 260 |
| RGD nanospacing influences MSC spreading and differentiation irrespective of surface stiffness. | ||||
| Both surface stiffness and nano-scale spatial organization of cell-adhesive ligands play key roles in regulating stem cell fate. | ||||
| MQMs: cell density and area as well as F-actin intensity are almost 30% higher on stiff surfaces compared to soft surfaces. | ||||
| collagen-GAG | 5.05–2.85 kPa | Carbodiimide cross-linking and benzophenone photoimmobilization chemistries | The stiffest surfaces guide the osteogenesis of ADSCs irrespective of the presence or absence of growth factors. | 261 |
| Softer surfaces needed biochemical cues to guide cell fate. | ||||
| Cell proliferation is enhanced on surfaces with moderate stiffness. | ||||
| MQMs: NA or difficult to summarize. | ||||
| Cellulose | 76–448 kPa | Changing glyoxal cross-linker concentration | Surface stiffness can regulate MG-63 morphology. | 262 |
| MQMs: NA or difficult to summarize. | ||||
| Silk fibroin | 3–7.4, 12–25.9, and 35.6–58.4 kPa | Using freezing | BMSC responses are sensitive to even subtle changes in the surface stiffness values. | 263 |
| Temperature | The ideal vascularization ability of the surface is in the 3–7.4 kPa stiffness range. | |||
| MQMs: 5.7 kPa stiffness is optimal for neo-vessel formation with a vessel density of 37, 70, and 45 vessels per mm2 after 1, 2, and 3 weeks, respectively, compared to other stiffness values. | ||||
| PLL/HA-SH | 60–210 kPa | Cross-linking through oxidation of thiol groups | Increasing the stiffness increases fibroblast adhesion. | 241 |
| MQMs: surface stiffness increases the cell metabolic activity and adhesion by 30% compared to soft surfaces. | ||||
| DEGDMA/nOM | 25–4700 kPa | Changing n-OM monomer concentration | Surface stiffness does not affect the gene expression of VIC αSMA. | 264 |
| Structural arrangement of αSMA is changed on stiffer surfaces. Increasing osteocalcin expression and nodule development on stiffer surfaces. | ||||
| MQMs: NA or difficult to summarize. | ||||
| PCL, PLA, PGA | 62, 128, and 204 MPa | By testing different materials with different stiffness properties | Surface stiffness and topographical cues influence MSC morphology and aggregation at the earlier phase of MSC chondrogenic differentiation. | 246 |
| Softer pillar surfaces stimulate the formation of hyaline-like cartilage with middle/deep zone cartilage features. | ||||
| Stiffer nanopillar areas increase the formation of hyaline/fibro/hypertrophic cartilage. | ||||
| MQMs: collagen type I expression is 2–3 times higher on stiff nanopillar surfaces compared to soft surfaces. | ||||
| MA/MMA | 0.1–310 MPa | Changing monomer concentration | Integrin subunit expression alters depending on the stiffness value of the surface and cell types. Surface stiffness values can determine and change the fate of MSCs. | 234 |
| MQMs: NA or difficult to summarize. | ||||
Navarrete et al.234 studied the substantial effects of biomaterial surface stiffness in determining the MSC fate by investigating the MSC differentiation to osteoblasts and chondrocytes as two narrowly interconnected cell phenotypes. They designed four methyl acrylate/methyl methacrylate scaffolds with elastic moduli ranging from 0.1 MPa to 310 MPa and then cultured cells on them. They reported that on softer surfaces, MSCs tend to increase the expression of chondrogenesis factors such as aggrecan, SOX9 (a chondrogenic transcription factor), type II collagen, and proteoglycan amount. However, the expression of osteogenesis factors including Runt-related transcription factor 2, ALP specific activity, osteocalcin, and osteoprotegerin decreases on softer surfaces. The expression of integrin subunits α1, α2, α5, αv, β1, and β3 is important in starting signaling pathways in response to the surface stiffness.234
Although it is clear that modifying the surface stiffness can affect cell responses, there are still some challenges, as described in the following paragraphs.
(i) The mismatch between surface stiffness and cells is a reason for foreign body responses. Moshayedi and co-workers235 investigated the reason for glial cell activation and encapsulation of implanted electrodes. They reported that nervous glial cells are highly sensitive to the surface mechanical properties and adjusting the stiffness leads to decreased adverse reactions.235 This is because of differences in the mechanical properties of various tissues (soft, like brain or ECM, to stiff, like bundled collagen or bone).236 At molecular levels, the stiffness of ECM components is also different from each other so that single collagen fibers are more rigid than fibrillary collagen structures.237
(ii) There are synergistic effects between stiffness, topography and/or chemistry of biomaterials so that surface stiffness also depends on the surface composition and topography.155 The surface stiffness of biomaterials can be adjusted through directly altering the polymer ratio and cross-linker solution or treatment temperature and/or period.155 More recently, researchers designed several covalently cross-linked hydrogels to investigate the synergistic effects of surface chemistry and stiffness in controlling cell adhesion and migration.238 They made some scaffolds by synthetic coupling of ECM proteins to the surface of hydrogels. The results revealed that the surface biochemistry of substrates could influence cell responses to surface stiffness.238
(iii) The surface chemistry of biomaterials could explain the contradictions between research results as different types of biomaterials have different chemistries and consequently different stiffness. Li et al.239 compared the roles of both bulk and interfacial stiffness of PDMS and polyacrylamide (PAAm) scaffolds in guiding A549 cell behaviors.239 They cultured cells on the surfaces of scaffolds with bulk stiffness ranging from 0.1 kPa to 40 kPa. On PAAm scaffolds, bulk stiffness directly affects the cell spreading speed. However, on PDMS scaffolds, the coated silica layer on the surface directs cell functions.239 Because of the synergistic effects of biochemical and mechanical cues on cell responses, there are some challenges in clarifying the individual effects of each surface feature on cell responses.
Nii et al.240 designed a 3D combinatorial hydrogel with individually adjustable biochemical and mechanical features to investigate the influence of interactive niche cues on the osteogenesis ability of adipose tissue-derived stem cells.240 Their results indicated that stiffness and biochemical cues interact in a non-linear manner, emphasizing their complex synergistic influence on cell behaviors.240
(iv) Most of the suggested approaches for stiffness generation and/or regulation are restricted to designing materials with static stiffness for dynamic cells. Wang et al.241 synthesized some polyelectrolyte multilayer films, whose mechanical properties are controlled dynamically through slight stimuli.241 They designed the films via different deposition of poly-L-lysine and thiol group treated hyaluronan (Fig. 12). This method increases the surface stiffness resulting in enhanced fibroblasts cell adhesion. However, using glutathione dynamically decreases surface stiffness leading to reduced cell adhesion.241
 | ||
| Fig. 12 A schematic representation of NIH/3T3 cell responses to cross-linking and decross-linking of poly-L-lysine (PLL) and thiol group treated hyaluronan (PLL/HA-SH) multilayer substrates. | ||
(v) The native ECM contains complex mechanical pathways, such as time-dependent dynamic manners and nonlinear stiffness. Deciding about the mechanotransduction pathways based on the hydrogels, as commonly used materials for investigating mechanical cues, which have simple linear elastic mechanics can lead to biomaterial failure after implantation.153 While cell spreading depends on the absolute stiffness of the surface, their alignment and migration depend on the stiffness gradient.242
Researchers typically pattern stiffness gradients by using different cross-linking densities, through either introducing chemical gradients in cross-linkers or different exposure of photosensitive cross-linkers.243 Evans et al.244 studied the migration and morphodynamics of Schwann cells on polyacrylamide scaffolds containing stiffness gradients on their surfaces.244 The cells could track the slope of stiffness gradients on the surfaces through the durotaxis mechanism, which supports the hypothesis that Schwann cells are extremely sensitive indicators of mechanical gradients.244 Additionally, scientists designed polyacrylamide hydrogels with stiffness gradients at their surface through controlling the differential diffusion distance of free cross-linkers and monomers into a prepolymerized hydrogel environment.245 They showed that lower gradients allow detecting more unknown stem cell responses, such as the concentration-dependent rather than switch-like reactions of mechanosensitive proteins (such as yes-associated protein) to some gradient amounts.245
(vi) We should also address the synergistic influence of other physicochemical properties with stiffness on cell responses. For example, Wu et al.246 studied the synergistic influence of surface nano-topography, chemistry and stiffness in controlling MSC chondrogenesis.246 They designed three polyesters (poly-epsilon-caprolactone, polylactic acid, polyglycolide) with different stiffness values and then generated nano-grating or pillar patterns of the same scale on surfaces. They also coated chondroitin sulphate on the surfaces to increase the chance of cell adhesion. They revealed that both surface stiffness and topographical features affect MSC morphology and aggregation. Softer pillar surfaces induce hyaline-like cartilage formation with middle/deep zone cartilage features; however, stiffer nanopillar surfaces stimulate hyaline/fibro/hypertrophic cartilage formation. Nano-grating of lower stiffness values causes fibro/superficial zone-like cartilage formation; nevertheless, greater stiffness with the same topography cannot stimulate chondrogenesis.246 Hence, different cell functions are affected by the simultaneous influence of various physicochemical properties, which can also up- or down-regulate each other's influence.246
(vii) As the mechanobiology field is still in its early stages, before making firm decisions about the role of stiffness and applying it, further biochemistry research on how cells translate mechanical signals to biochemical ones is essential. To investigate the mechanotransduction pathways involved in cell responses to materials, some studies tracked phenotypic changes of cells cultured on PDMS or acrylamide substrates with stiffness gradients by patterning ECM proteins.243,247 Tseng et al.243 developed an approach for having precise, decoupled control of the ECM pattern and local surface stiffness to cells by chemical modification of PDMS surfaces.243 After culturing MC-3T3 cells on PDMS surfaces with different stiffness gradients and ECM patterns (including X, square, and I), the actin cytoskeleton polarizes to make interactions with the PDMS surface.243
(viii) The accurate evaluation of cell responses to mechanical properties of the surface can provide useful information about mechanotransduction pathways involved in cell responses to the surface.
(ix) Not only cell type but also cell volume or size is a key player in determining cell responses to surface stiffness. Bao et al.248 cultured hMSCs in separate 3D micro niches with various volumes (2800, 3600, and 6000 μm3) on methacrylated hyaluronic acid hydrogels with different stiffness (5, 12, and 23 kPa).248 They revealed that cell volume has a strong influence on the hMSC responses to surface stiffness. Cells with ideal volume can form obvious stress fibers and focal adhesions on all surfaces with different stiffness. However, in small volumes, stiffness does not have any effects on stress fiber formation and yes-associated protein/PDZ-binding motif localization.248
9.2. Impact of biomaterial surface chemical properties on biological responses
In the following sections, we provide an overview of the current progress and challenges related to chemical surface modification of biomaterials for improving their interactions with cells and proteins. We also provide a concise overview of the most common chemical techniques used for applying these strategies.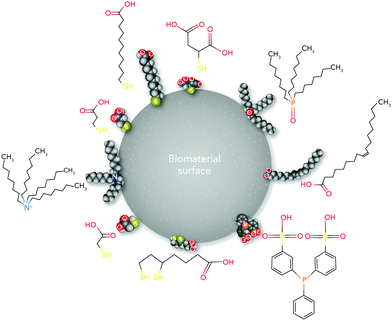 | ||
| Fig. 13 A schematic representation of different hydrophilic and hydrophilic functional groups used for adjusting the surface wettability. | ||
| Functional group | Materials | Other surface features or comment | Cell responses | Ref. |
|---|---|---|---|---|
| Abbreviations: dopamine and hexamethylendiamine (PDAM/HD), amine (NH2), carboxylic groups (COOH), human umbilical vein endothelial cells (HUVECs), self-assembled monolayers (SAMs), S-nitrosothiols (RSNOs), poly(etheretherketone) (PEEK), pre-osteoblast cell (MC3T3-E1), organic nanoparticles (ONPs), thiol (SH), mesoporous bioactive glass (MBG), human bone marrow-derived mesenchymal stem cells (hBMSCs), osteocalcin (OCN), bone sialoprotein (BSP), bone morphogenic protein 1 (BMP1), collagen type I (COL-1), gold nanoparticles (AuNPs), ultra-high molecular weight polyethylene (UHMWPE), sulfonate (SO3H), phosphorylcholine [(CH3)3N+CH2CH2PO4−] benzophenone [(C6H5)2CO], 3-methacryloyloxy-2-hydroxypropyl-4-oxybenzophenone (MHPBP), poly(2-methacryloyloxyethyl phosphorylcholine (MPC)-co-MHPBP) (PMH), human cervical cancer cells (HeLa), macrophage inflammatory protein-1α (MIP-1α), colony-stimulating factor (CSF), poly(3-hydroxybutyrate-co-3-hydroxyvalerate-co-2,3-dihydroxybutyrate) [PHBVDB], Arg-Gly-Asp (RGD), poly(ethylene glycol) (PEG), human dental pulp stem cells (hDPSCs), human lung cancer cell (HCC-15), alveolar type II epithelial cell (RLE-6TN), polystyrene (TCP), poly(L-lactide) (PLLA), octyl (C8H17), fetal bovine serum (FBS), sulfonate (SO3−), osteopontin (OPN), oligo[(polyethylene glycol)fumarate] (OPF), human breast cancer cell line (MDA-MB-231), B-cell lymphoma 2 (Bcl-2), single-walled carbon nanotubes (SWCNTs), human bronchial epithelial cell line (BEAS-2B), 3-aminopropyltriethoxysilane (APTES), major quantitative measurements (MQMs). | ||||
| NH2 | Stainless steel | PDAM/HD coating | NH2 improves HUVEC cell responses. | 286 |
| MQMs: (I) the ratio of apoptotic cells cultured on treated surfaces is ∼2.9% compared to NC (∼17.8%). (II) The cell migration is 3.9 times greater on treated surfaces compared to untreated surfaces. | ||||
| NH2 | MBG | NA | Higher expression of osteogenic genes and anti-inflammatory factors as well as fewer TRAP+ multinuclear cells of BMSCs and macrophages in modified surfaces compared to untreated surfaces. | 287 |
| MQMs: (I) after 4 weeks of implantation in the maxillary of rabbits, alizarin red labeling on treated surfaces is 1.01 ± 0.13% compared to untreated surfaces (0.70 ± 0.11%). (II) After 8 weeks healing, calcein labeling on treated surfaces is 3.62 ± 0.16% compared to untreated surfaces (2.89 ± 0.14%). | ||||
| NH2 | TCP, PLLA | Designing the surfaces in two different topography (flat and fibrous). | Surface topography has more effects on cell proliferation and gene expression of hMSCs than surface chemistry. | 288 |
| MQMs: (I) topography makes a significant difference (F = 39.27, df = 1, p < 0.001, GLM) in cell proliferation. (II) Chemistry does not significantly affect cell proliferation (F = 0.03, df = 1, p = 0.868, GLM). | ||||
| NH2 | MBG | APTES | NH2 improves MC3T3-E1 cell proliferation. | 289 |
| MQMs: cell proliferation on treated surfaces is almost twice that on untreated surfaces. | ||||
| NH2 | UHMWPE | Allylamine coating, using oxygen and nitrogen for making the surface more hydrophillic | NH2 improves human foreskin fibroblast stability and morphology. | 290 |
| MQMs: cell viability on treated surfaces is (∼70%) higher than NC. | ||||
| OH | PHBVDB | NA | OH treated surfaces do not show significant cytotoxicity for hMSCs. | 291 |
| MQMs: cellular metabolic activity on treated surfaces is almost twice that on untreated surfaces. | ||||
| SNO | Polyester | NA | Fibroblast cell response tests indicate a lack of toxic leachates at cytotoxic concentrations. | 292 |
| MQMs: cell viability on treated surfaces is 100% higher than negative control. | ||||
| SO3− | PEEK | NA | SO3− improves BMSC adhesion, spreading, proliferation, and osteogenic differentiation. | 293 |
| MQMs: cell adhesion and proliferation on treated surfaces are ∼1.5 times higher than that on untreated surfaces. | ||||
| PO4H2 | OPF | 0%, 20%, or 40% concentration of the functional group | The functional groups increase osteoblast precursor cell activities toward new bone formation. | 294 |
| MQMs: a higher bone volume on 20% (35 ± 13%) and 40% (34 ± 13%) phosphate surfaces compared to untreated surfaces (17 ± 10%). | ||||
| OH, CH3 | ONPs | Different sizes of functional groups | The surface functional groups play key roles in directing HeLa cellular uptake of nanoparticles under both serum and serum-free conditions. | 295 |
| The cell response trend can be predicted partly by surface lipophilicity. | ||||
| MQMs: the cellular uptake index can increase to ∼80% by increasing the carbon chain of ONPs. | ||||
| OH, COOH | SWCNTs | Different material lengths (1–3 μm and 5–30 μm) | The functional groups might have different effects on HepG2 responses in long and short SWCNTs. OH treated surfaces might be safer in comparison with other surfaces. | 296 |
| MQMs: NA or difficult to summarize. | ||||
| NH2, COOH | Polystyrene NPs | NA | The functional groups do not affect cell viability and the expression of type 1 macrophage markers. However, regarding type 2 macrophages, both surfaces hinder the expression of scavenger receptor CD163 and CD200R, as well as IL-10. | 297 |
| MQMs: (I) the exposure of type 1 macrophages to PS-COOH can significantly reduce the E. coli uptake by ∼30%; however, it does not have any effects on E. coli phagocytosis by type 2 macrophages. (II) PS–NH2 can significantly reduce the phagocytosis of both types of macrophages by ∼20%. | ||||
| NH2, SH | MBG | NA | The functional groups increase hBMSC cell adhesion, proliferation and differentiation. | 271 |
| MQMs: cell differentiation on the NH2-treated surface is almost 1.5 times higher than that on the SH-treated surface. | ||||
| OH, CH3, NH2 | Glass coverslip | Surfaces modified with silane. | Surfaces treated with CH3 and NH2 stimulate mitochondria-mediated apoptosis in MDA-MB-231 cells through suppressing the PTEN/PI3K/AKT pathway. | 298 |
| MQMs: almost 28.10% and 19.07% of the cells exhibit apoptotic behavior after two days of cell culture on the CH3- and NH2-treated surfaces, respectively, compared to untreated surfaces (13.85%) and OH-treated surface (12.82%). | ||||
| OH, CH3, NH2 | Au-sputtered silica wafers | SAM | Fibronectin adsorption force on SAMs follows a chemistry-dependence of –NH2 > –CH3 ≫ –OH. | 265 |
| Fibronectin adsorption force and conformation can control the late osteoblast adhesion and subsequent reorganization of adsorbed proteins. | ||||
| MQMs: the fibronectin adsorption force is 1.47 ± 0.43, 25.56 ± 7.47, and 14.10 ± 4.12 for OH-, NH2-, and CH3-treated surfaces respectively. | ||||
| CH3, NH2, COOH | Allylamine | Comparing surface modification with (i) functional groups, (ii) coating with COL-1 or immobilization of the integrin adhesion peptide sequence RGD, and (iii) treatment with plasma containing argon/oxygen gas | Only surfaces containing amino groups could chemically mask the microgrooves and block the microtopography-mediated guidance of MG-63 cells. | 299 |
| MQMs: NA or difficult to summarize. | ||||
| NH2, COOH, OH | AuNPs | Containing different surface charge | Positive charges increase hBMSC cellular uptake. | 272 |
| COOH decreases ALP activity and calcium deposition. | ||||
| MQMs: NA or difficult to summarize. | ||||
| NH2, COOH, OH | InP/ZnS quantum dots | NA | All the functional groups increase HCC-15 and RLE-6TN cell apoptosis and intracellular ROS generation. | 300 |
| All functional groups could enter the cells, with greater uptake efficiency for surfaces modified with COOH and NH2 at low amounts. | ||||
| Modifying surfaces with COOH and NH2 causes more toxicity than surfaces containing OH. | ||||
| MQMs: (I) the HCC-15 cellular uptake efficiency of COOH-, NH2-, and OH-treated surfaces is 87.4 ± 2.67%, 89.0 ± 2.15%, and 74.5 ± 1.89%, respectively. (II) And for RLE-6TN cells it is 67.1 ± 0.95%, 48.6 ± 2.03%, and 32.6 ± 2.14%, respectively. | ||||
| PO4H2, COOH, OH | PEEK | NA | The functional groups increase MC3T3-E1 adhesion, spreading, proliferation, and osteointegration. | 301 |
| MQMs: NA or difficult to summarize. | ||||
| PO4H2, COOH, OH | PEEK | Vinyl-terminated silanization layers generated on the hydroxylation-pretreated substrate surface. | Apatite forms homogeneously on the functionalized surface and strongly attaches to the surface. The functional groups improve MC3T3-E1 attachment, spreading and proliferation. | 302 |
| MQMs: cell adhesion and proliferation on treated surfaces is almost 1.25 times higher than that on untreated surfaces. | ||||
| CH3, NH2, COOH, OH | Au-coated glass slides | SAM | Endothelial cell migration depends on surface functional group types in the order CH3 > NH2 > OH > COOH. | 276 |
| MQMs: (I) cell migration distance of CH3, NH2, and COOH SAMs is 369.5 ± 27.9 μm, 325.6 ± 26.3 μm, and 213.6 ± 10.7 μm, respectively, NA for OH SAMs. (II) Wound repair duration of CH3, NH2, OH, COOH treated and untreated surface is 17.5 ± 1 h, 19 ± 1 h, 25 ± 1.5 h, 26.5 ± 1.5 h, and 22 ± 1.5 h, respectively. | ||||
| CH3, NH2, COOH, OH | PEG | Coating surfaces with alkanethiols with one of the functional groups and then experiencing 9 days of chondrogenic induction. | The functional groups direct MSC differentiation through controlling protein adsorption, nonspecific cell adhesion and cell spreading. | 303 |
| Free neutral surfaces (–CH3 and –OH) cause less protein adsorption, cell spreading and adhesion but more chondrogenic induction than charged surfaces (–COOH and –NH2). | ||||
| MQMs: NA or difficult to summarize. | ||||
| CH3, NH2, COOH, OH | Au-coated titanium implants | SAM | NH2 increases the hDPSC adhesion, proliferation, and osteo/odontogenesis differentiation compared to other functional groups. | 304 |
| MQMs: after 7 and 21 days, alizarin red labeling on NH2-treated surfaces is almost 1.5 times higher than other groups. | ||||
| OH, COOH, NH2, CONH2 | Carbon nanowall | Designing surfaces from hydrophobic to hydrophilic with the incorporation of different concentrations of carbon, oxygen and nitrogen functional groups | Changing surface chemistry by plasma treatment has effects on macrophage response in vitro, regardless of surface wettability features. | 305 |
| MQMs: NA or difficult to summarize. | ||||
| [(CH3)3N+CH2CH2PO4−], (C6H5)2CO | Photoreactive methacrylate | Using amphiphilic polymers PMH and poly(MPC-co-n-butyl methacrylate-co-MHPBP) | The functional groups increase HeLa cell responses to surfaces. | 306 |
| MQMs: NA or difficult to summarize. | ||||
| OH, COOH, biotin, azide, nitrilotriacetic acid | AuNPs | Conjugation to functional groups through using PEG | After 2 h incubation at physiological temperature, active BEAS-2B cellular uptake can occur only on citrate-stabilized and COOH-treated surfaces. | 307 |
| MQMs: NA or difficult to summarize. | ||||
The surface functional groups can direct cell functions through covalent conjugation with functional groups of cell surface lipids, proteins and glycans.273 Bygd et al.274 systematically studied how using various types of functional groups could affect macrophage reprogramming and polarization in vivo by working on poly(N-isopropylacrylamide-co-acrylic acid) nanoparticle surfaces.274 They also used a quantitative structured activity relationship (QSAR) method as an analytical predictive tool for measuring the macrophage responses to various surface chemistries. Based on the surface functional groups, they observed a spectrum of macrophage phenotypes.274
However, we should keep in mind that not only the surface properties of biomaterials but also the local cell microenvironment affects the proliferation of macrophages.275 Shen and colleagues studied the effects of different functional groups (CH3, NH2, COOH, OH) prepared by self-assembled monolayers on endothelial cell responses through studying the expression of key proteins in integrin-induced signaling pathways.276 By observing the differences in the expression of focal adhesion components and Rho GTPases, they showed that endothelial cell migration is highly dependent on the type of surface functional groups in the order CH3 > NH2 > OH > COOH.276
Rashad et al.277 designed wood-derived cellulose nanofibril hydrogels containing two different surface functional groups to address the influence of surface chemistry on fibroblast cell functions.277 They used 2,2,6,6-tetramethylpiperidine-1-oxy radial (TEMPO)-mediated oxidation or carboxymethylation pretreatments on the surface. The TEMPO-oxidized surface induces favorable cell morphology and spreading, whereas the carboxymethylated surface inhibits these processes.277
Here we summarize some of the recent challenges in this area:
(i) Using different functional groups on the surface of biomaterials can stimulate different biochemical signaling pathways.278 Zhang et al.278 investigated the potential pro-inflammatory influences of surface functional groups on human pulmonary epithelial cells and macrophages by using PEGylated CdSe/ZnS quantum dots (QDs) containing an amphiphilic polymer coating (PEG–pQDs) (Fig. 14A).278 The pro-inflammatory effects of this surface depend on the functional groups (–COOH, –NH2, –OH, and –OCH3) at the end of the PEG chain. COOH–PEG–pQDs cause the highest pro-inflammatory effects followed by NH2–PEG–pQDs, HO–PEG–pQDs and CH3O–PEG–pQDs. Fig. 14B shows that COOH containing areas internalize via lipid raft- and class A scavenger receptor-mediated endocytosis and subsequently stimulate the NF-κB signaling pathway. However, lipid raft-mediated endocytosis and stimulation of p38 MAPK/AP-1 signaling pathways are affected by surfaces containing NH2 and HO.278
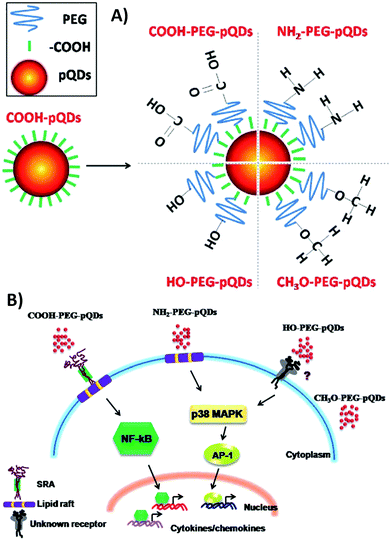 | ||
| Fig. 14 (A) A schematic representation of the structure of water-soluble CdSe/ZnS QDs bearing carboxyl groups (COOH–pQDs) and the derivative PEGylated pQDs. (B) The association between the terminal functional group-dependent endocytic pathways and the pro-inflammatory responses induced by PEGylated quantum dots. Reprinted from ref. 278. Copyright © 2013, Royal Society of Chemistry. | ||
Hence, researchers suggest using surfaces containing multiple functional groups to affect different biological pathways simultaneously. For instance, Wang et al.279 designed a poly(ether sulfone) (PES) surface containing multiple bio-functional groups such as sodium carboxylic, sodium sulfonic and amino groups to act as an antithrombotic bio-interface. They introduced functional groups onto the surface in three steps: (1) making PES with carboxylic groups (CPES) and water-soluble PES with sodium sulfonic and amino groups (SNPES); (2) presenting carboxylic groups onto the PES membrane by mixing CPES with PES; (3) and grafting SNPES onto CPES/PES membranes by coupling amino and carboxyl groups on the surface (Fig. 15A).279 Their results indicated that the treated surfaces could cause an excellent hindrance to platelet adhesion and activation, extend clotting times, and block blood-related complement and leukocyte-related complement receptor activation. Fig. 15B shows that owing to the synergistic enhancement of the functional groups, endothelial cell proliferation improves on treated surfaces.279
 | ||
| Fig. 15 (A) Fabricating poly(ether sulfone) (PES) with carboxylic (–COOH) groups (CPES) and water-soluble PES with sodium sulfonic (–SO3Na) groups and amino (–NH2) groups (SNPES) and grafting of SNPES. (B) Fluorescence staining (FITC) images of cultured vein endothelial cells on PES, CM-1, CM-2, CM-3, CM-4, GCM-1, GCM-2, GCM-3 and GCM-4 after 6 days (CM: carboxylated surface with the PES/CPES ratios of 10/0, 9/1, 8/2, 7/3 and 6/4 is termed PES, CM-1, CM-2 and CM-3, and CM-4, respectively; GCM: functionalized surfaces with sodium carboxylic (–COONa) groups, sodium sulfonic (–SO3Na) groups and amino (–NH2) groups, the grafted CMs with PES/CPES ratios of 10/0, 9/1, 8/2, 7/3 and 6/4 are named as PES, GCM-1, GCM-2, GCM-3, and GCM-4, respectively). Reprinted from ref. 279. Copyright © 2017, Royal Society of Chemistry. | ||
(ii) Similar to physical properties, research groups suggest using gradients of functional groups on the biomaterial surface to more precisely control biological mechanisms.280 Liu et al.280 designed a surface chemical gradient of amine functional groups through tuning the gas composition of 1,7-octadiene (OD) and allylamine of plasma phase.280 Under standard culture conditions (with serum), hASC adhesion and spreading area improve toward the allylamine side of the gradient surface. However, there is no difference in cell behaviors in the absence of serum, which supports the idea that surface functional groups affect hASC response through cell-adhesive serum proteins, rather than directly influencing cell functions. In addition, osteogenic differentiation is enhanced on the allylamine side of the gradient, while the adipogenic differentiation is reduced. Differences between the cell differentiation in different chemical gradients of surfaces disappear via blocking the extracellular signal-regulated kinase 1/2 signaling pathway activation via PD98059 (a specific inhibitor of the mentioned signaling pathway).280
(iii) The presence or absence of protein serum in the experimental media can affect cell responses and research outcomes. Shahabi et al.281 studied the cellular uptake of five various single or multifunctionalized fluorescent silica nanoparticles (FFSNPs) to address the role of surface charge in directing cell responses by using different concentrations of sulfonate and amino groups (Fig. 16).281 They set the zeta potential values of the surfaces from extremely positive to extremely negative, whereas other surface properties remained nearly constant. Depending on the surface charge and on the presence or absence of protein serum, two reverse trends for FFSNP cellular uptake exist. In the absence of serum, human osteoblasts can better accumulate positively charged nanoparticles than negatively charged surfaces. However, in the serum-containing medium, osteoblasts can better internalize anionic particles. Under physiological conditions, sulfonate-functionalized silica nanoparticles are the preferred choice to have a high rate of nanoparticle internalization.281
 | ||
| Fig. 16 (A) An illustration of the synthesis and concurrent functionalization of various single or multifunctionalized fluorescent silica nanoparticles (FFSNPs) by 3-aminopropyl-triethoxysilane (APTES) and 3-(trihydroxysilyl)-1-propanesulfonic acid (HSPSA). (B) An illustration of the designed particles by modulating the primary molar ratio of amino and sulfonate functional groups. Reprinted from ref. 281, copyright © 2013, Royal Society of Chemistry. (DOI: http://10.1021/acsami.5b01900). Further permissions related to the material excerpted should be directed to ACS. | ||
(iv) Hasan et al.282 studied the effects of media conditions on protein and L929 mouse fibroblast cell responses to five dissimilar nano-scaled surfaces treated with functional groups including amine, octyl, mixed, hybrid, and carboxylic.282 They studied the protein and cell responses under three dissimilar conditions consisting of (1) foetal bovine serum (FBS) in media, (2) pre-adsorbed FBS on surfaces, and (3) partial media without FBS. Regardless of the functional groups used on the surfaces, surfaces with pre-adsorbed FBS show the highest L929 fibroblast adhesion rate and cell spread area. However, surfaces placed in partial media show minimum adhesion rate, poor cell spreading and inappropriate morphology.282
(v) The functional group density can also influence protein adsorption mechanisms.283 Meder et al.283 studied the adsorption of three model proteins, bovine serum albumin, lysozyme and trypsin, on colloidal alumina surfaces. They functionalized the surfaces with SO3H in densities ranging from 0 to 4.7 SO3H nm−2.283 Their results indicated that the functional group surface density affects the adsorption of all three proteins. Simply changing the density of functional groups on the surface can cause a continuous tuning of protein adsorption from nearly no adsorption to a theoretical monolayer.283
(vi) Although there are different strategies for using functional groups to direct cell responses, the biochemical signaling pathways which respond to each functional group are not yet fully detected.
(vii) Other factors such as cell lines, topography, stiffness as well as the complex ECM and physiological metabolism of cells may also be involved in directing cell responses to surface functional groups.268 Researchers designed a series of model surfaces with controlled surface nanotopography ranging from 16, 38, to 68 nm.284 They functionalized surfaces with amine, carboxyl, or methyl groups to investigate the primary neutrophil and macrophage responses. In these chemically modified surfaces, the surface nanotopography can decrease matrix metallopeptidase 9 expression in neutrophils as well as the concentration of IL-6 and IL-1β in macrophages.284 Surface chemistry and nanotopography can, in a synergistic manner, control the osteo-immune environment functions such as the production of inflammatory cytokines, osteoclastic activities, as well as osteogenic, angiogenic, and fibrogenic factors.285
(viii) Table 3 shows that CH3, NH2, COOH, and OH are the commonly used functional groups used for modifying the surface chemistry. Here we suggest also considering the other existing functional groups present in nature.
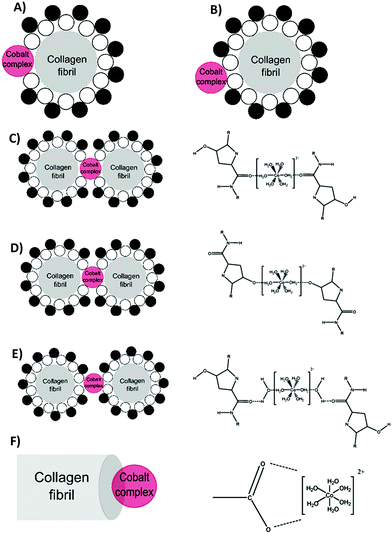 | ||
| Fig. 17 A schematic representation of the binding sites of a cobalt complex with a collagen fibril. (A) The direct interaction of the cobalt complex with the collagen fibril. (B) The interaction of the cobalt complex with the non-freezing water layer surrounding the collagen fibril. Suggested binding sites of a cobalt complex with a collagen fibril, where interactions occur with the carboxylic group (C), hydroxyl group (D), forming a water bridge (E), or with the hydroxyl group in the carboxy terminus (F). Reprinted from ref. 309, copyright © 2018, Royal Society of Chemistry. Further permissions related to the material excerpted should be directed to the ACS. | ||
| Materials | Doped ions | Other surface modifications | Main outcomes | Ref. |
|---|---|---|---|---|
| Abbreviation: poly ether ether ketone (PEEK), bone mesenchymal stem cells (bMSCs), plasma polymerized allylamine film (PPAAm), polyurethane (PU), extracellular matrix (ECM), primary human nasal epithelial cell (HNEpC), Runt-related transcription factor 2 (RUNX2), poly(lactic acid) (PLA), pre-osteoblast line (MC3T3), bovine serum albumin (BSA), cobalt–chromium (Co–Cr), human osteoblast-like cell line (SAOS-2), diamond-like carbon (DLC), human umbilical vein endothelial cells (HUVECs), polydimethylsiloxane (PDMS), bone morphogenetic protein 2 (BMP2), vascular endothelial growth factor (VEGF), major quantitative measurements (MQMs). | ||||
| PEEK | Calcium | NA | Increasing rat bMSC adhesion, proliferation, and osteo-differentiation. | 324 |
| MQMs: after 7 days of cell culture, cell proliferation on treated surfaces is almost 40% higher than that of untreated surfaces. | ||||
| Nitrogen | NA | Increasing the MG-63 responses and antibacterial activity of PEEK. | 325 | |
| MQMs: after 3 days of cell culture, cell proliferation on treated surfaces could be almost twice that on untreated surfaces. | ||||
| Nitrogen | Coating tropoelastin | Increasing SAOS-2 cell attachment, spreading, proliferation, and bone nodule formation. | 326 | |
| MQMs: (I) after 7 days of cell culture, cell proliferation on PIII treated surfaces is 70 ± 8% higher than that on untreated surfaces. (II) After 6 days in osteogenic media, osteopontin expression on PIII treated surfaces, with and without tropoelastin, is 6-fold higher than that on untreated surfaces. | ||||
| Titanium | Copper | Coating PPAAm | Decreasing the inflammatory responses of macrophages, antigen-presenting cells, and T lymphocytes to treated surfaces. | 327 |
| MQMs: (I) decreasing the number of activated NK cells on copper-treated surfaces from a median of 0.91 × 10−3 on day 7 to 0.46 × 10−3 on day 56. (II) After 56 days, higher numbers of mast cells on copper-treated surfaces with median 2.29 × 10−3 compared to Ti–Cu–PPAAm surfaces with median 1.97 × 10−3. | ||||
| Oxygen | NA | Improving fibronectin adsorption as well as hbMSC cell adhesion, migration, proliferation, mineralization, and differentiation. | 314 | |
| MQMs: (I) after 7 days of cell culture, cell proliferation on treated surfaces with different oxygen concentrations has a median of 0.840–1.041 compared to untreated surfaces with a median of 0.737. (II) Collagen type I expression on treated and untreated surfaces has a median of 129.70–154.91 and 100.00, respectively. | ||||
| Oxygen | NA | Increasing responses of blood cells and antibacterial activities. | 328 | |
| MQMs: after 10 min, the optical density of blood (related blood clot formation) on treated surfaces is 50% less than that of the untreated group. | ||||
| Nitrogen | NA | Increasing blood clot formation, the adhesion of platelets, as well as adhesion and proliferation of HNEpC cells. | 329 | |
| MQMs: NA or difficult to summarize. | ||||
| Nitrogen | NA | Increasing hbMSC cell adhesion, proliferation, and mineralization. | 330 | |
| MQMs: after 1 h incubation, the cell spreading area order of groups is high-dose oxygen > low-dose oxygen> untreated surfaces. After 7 days, cell proliferation on treated surfaces is 1.3-fold higher than that on untreated surfaces. | ||||
| Calcium & magnesium | NA | Increasing initial hbMSC cell attachment in ion-doped surfaces. | 331 | |
| There is no difference in cell proliferation among different groups. | ||||
| RUNX2 expression is greater on the magnesium ion-implanted surface. | ||||
| Osteocalcin expression is less on the calcium ion-implanted surface. | ||||
| MQMs: NA or difficult to summarize. | ||||
| Magnesium | NA | Increasing the number of type 2 macrophages and anti-inflammatory cytokine expression. | 322 | |
| MQMs: (I) after 4 days, the percentage of CD206, as a surface marker of type 2 macrophages, has the order Ti < Mg30 < Mg90 < Mg120. (II) The expression of C–C motif chemokine receptor 7, as the surface marker of type 2 macrophages, has the order Ti > Mg30 > Mg90 > Mg120. | ||||
| Nitrogen & copper | NA | Increasing angiogenic abilities of treated surfaces (especially dual ion treated surfaces) in response to HUVECs. | 332 | |
| MQMs: the number of cell migration on dual ion treated surfaces is almost 1.5-fold higher than that of untreated surfaces. | ||||
| Silver | NA | Increasing implant stability in vivo as well as MG-63 cell responses toward neo-tissue formation, bone mineral density, and trabecular pattern. | 333 | |
| MQMs: bone implant contact for 30 min-, 60 min-, 90 min-Ag PIII treated surfaces as well as untreated surfaces are 73.18 ± 5.23, 69.92 ± 4.10, 66.05 ± 3.97, and 61.99 ± 4.66, respectively. | ||||
| Titanium-nickel | Carbon | Coating DLC | Increasing MG-63 cell morphology, viability and spreading. | 334 |
| MQMs: after 24 h cell incubation, the optical density value of treated surfaces is ∼1.8-fold higher than that of untreated surfaces. | ||||
| PU | Nitrogen | Coating collagen | Decreasing acute inflammatory responses (macrophages at day 28) in ion-collagen modified surfaces. | 335 |
| MQMs: after 1, 3, 7, 14 or 28 days, the cell numbers in the surrounding capsule of treated implants is 42%, 41%, 51%, 43%, and 45% higher than that of untreated surfaces. | ||||
| Nitrogen | NA | Increasing endothelial cell attachment and proliferation. | 336 | |
| MQMs: after 5 days of cell proliferation, the average cell density on treated surfaces is almost 3-fold higher than that of untreated surfaces. | ||||
| Polystyrene | Nitrogen & oxygen | NA | Treated surfaces can be used for printing multiprotein micropatterns on the surface to investigate local effects on mouse pancreatic β cell morphology and protein production. | 337 |
| MQMs: after 11 days, the amount of immobilized BSA on treated surfaces is 30% lower than that of untreated surfaces. | ||||
| Co–Cr | Tantalum | NA | Increasing endothelialization, platelet activation, and blood coagulation. | 338 |
| MQMs: after 1 day cell culture, the cell spreading area and surface coverage of treated surfaces are 195% and 209%, respectively, higher than those of untreated surfaces. | ||||
| Oxygen | NA | Increasing endothelial cell viability after 7 days. | 339 | |
| MQMs: NA or difficult to summarize. | ||||
| Nitrogen | NA | Increasing the MSC osteogenic differentiation. | 340 | |
| MQMs: increasing integrin-binding sialoprotein expression and mineralization levels on treated surfaces 30-fold higher than those of untreated surfaces after 21 days of cell incubation. | ||||
| Stainless steel | Nitrogen | Coating hydroxyapatite | Increasing hydroxyapatite growth and human oral fibroblast viability. | 341 |
| MQMs: after 24 h cell culture, the percentage of cell growth on treated surfaces can be twice that of untreated surfaces. | ||||
| Silver | NA | Increasing antibacterial activity and osteogenic differentiation of hbMSCs. | 316 | |
| MQMs: NA or difficult to summarize. | ||||
| Silicone | Tantalum | NA | Increasing hydrophilicity and human primary dermal fibroblast affinity. | 342 |
| Decreasing fibrous capsule formation and contracture in vivo. | ||||
| MQMs: after 4 days, the cell spreading area on treated surfaces is more than twice that of untreated surfaces. | ||||
| PLA | Tantalum | NA | Increasing MC3T3 cell adhesion, osseointegration and ontogenesis. | 321 |
| MQMs: ALP activity on Ta-implanted PLA surfaces is 3.5 and 4.3 times more than Ta-coated and untreated PLA surfaces, respectively. | ||||
| PDMS | Oxygen | NA | Increasing Chinese hamster ovarian cell responses to surfaces. | 343 |
| MQMs: after 2 days of cell incubation, the relative dead/live cells ratios are 1 ± 0.037, 0.653 ± 0.026, and 0.425 ± 0.033 on untreated, plasma treated, and PIII treated surfaces, respectively. | ||||
Engineers use ion implantation, a standard technique in semiconductor processing, for improving the surface cell adhesion or wettability through modulating surface mechanical properties, wear resistance, and corrosion resistance.312,313 Ion implantation can improve the surface properties of metallic implants, polymers and ceramics.314–318 This technique includes the bombardment of ionized species and their implantation into the topmost layers of a solid material.319 It needs an ion generation source, an electrostatic acceleration system, and a vacuum chamber. Physical sources in a discharge chamber make ions and then precursors convert them into vapor.
The charge/mass selective mode and linear acceleration mode are the key operative modes of ion implantation. In the charge/mass selective mode, the ionized species are pre-accelerated prior to entering a quadrupole magnet. The filtered beam is then postaccelerated and concentrated onto the biomaterial surface. However, in the linear acceleration type, all ionized species in the discharge chamber speed toward the biomaterial surface.319
Ion implantation has several advantages including making a treated surface without delamination problems or changing the bulk properties of the biomaterial.320 Any kind of dopant can be introduced into any solid biomaterial in the defined areas of the surface, and the process temperature is low. Engineers use this approach for transferring energy into the surface layer of biomaterials, which results in changing the surface properties without any variations in the chemical state of biomaterials.320
However, at the time of ion bombardment all targeted areas need to be orthogonally exposed to the ion beam, which is a big limitation when it comes to surfaces with complex surface geometries. Hence, plasma-immersion ion implantation (PIII) is an effective approach in ion bombardment of inhomogeneous surfaces.313 Park et al.321 investigated the effects of tantalum ion immersion on the poly(lactic acid) (PLA) surface for improving its osteoblast affinity by using PIII in combination with conventional direct current magnetron sputtering. They revealed that tantalum ion-doped surfaces have twice the surface roughness and improve adhesion stability in comparison with tantalum-coated PLA surfaces. Tantalum doped ions improve the osseointegration and osteogenesis of implanted PLA surfaces in vivo through improving surface hydrophilic properties.321
Researchers also studied macrophage polarization responses to titanium implants doped with magnesium (0.1–0.35%).322 Doping magnesium ions on the surface causes a greater expression of type 2 macrophages, anti-inflammatory cytokines (such as IL-4 and IL-10) as well as genes encoding bone morphogenetic protein 2 (BMP2) and VEGF.322 There are still challenges about using ions on biomaterial surface for improving cell responses:
(i) As different ions can affect cell functions differently, simultaneously doping different types of ions on the surface of biomaterials is effective for providing multifunctional materials. Yu et al.323 doped both zinc and magnesium ions on the titanium surfaces by using PIII. They detected that the dual implantation of ions enhances rat bMSCs’ initial adhesion and spreading through the upregulation of the gene expression of integrin α1 and integrin β1.323 Zn/Mg-PIII can also increase zinc and magnesium ion concentrations in the cells via enhancing the influx of both ions and hindering the outflow of zinc ions. Upregulating the expression of magnesium transporter 1 in human umbilical vein endothelial cells improves the magnesium ion influx, which leads to promotion of angiogenesis.323
(ii) The concentration of doped ions plays a key role in determining their influences on surface properties and biological pathways.309 This indicates the crucial demand for precise systematic studies to find the optimal concentration of ions depending on the purpose.
Surface functionalization with cell-specific molecules such as antibodies and receptor-targeting peptides improves the biomaterial surface affinity and specificity to the cell membrane.348 Interactions between biomaterial surface and proteins can be extremely site-specific or non-specific (Fig. 18).
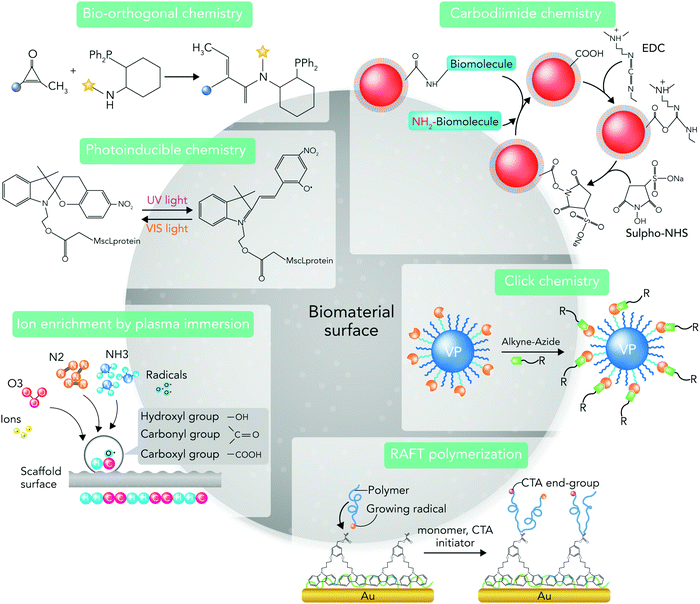 | ||
| Fig. 18 The commonly used chemical strategies for engineering the biomaterial surface to manipulate biological responses. | ||
Adsorption/physisorption is the mainly used strategy for non-site-specific conjugation of proteins to the surface, in which the protein–surface interactions are mainly directed through hydrophobic, hydration and electrostatic forces.349 The incubation of a designed biomaterial with the targeted protein in buffer solutions is the main principle of this strategy. It is the most straightforward strategy for tethering proteins on the biomaterial surface.344,350,351 However, non-covalently attached proteins can detach from the biomaterial surface after implantation and lose their functionality.352 After implantation, the attached proteins can be replaced by adsorbed blood serum proteins through the Vroman effect.353
Therefore, there are several strategies for enhancing protein adsorption to surfaces including plasma treatments or using a transitional “sticky” layer such as mussel adhesive proteins.354 Plasma treatment can induce different functional groups on the surface, which improves the protein conjugation.355 Because aldehyde groups can trigger nucleophilic groups in lysine, arginine, asparagine and glutamine residues to make covalent imine bonds, plasma polymers, which have aldehyde groups on their surface, provide a reactive surface for covalent binding of proteins.356
In addition, engineers immobilize several proteins on the biomaterial surface by using PIII without changing their conformation. PIII is a one-step immobilization method, which through vapor-phase precursors produces radical species for covalently bonding with amino acid residues in proteins.357,358 Using PIII, Tan et al.359 developed a bioactive vascular graft coating with the macrophage polarizing cytokine IL-4 to control the macrophage phenotype and subsequent local inflammatory responses.359 When subcutaneously implanted in mice, they observed that bioactive IL-4 surfaces could enhance the polarization of macrophages to their anti-inflammatory M2 phenotype. The functionalized surfaces could also positively regulate the local cytokine environment and reduce the foreign body responses.359
Mussel adhesive proteins are a coating layer for intermediating the biomolecule–surface interactions through their (S)-2-amino-3-(3,4-dihydroxyphenyl)propanoic acid containing a benzene ring with two neighboring hydroxyl groups.360 However, due to the instability of non-specific attachment approaches, chemical conjugation methods have gained more attention for protein tethering on the biomaterial surface. In site-specific approaches, protein conjugation occurs through interactions between specific chemical groups in protein molecules and biomaterial surface, which leads to proper protein orientation with high stability on the surface.344
Primary amine, carboxyl, sulfhydryl and carbonyl are the main chemical groups used for improving cell–biomaterial interactions.347 Controlling chemo- and regio-selectivity and also the selection of treatment methods based on the surface properties of both protein and biomaterial are the main requirements of using site-specific strategies.361
Antibody conjugation on the biomaterial surface demands one of three functionalities in the antibody: (i) lysine amino acids, (ii) 12 cysteine residues, or (iii) 2–5 carbohydrate moieties in the Fc stem.362 Amine conjugation occurs through an amine functionality, which is available on the surface of proteins or biomaterials.363 Using covalent cross-linkers is one of the main strategies for establishing chemical interactions between proteins and cell surface, due to their ability to control the immobilization and accessibility of substrate surface biomolecules.364
Among cross-linkers, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide (EDC/NHS) can more efficiently make chemical interactions between amine functional groups of biomolecules and carboxylic groups of the biomaterial surface.365 The cross-linkers’ non-cytotoxicity and water solubility of waste products make EDC/NHS a favorable candidate for mediating chemical interactions between cells and biomaterial surface.366
Additionally, the interaction between maleimide chemical compound (H2C2(CO)2NH) and thiol (R–SH, where R represents an alkyl or aryl group) attracts much attention for establishing cell–biomaterial interactions. As the maleimide chemical compound can selectively react with cysteine residues in the protein, researchers use the thiol–maleimide conjugation reaction or immobilization of thiol-containing surfaces.273,367 Cell surface thiols are present in oxidized disulfide bridges or reduced thiol group formations and can be labeled with a broad number of accessible reagents.368,369 Some studies suggest using mono- and di-bromomaleimides as an additional reversible cysteine modification treatment on maleimide-based conjugation reaction.370
Reversible addition fragmentation chain transfer (RAFT) as a living radical polymerization reaction can also conjugate biomolecules to polymeric biomaterials.371,372 RAFT-mediated bioconjugation can increase the chance of having a well-defined, site-directed bioconjugate architecture. The combination of “Click” chemistry reactions with RAFT increases the effectivity and selectivity of protein conjugation on the substrate surface without interfering with the protein functionality.344,373
“Click” chemistry is defined as a class of small molecule chemical reactions, which joins a biomolecule and a reporter molecule, allowing coupling of substrates of choice with specific biomolecules. The reactions are fast, spontaneous, flexible, and very selective.374 Over the past two decades, copper-based “click” chemistry reactions have been a common strategy for synthesizing hydrogels.375 In addition, bio-orthogonal “click” chemistry is used to describe the way of generating products by joining small biomolecules such as proteins that occur in the presence of macromolecules such as proteins or cells.375 Bio-orthogonal “click” chemistry is recognized as a promising molecular labeling approach, which does not affect normal biochemical processes.376 Using these approaches, researchers can synthesize novel polymers and multifunctional hydrogels for biomedical applications.377,378 “Click” chemistry synthesis occurs under stable physiological conditions, with highly stereo-specific and simple product separation. It does not produce any toxic end products. It is also insensitive to oxygen and water.375
Owing to its copper ion toxicity and ROS generation, copper-based “click” chemistry has been recently replaced with copper-free “click” chemistry strategies.379 Copper-free “click” chemistry proceeds at a lower activation barrier and is free of cytotoxic catalysts or end products after gel formation.374,375 There are many copper-free “click” chemistry strategies for designing hydrogels such as strain-promoted azide–alkyne cycloaddition “click” hydrogels,380 Diels–Alder “click” chemistry hydrogels,381 thiol–ene,382 oxime,383 and thiol–yne.384 The available “click” chemistry-based functional hydrogels play key roles in fabricating 3D tissue and organ models by using 3D bioprinting.385,386
“Click” chemistry reactions improve biomolecule–surface conjugation in a controlled manner.387 However, choosing or introducing suitable functional groups on proteins to mediate their attachment in a controlled manner without influencing their activity is still a big challenge in this field.344,361
Interfacial free energy between a solid and a liquid is a material's property that is determined by its surface structure and chemical composition. It measures the disruption of intermolecular bonds, which occurs while creating a surface.388,389 Transferring an atom from the bulk of materials to the surface in response to liquids changes the surface energy value. The migrated atoms at the surface have fewer nearest neighbors than the same atoms positioned in the bulk. Consequently, the surface atoms have a greater energy state than atoms in the bulk, a phenomenon known as coordinative unsaturation of bonds.389
Thus, the type and amount of existing dangling bonds at the surface represent changes in the surface free energy value, which can be primary-(ionic, covalent, and metallic) as well as secondary-(van der Waals) type bonds.389 If the dangling bonds are of the secondary type, the surface free energy is low and has a non-polar nature. However, if the bonds are mainly of primary type, a substantial Lewis acid and base contribute to the total surface free energy and its value is high.389 Most surfaces have a combination of all these chemical bonds, which makes the surface interactions with the biological compounds complex.389
The surface energy value for high energy surfaces (e.g. metals and oxides) can be in the range from 500 to 5000 mN m−1; however, for low energy surfaces (such as molecular crystals and polymers) it is from 5 to 50 mN m−1.389 Researchers commonly modify the surface free energy through changing the crystallographic termination of surfaces or adding ions using plasma treatments.391
The common technique for measuring the biomaterial surface energy and wettability is contact angle measurement.392–396 The equilibrium state for liquids on a surface is reliant on both the thermodynamic equilibrium at interfaces and the total length/area of the phases in contact. Consequently, contact angle measurements can provide information about both surface free energy and wettability through the calculation of surface free energy from droplet geometry, or surface geometry from surface free energy.397–399 Changes in both surface chemical composition and wettability can affect the obtained surface energy value.400–402 Therefore, researchers should know the surface roughness and wettability before characterizing the surface energy using contact angle measurements.
As mentioned above, the intermolecular bonds at a biomaterial surface determine its surface free energy values. After biomaterial implantation, these chemical bonds interact with small molecules such as proteins. The chemical bonds also determine which molecules firstly adsorb, their orientation, conformation and bioactivity.389
Measuring wettability is a common method to determine the adsorption potential of proteins to biomaterial surfaces. However, because the relative wettability of many surfaces can be equal while their surface chemistries are different, these terms cannot precisely determine the surface interactions with proteins and cells.389 It is commonly accepted that surfaces with high surface energy values and wettability enhance cell responses.403–408 However, there is no direct correlation between hydrophilicity and surface free energy. Thus, investigating the effects of surface free energy on protein adsorption and cell responses, irrespective of surface wettability, is important.389
9.3. Nanofunctionalization of biomaterial surface
The ECM is composed of multifunctional nanostructures. Binding of cells to the ECM plays a vital role in regulating cell signaling pathways.409 Therefore, bio-inspired nanofunctionalization, which combines biomimicry and nanotechnology, has attracted considerable attention as a promising strategy to modify the biomaterial surface.410–412 Using patterning approaches, biomedical engineers can accurately position different biomolecules on a nano-scale surface to achieve site-specific attachment of biomolecules in some areas, while reducing undesirable surface interactions in other areas.413 Techniques for producing nanostructures are commonly divided into two categories: (1) in situ surface nanofunctionalization, (2) nanocoating and film deposition. Electron beam, laser etching, acid and alkali treatments, anodic oxidation, and ion implantation are the most common in situ nanofunctionalization techniques.413,414 Many nanocoating and film deposition techniques are also available including plasma spraying, plasma-immersion ion implantation and deposition, chemical or physical vapor deposition, cold spraying, lithography and self-assembly.413,414By combing the above-mentioned techniques, more complex hybrid nanostructures can be designed.413 For instance, Wang et al.415 studied the synergistic effects of micro/nanostructure and bioactive ions on murine osteoblast responses to titanium surfaces containing bioactive ions (Zn2+ and Sr2+). They created surfaces by using a combination of sandblasting, acid etching, alkali-heat treatment, and ion exchange techniques. Compared to polished titanium surfaces, the micro/nanostructured functionalized surfaces could significantly enhance cell spreading, proliferation, and differentiation.415 Furthermore, Frey et al.416 used micelle nanolithography and soft micro-lithography to develop PEG-based hydrogels with a micro-grooved surface. They also incorporated gold nanoparticles on the surface to improve the binding of adhesive ligands. Compared to conventional micro-grooved surfaces, the nanofunctionalized surface could improve human fibroblasts’ contact guidance and regulate cell signaling more accurately.416
Christo et al.417 developed a biomaterial surface with controlled nanotopography and chemistry by combining plasma polymerization and electrostatic self-assembly techniques to evaluate the inflammasome responses.417 They assessed the innate immune responses by using bone marrow derived macrophages harvested from genetically engineered mice deficient in apoptosis-associated speck-like protein containing CARD, NLRP3 and AIM2 inflammasome components. Their results showed that the macrophage adhesion changes on all controlled nanotopography surfaces irrespective of their surface chemistry or nanotopography scale. Although other studies reported that different surface chemistries influence the initial binding of serum proteins and cell attachment,418,419 Christo and colleagues could not detect any difference between groups. However, both chemistry and nanotopography could change the macrophage functionality in the absence of main inflammasome components suggesting that these components could be key players in macrophage responses to surface nanofunctionalization.417
In another study, Marzaioli et al.420 studied the effects of silica nanoparticle functionalization on inflammasome signaling pathways using murine bone marrow-derived dendritic cells and a mouse model of mild allergic inflammation.420 They showed that non-functionalized surfaces activated the NLRP3 inflammasome response leading to the expression of inflammatory cytokines and chemokines. However, surface functionalization with phosphonate or amino groups reduced the inflammasome activation.420
Owing to their chemical and structural features, proteins, peptides, and ligands play key roles in surface nanofunctionalization. Researchers commonly design biomaterials with integrin-specific ligands on their nanostructured surface for enhancing cell responses.421 In addition, the ligand clustering on biomaterial surface can regulate intracellular signaling events that affect cellular phenotype. Karimi et al.421 developed RGD-functionalized copolymers using RAFT polymerization to study the effects of nano-scale clustering of integrin-binding ligands on endothelial cell functions (Fig. 19).421 They used the synthesized copolymers to prepare random and nano-clustered surfaces spanning different global and local RGD densities. Their results indicated that nano-clustering ligands on the surface could promote endothelial cell adhesion and migration.421
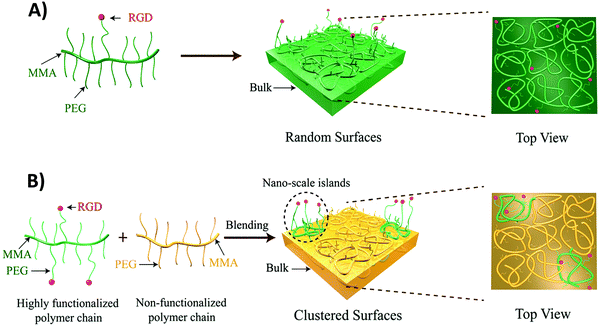 | ||
| Fig. 19 Fabricating random and clustered surfaces using nanofunctionalization strategies. (A) Random surfaces were developed by film casting lightly functionalized polymer chains. (B) Clustered surfaces were developed by film casting blends of highly functionalized polymer chains (green) with non-functionalized polymer chains (orange) to fabricate nano-scale islands of high peptide density defined by the random coils of the polymer molecule. Reprinted from ref. 421. Copyright © 2017, Royal Society of Chemistry. | ||
Proteins can be also patterned onto a surface by microcontact printing or dip-pen nanolithography.422–424 After immobilizing biomolecules, the surface arrays can display a selective capture of proteins from a mixture like serum.425 Additionally, when the captured proteins are patterned on a desirable surface, the specific bound target proteins can be transferred to another surface by microcontact printing, which is a simple method for fabricating surface arrays of different proteins.426
Despite the substantial progress in this field, there are still some challenges to be solved. Thoroughly investigating the thermodynamics and kinetics of protein adsorption on nanostructures is vital.427 In addition, proteins have unfolding potential upon adsorption on nanostructures leading to tissue damage. Researchers should more profoundly study both unfolding and stabilization potential of proteins on nanostructures before their implantation in the body.427
10. Chemical surface analysis
Owing to the advances in analytical chemistry, biomedical researchers have achieved in depth information about the surface composition and structure of both biomaterials and biological molecules.428–430 The commonly used analytical techniques for biomaterial surface characterization include secondary-ion mass spectrometry (SIMS), time of flight-SIMS (ToF-SIMS), X-ray photoelectron spectroscopy (XPS), auger electron spectroscopy (AES), (Raman), infrared (IR) spectroscopy, X-ray scattering and spectroscopy techniques, scanning electron microscopy-energy dispersive X-ray (SEM-EDX), atomic force microscopy (AFM) and streaming potential measurements/electroosmosis.431–434Among these techniques, XPS, ToF-SIMS, Raman, and IR spectroscopy have gained substantial attention for characterizing the surface composition after applying surface treatments.431,434,435 XPS and ToF-SIMS have been the main techniques used for evaluating the biological responses to surface chemistry because of several advantages. (I) These techniques can provide information about the interfacial intermolecular forces that control the biomaterial–biomolecule interactions through analyzing the surface composition and structure over the depth scales of a few nanometers.434 (II) They can be useful for detecting surface contamination, which can be critical when interpreting cell responses to biomaterials. (III) They can provide high quality images of the spatial distribution of surface composition by combining imaging software technologies with narrowly focused ion guns. (IV) The recent progress in imaging technologies has led to using these techniques in the nanotechnology field for analyzing the designed patterned surfaces with more sensitivity and resolution.434,436
These two techniques are complementary and both are required to thoroughly analyze the physicochemical composition of biomaterial surface, because they can overcome each other's drawbacks.436 For instance, XPS cannot differentiate isotopes, while ToF-SIMS can do it. Although quantification is commonly done by XPS, it is difficult to do it by using ToF-SIMS.436
Oteri et al.428 designed a biomaterial based on calcium triphosphate and hydroxyapatite microgranules for bone regeneration.428 They used ToF-SIMS and XPS to analyze the chemical composition and the micromorphological structure of the surface and confirm its osteoconductivity.428 In another study, Stevanovic et al.437 coated a layer of chitosan hydroxyapatite and gentamicin on pure titanium plates.437 They used XPS and FTIR to confirm that the coating layer was connected to the titanium surface through intermolecular hydrogen bonds.437 Moreover, Metoki et al.438 studied the calcium phosphate electrodeposition on a titanium alloy covered with SAMs regarding the chain length, end-group charge, and anchoring group.438 They used ToF-SIMS to confirm that calcium-rich phases were formed primarily in the presence of SAMs.
Analytical chemistry can also be useful to provide information about the chemical characterization of surface bound proteins.430,433,435 Proteins are recognized as key players in determining biomaterial–cell interactions and many biological pathways.14 The protein structure, which is the 3D arrangement of atoms in an amino acid-chain, can break down into primary structural units, secondary structural units, tertiary structural units, and quaternary structural units.433 However, because proteins are extremely flexible and can easily restructure when they meet a surface, analyzing the surface structure after binding to large proteins is challenging and requires combining experimental and simulation techniques.439 Furthermore, analyzing the well-defined, smaller subunits of larger proteins (such as peptides) could be a promising solution toward developing better methodologies for this purpose.433 XPS, surface plasmon resonance, and quartz-crystal microbalance with dissipation can be used for characterizing peptides and protein surface coverage in single component protein solutions.433 However, on surfaces covered with a mixture of proteins, these techniques can only determine the total protein surface coverage.
ToF-SIMS can determine the unique signatures from different proteins adsorbed on these surfaces.440 Nevertheless, when the protein films become more complex, ToF-SIMS can only provide a qualitative measurement about the total composition of the films.433,441 ToF-SIMS can also provide information about the overall protein orientation using selected peaks from asymmetrically distributed amino acids in the protein structure.442,443 Additionally, ToF-SIMS and sum frequency generation techniques can give molecular level details about the orientation of surface bound peptides and proteins.444 The obtained molecular level data from these techniques can be used to govern the molecular dynamics simulations, which provide atomic-level structural information about surface bound peptides.445
For instance, Foster et al.435 studied the adsorption of single-component bovine serum albumin, bovine fibrinogen, and bovine immunoglobulin G films as well as multicomponent bovine plasma films onto two different gold surfaces.435 They used a multitechnique method based on XPS, ToF-SIMS, principal component analysis (PCA), and visual molecular dynamics (VMD) to obtain new information about the structure of proteins. Using XPS, they could evaluate the adsorption isotherms and show the effects of protein solution amount on the surface nitrogen composition. Using a combination of ToF-SIMS, PCA, and VMD, they could also provide some information about the differences in adsorbed protein structure on different surfaces. They finally analyzed the amino acid distributions in the adsorbed proteins by using PCA and VMD.435
Even though substantial progress has been made in the analytical chemistry of biomaterial and biomolecule surfaces, there are still many challenges that should be addressed. Over the past few years, researchers have suggested new strategies to improve ToF-SIMS applicability such as applying multivariate analysis methods for data mining, as well as using cluster-ion beams and metal-assisted SIMS.436 Nevertheless, all the current available techniques have their own drawbacks, which need to be addressed. For instance, both XPS and ToF-SIMS measure in dry state, need vacuum, and are contamination sensitive.434 ToF-SIMS has also inadequate optical capabilities and problems in collecting positive or negative ion data.431 XPS cannot detect hydrogen and helium.434 The current techniques cannot provide atomic-level structural information about biomolecules.433 Future research dealing with analytical techniques should emphasize on the combination of most developed imaging and analytical chemistry techniques to go beyond the current limitations.
11. Using the 3D bioprinting technique for improving cell–biomaterial interactions
Over the past few decades, 3D bioprinting technology has emerged as a promising approach for designing patient-specific scaffolds by depositing biomaterials and living cells layer-by-layer based on a digital model.446–448 It draws knowledge from developmental biology, chemistry, computer science, and materials science to fabricate biological substitutes mimicking their native counterparts.449 3D bioprinting techniques can precisely print and control the geometrical localization of living cells into biomaterials.446 Therefore, researchers use 3D printed tissues in many biomedical engineering fields including tissue engineering and regenerative medicine,450 transplantation,451 drug testing and high-throughput screening,452 and cancer research.453 The 3D bioprinted tissue models can be promising substitutes for the current 2D cell culture and animal models to test drugs and biomaterials in vitro.446Materials used for constructing 3D bioprinted tissues include biomaterials, living cells, drugs, growth factors, and genes.446 Thermoplastic polymers, hydrogels, and decellularized extracellular matrix (dECM) are three main types of biomaterials used in combining cells and biomaterials field research.448,454,455 Thermoplastic polymers such as PCL, PU, and PLA can be used as structural supports owing to their high mechanical properties.448 However, printing thermoplastic polymers needs either high temperatures or toxic solvents, leading to low cytocompatibility. Furthermore, combining these polymers with cell-supportive hydrogels is a challenge in the printing process.448
As reviewed by Gopinathan et al.,375 owing to the emergence of “click” chemistry strategies, many natural and synthetic hydrogels are currently available to 3D bioprint tissue models.375 Natural hydrogels such as chitosan, collagen, alginate, and gelatin can provide a native ECM-like microenvironment for cells.456 However, compared to natural hydrogels, the mechanical properties and cell-adherent characteristics of synthetically derived hydrogels (such as methacrylated gelatin, PEG, and polyoxyethylene–polyoxypropylene triblock copolymers) can be more easily manipulated.448 Although increasing the chemical concentrations and crosslink capacities leads to better printability and shape fidelity, they can cause smaller pore size and lower cell viability.457,458
Because dECM has the ECM constituents, it can overcome these challenges.459 However, its low post-printing shape fidelity and ethical issues should be solved before translating the designed models to clinical settings.448 Designing composite biomaterials can be a solution to overcome the weakness of each component regarding mechanical strength, printability, biocompatibility, and gelation properties.448
Although bioprinting has been a powerful tool for creating 3D tissue models, there are still several challenges that we should address, as follows:
(i) Constructing whole organs. Organs are complex structures containing different cell types and gradients of physicochemical properties.460 In addition, native organs have adequate mechanical strength to keep their shape and integrity over time.461 Therefore, researchers should enhance the printing resolution to create 3D organ models with internal complex networks.446
(ii) Vascularization. Vascular networks are essential for providing passage to nutrients, oxygen, and metabolic wastes.448 Fabricating scale-up tissues or organs with a functional vascular network that enables effective local innervation remains a critical challenge for the field.
(iii) Cell culture technique. Cells are one of the key elements of bio-inks, which should be cultured in large numbers. By using the current available techniques, cell culture expansion processes take from weeks to months for each cell type.462 It is vital to develop new techniques, which are capable of speeding up the cell expansion time without distorting cells.446
(iv) Fabricating functional tissues. Cell responses, scaffold stability, and ECM deposition are three main elements of a functional tissue construct.463,464 The 3D ECM microenvironment should allow differentiation and proliferation of printed cells. Designing new biomaterials and modifying the properties of available ones are essential to facilitate ECM–cell signaling.446 In addition, controlling the biomaterial degradation rate is crucial to make sure the synthetic ECM degradation rate is proportional to the native ECM production.465
(v) Improving the mechanical strength of hydrogels as synthetic ECMs. There are two main strategies for enhancing the mechanical strength of hydrogels. (I) Decreasing the construction time can enhance the mechanical strength of printed structures. However, slower printing speed leaves the cells longer inside the ink, which may not be beneficial for cells. To overcome this issue, researchers introduced the 4D bioprinting concept. It allows a 3D printed structure to change its configuration and/or function over time in response to external stimuli such as temperature, light, and water, which makes 3D printing alive.446,466,467 (II) Improving the mechanical strength of hydrogels by co-printing them with tough degradable biomaterials.462
12. State-of-the-art of evaluation of biological responses and future perspective
Based on the International Organization for Standardization (ISO) and national standards, we must prove the biocompatibility of any biomaterial or medical device by doing a series of tests regarding its genotoxicity, carcinogenicity, toxicity, sensitization, as well as acute and chronic systemic toxicity prior to doing any clinical trials in humans.468,469 Since 1992, ISO has published and modified a series of international biocompatibility standards for medical devices (ISO 10993), as a living and regularly evolving document, which provides the general principles that we should bear in mind during both in vitro and in vivo evaluation of material–tissue interfaces.57,470–472 Despite applying a wide range of chemical analyses for evaluating the biocompatibility of biomaterials based on ISO standards, there are still some challenges in the evaluation of biomaterials by following the standards.In 2010, Poly Implant Prothèse (PIP), a French manufacturer of silicone gel breast implants, went bankrupt after using low-grade industrial silicone gel in their products.473 It is clear that their implants should not have passed the defined biocompatibility tests of ISO standards 10993. Consequently, the European governments and competent authorities set an additional approval process named as Medical Device Regulation (MDR 2017/745) to address the biocompatibility evaluations of medical devices.473
The main purpose of MDR is to improve both the report quality and clinical trials data availability, rather than changing ISO standards. In 2021, the results of clinical trials will be available for the public on the European database for medical devices (EUDAMED),474,475 which could be a helpful tool for the rapid reviewing of available medical devices’ biocompatibility properties.476 As it would be more expensive to bring new medical devices to the market, one could argue that the new MDR will reduce the number of novel biomaterials in the future.477 Thus, it is crucial to ask if there are any alternatives for improving the translation reliability between “pre-market” clinical trial requirements and the “post-market surveillance”. The “post market surveillance” is defined as the clinical follow-up of the Conformité Européenne (CE) approved medical devices, whereas the “pre-market clinical trials” are the required testing prior to CE mark. There is still a question that if the new MDR will not amend the in vitro biocompatibility tests, what would be the alternatives to determine the cellular and molecular pathways and responses to new biomaterials more accurately than ISO standards?478
Several studies pointed out the weakness of ISO 10993-5. The cell lines are commonly tested on extracts. Obviously, using extracts cannot provide us valid information about the biochemical transduction pathways and signals, which affect the biocompatibility of materials. Additionally, extracts of biomaterials may show cytotoxic effects on the cultured cells caused by changes in the ionic composition of the medium.
Although using cell lines might provide reproducible results, it is difficult to predict the cells’ biochemical and biophysical signals in response to implanted biomaterials in the body based on these results. The biochemical transduction pathways cannot be examined and detected precisely through using such immortalized cell lines.479–482
Even though cytotoxicity is one of the most important indicators for biomaterial evaluation, all present cytotoxicity test methods have certain drawbacks.468 Considering the other potential key biochemical signals which might be involved in biological responses to a material and can affect the evaluation criteria of material safety in the body is important.12
Traditionally biocompatibility was investigated by considering the wound healing processes that occur after biomaterial implantation. The in vivo evaluation of biological responses to biomaterials involves implanting biomaterials in the targeted tissues at certain time points and then studying the histological changes in the implanted tissue and its surroundings. However, in the case of tissue-regenerated biomaterials, one would keep in mind not only wound healing processes, but also several other physiological, mechanical and biochemical pathways, which might be involved in determining the material's success or failure. In addition, there is a lack of enough valid chemical techniques for evaluating the biochemical features of surface biocompatibility in vivo. The main biochemical signaling pathways involved in host responses should be evaluated based on the properties of biomaterials and targeted cells.11 As the biological responses to biomaterials is dependent on the cell type used, we emphasize using suitable cell lines for in vitro cytotoxicity testing.57
Regarding animal studies, the choice and design of animal models for testing biomaterials are complex and there are few guidelines. To evaluate both intended and unintended effects, we use several criteria to evaluate implants, including biological relevance, biofunctionality, biocompatibility/safety, and clinical relevance/efficacy. Safety studies often use smaller species (e.g. rodents and rabbits) to detect local tissue damage and systemic toxicity from degradable or leachable products.483,484 However, tissue reactions to specific biomaterials are tested typically in larger animals to provide more detailed biological information.476,484 The anatomy, physiology and pathogenicity of experimental models should relate as much as possible to those of patients in order to demonstrate the safety and efficacy of new biomaterials.485
Knock-out methods are among the most common systems used for investigating molecular pathways in animal models. However, we should consider that these knock-out animal models are not able to target several natural pathways that might be alongside responsible for one mechanism.57 There is an argument in the literature over communal animal implant models for evaluating host material response and trying to link common host responses across various species in response to biomaterials. Because of the complexity of the human biological pathways (including biochemical, biophysical, mechanical and physiological pathways), choosing the right animal model based on the aim of the study is a big issue in this field. Hence, as Grainger486 stated in his review “Surprisingly, little consensus or consistency is found in published literature for host-implant integration metrics for implant healing versus foreign body response.” The translation of preclinical animal data into successful clinic products is typically poor.487–489
Consequently, we should use different types of animal models for each newly designed biomaterial to mimic all the potential biological pathways involved in human body responses to biomaterials, which increases the ethical issues and complexity in this field. If we assess the preclinical animal data shortcomings more systematically, we can achieve more reliable preclinical in vivo data. Typical deficiencies that must be overcome are poor design of animal models, acute models opposed to chronic ones, poor blinding and randomization, and statistically underpowered studies.490
Over the past few years, researchers have suggested using organoids as an alternative technology for biocompatibility assessment.491 An organoid is a 3D structure, grown from stem and/or progenitor cells, consisting of organ-specific cell types, which organizes itself through cell sorting and spatially restricted lineage commitment. These models can provide systematic information about chemical and genetic perturbation of 3D tissue. Such models have substantial potential in bridging the gap between in vitro biocompatibility and animal models and might be a valid alternative to in vivo animal studies. The great advantage of using organoids could be discovering chemical-transduction pathways, which cannot be achieved by current in vivo techniques. Conventional in vitro cell cultures are based on 2D systems, which often fail to induce full proliferation and differentiation potential of cells. There is a massive discrepancy between 2D and 3D in vitro cell culture environments, where the latter system allows cell-to-cell and cell-to-biomaterial interactions, more similar to the body environment. This disparity between 2D and 3D systems makes it difficult for 2-D cell-based in vitro models to be reliable for investigating disease mechanisms and drug screening.492
Although it is doubtful that organoids will completely replace animal studies, we should evaluate the obvious advantages of using organoids in biocompatibility tests.493 As Bredeboord et al.494 stated: “We suggest that the use of organoids is complementary to, rather than in competition with, these classical research methodologies”.494
In silico bioinformatics are computational frameworks, which are currently playing a key role in the discovery and validation of new chemical identities. Therefore, using these computational models for also predicting the biochemical responses to biomaterials could be a revolution in biological response evaluation. Such computational models of tissue engineering processes attracted much attention as new non-invasive evaluation techniques, which can decrease or replace in vivo animal studies without having any ethical issues.495–498 Although these computational models might strongly help in evaluating and predicting biomaterial–host complex interactions and decrease the number of unnecessary animal studies, without doing any animal studies we cannot make well-founded decisions regarding the safety of biomaterials and directly implant them in the human body with confidence.12
Considering more quantitative and statistical analyses can help us to make conclusions that are more convincing. Due to the immense development of new biomaterials for tissue engineering applications, researchers design more and more tissue-engineered templates with different chemistry. More reliable analytical techniques should be established alongside to provide atomic-level structural information about biomolecules. Combining imaging and analytical chemistry techniques can be a promising strategy to go beyond the current limitations of analytical chemistry.
13. Conclusions
It is undeniable that the medicine world owes chemistry science for the current existence of promising biomaterials as therapeutic solutions. By using the chemical principles present in nature, researchers have made substantial progress in biomaterials and tissue engineering fields. Nevertheless, there are still critical challenges regarding their biocompatibility that need to be further studied and are typically neglected when describing new therapeutic strategies using biomaterials. The biological responses to biomaterials should not be defined as only not having any adverse effects. They should have a strong affinity for targeted cells to stimulate neo-tissue formation, which depends on the chemical characteristics of both the biomaterials and biological environments. The host responses to biomaterials mainly originate from biochemical signaling pathways and factors. As our knowledge in the biochemistry field has grown tremendously since the first definition of biocompatibility, we should update our definitions about biological responses to biomaterials. In addition, from the materials engineering point of view, biomaterial surface physicochemistry can profoundly affect biophysical and biochemical responses. Providing more reliable analytical techniques and standards for biochemical interface characterization of biomaterials is highly important. Nevertheless, the chemical basis of biological responses to biomaterials is a domain where much remains to be studied.Abbreviations
| R a/Sa | Roughness value |
| PDMS | Polydimethylsiloxane |
| NIPAM | N-Isopropylacrylamide |
| HepG-2 | Hepatocellular carcinoma cell line |
| PAAm | Polyacrylamide |
| PES | Poly(ether sulfone) |
| CPES | PES with carboxylic groups |
| SNPES | PES with sodium sulfonic and amino groups |
| OD | 1,7-Octadiene |
| FBS | Foetal bovine serum |
| PIII | Plasma-immersion ion implantation |
| PLA | Poly(lactic acid) |
| BMP2 | Bone morphogenetic protein 2 |
| RAFT | Reversible addition fragmentation chain transfer |
| ECM | Extracellular matrix |
| IL | Interleukin |
| PHSRN | Proline–histidine–serine–arginine–asparagine |
| RGD | Arginine–glycine–aspartic acid |
| PAMPs | Pathogen associated molecular patterns |
| DAMPs | Damage-associated molecular patterns |
| TLRs | Toll-like receptors |
| PRRs | Pattern recognition receptors |
| NLR | NOD-like receptor |
| ALP | Alkaline phosphatase |
| MSCs | Mesenchymal stem cells |
| PGE2 | Prostaglandin E2 |
| Tregs | T regulatory lymphocytes |
| hMSCs | Human-MSCs |
| VEGF | Vascular endothelial growth factor |
| FAK | Focal adhesion kinase |
| RANKL | Receptor activator of nuclear factor kappa-B ligand |
| SIMS | Secondary-ion mass spectrometry |
| ToF-SIMS | Time of flight-SIMS |
| XPS | X-ray photoelectron spectroscopy |
| IR | Infrared |
| SEM-EDX | Scanning electron microscopy-energy dispersive X-ray |
| PC | Principal component analysis |
| VMD | Visual molecular dynamics |
| FBGC | Foreign body giant cell |
| AIM2 | Absent in melanoma 2 |
| CAV1 | Caveolin 1 |
| ROS | Reactive oxygen species |
| TRPM | Transient receptor potential melastatin |
| NMDA | N-Methyl-D-aspartate |
| ATP | Adenosine triphosphate |
| P2XRs | P2X receptors |
| KCa3.1 | Ca2+-Activated K+ channel |
| cGMP | Cyclic guanosine monophosphate |
| AFM | Atomic force microscopy |
| PDMAEMA | Poly(2-(dimethylamino)ethyl methacrylate) |
| ATRP | Atom transfer radical polymerization |
| BSA | Bovine serum albumin |
| QCM-D | Quartz crystal microbalance with dissipation |
| SIMS | Secondary-ion mass spectrometry |
| ToF-SIMS | Time of flight-SIMS |
| XPS | X-ray photoelectron spectroscopy |
| IR | Infrared |
| SEM-EDX | Scanning electron microscopy-energy dispersive X-ray |
| PCA | Principal component analysis |
| VMD | Visual molecular dynamics |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a project “Promoting patient safety by a novel combination of imaging technologies for biodegradable magnesium implants, MgSafe” funded by European Training Network within the framework of Horizon2020 Marie Skłodowska-Curie Action (MSCA) grant number no. 811226 (http://www.mgsafe.eu). Eduardo A. Silva was supported by the American Heart Association grant # 19IPLOI34760654/Eduardo Silva/2019. We would like to thank Prof. Silvia M. Rogers, the head of MEDWRITE Company, Basel, Switzerland, who kindly helped us with her professional comments on the scientific writing part of this paper. We also thank Dr Matilde Bongio (http://www.matildebongio.com), who designed some informative illustrations for this paper.References
- F.-M. Chen and X. Liu, Prog. Polym. Sci., 2016, 53, 86–168 CrossRef CAS PubMed.
- M. Rahmati, C. P. Pennisi, A. Mobasheri and M. Mozafari, Cell Biology and Translational Medicine, Springer, 2018, vol. 3, pp. 73–89 Search PubMed.
- A. Khademhosseini and R. Langer, Nat. Protoc., 2016, 11, 1775–1781 CrossRef CAS PubMed.
- C. P. Bergmann and A. Stumpf, Dental Ceramics, Springer, 2013, pp. 55–65 Search PubMed.
- M. Rahmati, C. P. Pennisi, E. Budd, A. Mobasheri and M. Mozafari, Cell Biology and Translational Medicine, Springer, 2018, vol. 4, pp. 1–19 Search PubMed.
- S. J. Lee, J. J. Yoo and A. Atala, Clinical Regenerative Medicine in Urology, Springer, 2018, pp. 17–51 Search PubMed.
- S. N. Alhosseini, F. Moztarzadeh, M. Mozafari, S. Asgari, M. Dodel, A. Samadikuchaksaraei, S. Kargozar and N. Jalali, Int. J. Nanomed., 2012, 7, 25 CAS.
- V. Shabafrooz, M. Mozafari, D. Vashaee and L. Tayebi, J. Nanosci. Nanotechnol., 2014, 14, 522–534 CrossRef CAS PubMed.
- K. Nazemi, F. Moztarzadeh, N. Jalali, S. Asgari and M. Mozafari, BioMed Res. Int., 2014, 2014, 898930, DOI:10.1155/2014/898930.
- D. F. Williams, Biomaterials, 2014, 35, 10009–10014 CrossRef CAS PubMed.
- D. F. Williams, ACS Biomater. Sci. Eng., 2016, 3, 2–35 CrossRef.
- D. F. Williams, Tissue Eng., Part C, 2017, 23, 926–937 CrossRef PubMed.
- D. F. Williams, Prog. Biomed. Eng., 2019, 1, 013001 CrossRef.
- M. Rahmati and M. Mozafari, Mater. Today Commun., 2018, 17, 527–540 CrossRef CAS.
- C. J. Kirkpatrick, Regener. Biomater., 2015, 2, 267–272 CrossRef PubMed.
- K. Sadtler, A. Singh, M. T. Wolf, X. K. Wang, D. M. Pardoll and J. H. Elisseeff, Nat. Rev. Mater., 2016, 1, 16040 CrossRef CAS.
- B. Reid, M. Gibson, A. Singh, J. Taube, C. Furlong, M. Murcia and J. Elisseeff, J. Tissue Eng. Regener. Med., 2015, 9, 315–318 CrossRef CAS PubMed.
- H. Rostam, S. Singh, N. Vrana, M. Alexander and A. Ghaemmaghami, Biomater. Sci., 2015, 3, 424–441 RSC.
- A. Albanese, P. S. Tang and W. C. Chan, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed.
- S. Bajpai, N. Y. Kim and C. A. Reinhart-King, Int. J. Mol. Sci., 2011, 12, 8596–8609 CrossRef CAS PubMed.
- F. M. Chen and X. Liu, Prog. Polym. Sci., 2016, 53, 86–168 CrossRef CAS PubMed.
- E. S. Place, N. D. Evans and M. M. Stevens, Nat. Mater., 2009, 8, 457–470 CrossRef CAS PubMed.
- B. Levi and M. T. Longaker, Stem Cells, 2011, 29, 576–582 CrossRef CAS PubMed.
- B. Lindroos, R. Suuronen and S. Miettinen, Stem Cell Rev. Rep., 2011, 7, 269–291 CrossRef PubMed.
- H. Mizuno, M. Tobita and A. C. Uysal, Stem Cells, 2012, 30, 804–810 CrossRef CAS PubMed.
- S. Pina, J. M. Oliveira and R. L. Reis, Adv. Mater., 2015, 27, 1143–1169 CrossRef CAS PubMed.
- D. I. Braghirolli, D. Steffens and P. Pranke, Drug Discovery Today, 2014, 19, 743–753 CrossRef CAS PubMed.
- M. J. Webber, E. A. Appel, E. Meijer and R. Langer, Nat. Mater., 2016, 15, 13–26 CrossRef CAS PubMed.
- F. Pati, D.-H. Ha, J. Jang, H. H. Han, J.-W. Rhie and D.-W. Cho, Biomaterials, 2015, 62, 164–175 CrossRef CAS PubMed.
- G. S. Boersema, N. Grotenhuis, Y. Bayon, J. F. Lange and Y. M. Bastiaansen-Jenniskens, BioRes. Open Access, 2016, 5, 6–14 CrossRef CAS PubMed.
- N. M. Seale, Y. Zeng and S. Varghese, Developmental Biology and Musculoskeletal Tissue Engineering, Elsevier, 2018, pp. 207–223 Search PubMed.
- M. Nightingale and M. R. Labrosse, J. Biomech., 2018, 79, 207–211 CrossRef PubMed.
- P. Zarrintaj, A. Urbanska, S. S. Gholizadeh, V. Goodarzi, M. R. Saeb and M. Mozafari, J. Colloid Interface Sci., 2018, 516, 57–66 CrossRef CAS PubMed.
- M. Gholipourmalekabadi, A. Samadikuchaksaraei, A. M. Seifalian, A. Urbanska, H. Ghanbarian, J. G. Hardy, M. Omrani, M. Mozafari, R. L. Reis and S. C. Kundu, Biomed. Mater., 2018, 13(3), 035003 CrossRef PubMed.
- A. S. Ayoub and L. A. Lucia, Introduction to renewable biomaterials: first principles and concepts, John Wiley & Sons, 2017 Search PubMed.
- M. Sekuła and E. K. Zuba-Surma, Biomaterials in Regenerative Medicine, InTech, 2018 Search PubMed.
- P. Ducheyne, Comprehensive biomaterials, Elsevier, 2015 Search PubMed.
- S. J. Hollister, Nat. Mater., 2005, 4, 518–524 CrossRef CAS PubMed.
- R. C. Dutta and A. K. Dutta, Biotechnol. Adv., 2009, 27, 334–339 CrossRef CAS PubMed.
- X. Gu, F. Ding and D. F. Williams, Biomaterials, 2014, 35, 6143–6156 CrossRef CAS PubMed.
- J. J. Rice, M. M. Martino, L. De Laporte, F. Tortelli, P. S. Briquez and J. A. Hubbell, Adv. Healthcare Mater., 2013, 2, 57–71 CrossRef CAS PubMed.
- M. A. Meyers, J. McKittrick and P. Y. Chen, Science, 2013, 339, 773–779 CrossRef CAS PubMed.
- S. Mitragotri and J. Lahann, Nat. Mater., 2009, 8, 15–23 CrossRef CAS PubMed.
- J. M. Holzwarth and P. X. Ma, Biomaterials, 2011, 32, 9622–9629 CrossRef CAS PubMed.
- F. Edalat, I. Sheu, S. Manoucheri and A. Khademhosseini, Curr. Opin. Biotechnol., 2012, 23, 820–825 CrossRef CAS PubMed.
- A. Ranella, M. Barberoglou, S. Bakogianni, C. Fotakis and E. Stratakis, Acta Biomater., 2010, 6, 2711–2720 CrossRef CAS PubMed.
- H.-I. Chang and Y. Wang, Regenerative medicine and tissue engineering-cells and biomaterials, InTechOpen, 2011 Search PubMed.
- M. P. Calatayud, B. Sanz, V. Raffa, C. Riggio, M. R. Ibarra and G. F. Goya, Biomaterials, 2014, 35, 6389–6399 CrossRef CAS PubMed.
- M. Hoefling, F. Iori, S. Corni and K.-E. Gottschalk, Langmuir, 2010, 26, 8347–8351 CrossRef CAS PubMed.
- F. Meder, T. Daberkow, L. Treccani, M. Wilhelm, M. Schowalter, A. Rosenauer, L. Mädler and K. Rezwan, Acta Biomater., 2012, 8, 1221–1229 CrossRef CAS PubMed.
- K. N. Ekdahl, J. D. Lambris, H. Elwing, D. Ricklin, P. H. Nilsson, Y. Teramura, I. A. Nicholls and B. Nilsson, Adv. Drug Delivery Rev., 2011, 63, 1042–1050 CrossRef CAS PubMed.
- J. M. Anderson and K. M. Miller, Biomaterials, 1984, 5, 5–10 CrossRef CAS PubMed.
- J. M. Anderson, Cardiovasc. Pathol., 1993, 2, S33–S41 CrossRef.
- R. Marchant, A. Hiltner, C. Hamlin, A. Rabinovitch, R. Slobodkin and J. M. Anderson, J. Biomed. Mater. Res., 1983, 17, 301–325 CrossRef PubMed.
- D. F. Williams, Biomaterials, 2008, 29, 2941–2953 CrossRef CAS PubMed.
- M. Rahmati and M. Mozafari, ACS Biomater. Sci. Eng., 2019, 6(1), 4–20 CrossRef.
- J. M. Anderson, Regener. Biomater., 2016, 3, 73–77 CrossRef CAS PubMed.
- D. F. Williams, Biomaterials, 2008, 29, 2941–2953 CrossRef CAS PubMed.
- P. M. Kou and J. E. Babensee, J. Biomed. Mater. Res., Part A, 2011, 96, 239–260 CrossRef PubMed.
- Z. Sheikh, P. J. Brooks, O. Barzilay, N. Fine and M. Glogauer, Materials, 2015, 8, 5671–5701 CrossRef CAS PubMed.
- J. M. Morais, F. Papadimitrakopoulos and D. J. Burgess, AAPS J., 2010, 12, 188–196 CrossRef CAS PubMed.
- R. A. Latour, Encycl. Biomater. Biomed. Eng., 2005, 1, 270–284 Search PubMed.
- S. L. Hirsh, D. R. McKenzie, N. J. Nosworthy, J. A. Denman, O. U. Sezerman and M. M. Bilek, Colloids Surf., B, 2013, 103, 395–404 CrossRef CAS PubMed.
- Q. Wei, T. Becherer, S. Angioletti-Uberti, J. Dzubiella, C. Wischke, A. T. Neffe, A. Lendlein, M. Ballauff and R. Haag, Angew. Chem., Int. Ed., 2014, 53, 8004–8031 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- Z. Xia and J. T. Triffitt, Biomed. Mater., 2006, 1, R1 CrossRef CAS PubMed.
- L. Tang and J. W. Eaton, Am. J. Clin. Pathol., 1995, 103, 466–471 CrossRef CAS PubMed.
- Y. Fong, L. L. Moldawer, G. T. Shires and S. F. Lowry, Surg., Gynecol. Obstet., 1990, 170, 363–378 CAS.
- G. F. Pierce, T. A. Mustoe, B. W. Altrock, T. F. Deuel and A. Thomason, J. Cell. Biochem., 1991, 45, 319–326 CrossRef CAS PubMed.
- Y. Onuki, U. Bhardwaj, F. Papadimitrakopoulos and D. J. Burgess, J. Diabetes Sci. Technol., 2008, 2(6), 1003–1015 CrossRef PubMed.
- J. M. Anderson, A. Rodriguez and D. T. Chang, Semin. Immunol., 2008, 20(2), 86–100 CrossRef CAS PubMed.
- J. M. Anderson, ASAIO Trans., 1988, 34, 101–107 CrossRef CAS PubMed.
- D. P. Vasconcelos, A. P. Aguas, M. A. Barbosa, P. Pelegrin and J. N. Barbosa, Acta Biomater., 2019, 83, 1–12 CrossRef CAS PubMed.
- B. N. Brown and S. F. Badylak, Acta Biomater., 2013, 9, 4948–4955 CrossRef CAS PubMed.
- B. D. Ratner and S. J. Bryant, Annu. Rev. Biomed. Eng., 2004, 6, 41–75 CrossRef CAS PubMed.
- S. A. Eming, T. Krieg and J. M. Davidson, J. Invest. Dermatol., 2007, 127, 514–525 CrossRef CAS PubMed.
- A. Vishwakarma, N. S. Bhise, M. B. Evangelista, J. Rouwkema, M. R. Dokmeci, A. M. Ghaemmaghami, N. E. Vrana and A. Khademhosseini, Trends Biotechnol., 2016, 34, 470–482 CrossRef CAS PubMed.
- S. F. Badylak, Science, 2016, 352, 298 CrossRef CAS PubMed.
- S. A. Eming, M. Hammerschmidt, T. Krieg and A. Roers, Semin. Cell Dev. Biol., 2009, 20(5), 517–527 CrossRef CAS PubMed.
- N. A. Hotaling, L. Tang, D. J. Irvine and J. E. Babensee, Annu. Rev. Biomed. Eng., 2015, 17, 317–349 CrossRef CAS PubMed.
- E. Mariani, G. Lisignoli, R. M. Borzi and L. Pulsatelli, Int. J. Mol. Sci., 2019, 20, 636 CrossRef CAS PubMed.
- P. Krzyszczyk, R. Schloss, A. Palmer and F. Berthiaume, Front. Physiol., 2018, 9, 419 CrossRef PubMed.
- P. J. Delves, S. J. Martin, D. R. Burton and I. M. Roitt, Essential immunology, John Wiley & Sons, 2017 Search PubMed.
- R. F. Diegelmann, Front. Biosci., 2004, 9, 283–289 CrossRef CAS PubMed.
- H. P. Felgueiras, N. S. Murthy, S. D. Sommerfeld, M. M. Bras, V. Migonney and J. Kohn, ACS Appl. Mater. Interfaces, 2016, 8, 13207–13217 CrossRef CAS PubMed.
- M. W. Mosesson, J. Thromb. Haemostasis, 2005, 3, 1894–1904 CrossRef CAS PubMed.
- B. O. O. Boni, L. Lamboni, T. Souho, M. Gauthier and G. Yang, Mater. Horiz., 2019, 6, 1122–1137 RSC.
- L. W. Qian, A. B. Fourcaudot, K. Yamane, T. You, R. K. Chan and K. P. Leung, Wound Repair Regen., 2016, 24, 26–34 CrossRef PubMed.
- M. E. Scarritt, R. Londono and S. F. Badylak, The Immune Response to Implanted Materials and Devices, Springer, 2017, pp. 1–14 Search PubMed.
- T. J. Koh and L. A. DiPietro, Expert Rev. Mol. Med., 2011, 13, e23 CrossRef PubMed.
- D. Ramnath, E. E. Powell, G. M. Scholz and M. J. Sweet, Semin. Cell Dev. Biol., 2017, 61, 22–30 CrossRef CAS PubMed.
- M. Karin and H. Clevers, Nature, 2016, 529, 307–315 CrossRef CAS PubMed.
- Z. Julier, A. J. Park, P. S. Briquez and M. M. Martino, Acta Biomater., 2017, 53, 13–28 CrossRef CAS.
- C. Esche, C. Stellato and L. A. Beck, J. Invest. Dermatol., 2005, 125, 615–628 CrossRef CAS.
- G. Y. Chen and G. Nunez, Nat. Rev. Immunol., 2010, 10, 826–837 CrossRef CAS PubMed.
- P. Scaffidi, T. Misteli and M. E. Bianchi, Nature, 2002, 418, 191–195 CrossRef CAS PubMed.
- F. J. Quintana and I. R. Cohen, J. Immunol., 2005, 175, 2777–2782 CrossRef CAS PubMed.
- M. J. Bours, E. L. Swennen, F. Di Virgilio, B. N. Cronstein and P. C. Dagnelie, Pharmacol. Ther., 2006, 112, 358–404 CrossRef CAS PubMed.
- H. Kono and K. L. Rock, Nat. Rev. Immunol., 2008, 8, 279–289 CrossRef CAS PubMed.
- M. Lamkanfi and V. M. Dixit, Cell, 2014, 157, 1013–1022 CrossRef CAS PubMed.
- B. K. Davis, H. Wen and J. P. Ting, Annu. Rev. Immunol., 2011, 29, 707–735 CrossRef CAS.
- J. von Moltke, J. S. Ayres, E. M. Kofoed, J. Chavarria-Smith and R. E. Vance, Annu. Rev. Immunol., 2013, 31, 73–106 CrossRef CAS.
- A. Denes, G. Coutts, N. Lenart, S. M. Cruickshank, P. Pelegrin, J. Skinner, N. Rothwell, S. M. Allan and D. Brough, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4050–4055 CrossRef CAS PubMed.
- Y. Qu, S. Misaghi, K. Newton, A. Maltzman, A. Izrael-Tomasevic, D. Arnott and V. M. Dixit, J. Exp. Med., 2016, 213, 877–885 CrossRef CAS PubMed.
- W. K. Ip and R. Medzhitov, Nat. Commun., 2015, 6, 6931 CrossRef CAS PubMed.
- T. Strowig, J. Henao-Mejia, E. Elinav and R. Flavell, Nature, 2012, 481, 278–286 CrossRef CAS PubMed.
- Y. He, M. Y. Zeng, D. Yang, B. Motro and G. Núñez, Nature, 2016, 530, 354 CrossRef CAS PubMed.
- H. Shi, Y. Wang, X. Li, X. Zhan, M. Tang, M. Fina, L. Su, D. Pratt, C. H. Bu, S. Hildebrand, S. Lyon, L. Scott, J. Quan, Q. Sun, J. Russell, S. Arnett, P. Jurek, D. Chen, V. V. Kravchenko, J. C. Mathison, E. M. Moresco, N. L. Monson, R. J. Ulevitch and B. Beutler, Nat. Immunol., 2016, 17, 250–258 CrossRef CAS.
- S. Jha, W. J. Brickey and J. P.-Y. Ting, Inflammasomes in myeloid cells: warriors within, Myeloid Cells in Health and Disease: A Synthesis, American Society for Microbiology, Washington, DC, 2017, ch. 17, pp. 305–324 Search PubMed.
- D. Fong, P. Gregoire-Gelinas, A. P. Cheng, T. Mezheritsky, M. Lavertu, S. Sato and C. D. Hoemann, Biomaterials, 2017, 129, 127–138 CrossRef CAS PubMed.
- J. I. Andorko and C. M. Jewell, Bioeng. Transl. Med., 2017, 2, 139–155 CrossRef PubMed.
- P. Broz and V. M. Dixit, Nat. Rev. Immunol., 2016, 16, 407 CrossRef CAS PubMed.
- C. M. Artlett, J. Pathol., 2013, 229, 157–167 CrossRef CAS.
- H. H. Simms, R. D'Amico and K. I. Bland, Ann. Surg., 1997, 225, 757–763 CrossRef CAS ; discussion 763–755.
- J. M. Anderson and S. Jiang, The Immune Response to Implanted Materials and Devices, Springer, 2017, pp. 15–36 Search PubMed.
- D. El Kebir and J. G. Filep, Cells, 2013, 2, 330–348 CrossRef CAS PubMed.
- K. R. Martin, D. Ohayon and V. Witko-Sarsat, Swiss Med. Wkly., 2015, 145, w14056 Search PubMed.
- N. X. Landen, D. Li and M. Stahle, Cell. Mol. Life Sci., 2016, 73, 3861–3885 CrossRef CAS PubMed.
- W. G. Brodbeck, M. Macewan, E. Colton, H. Meyerson and J. M. Anderson, J. Biomed. Mater. Res., Part A, 2005, 74, 222–229 CrossRef PubMed.
- M. Aziz, N. E. Holodick, T. L. Rothstein and P. Wang, Immunol. Res., 2015, 63, 153–166 CrossRef CAS PubMed.
- F. Alegre, P. Pelegrin and A. E. Feldstein, Semin. Liver Dis., 2017, 37(02), 119–127 CrossRef CAS PubMed.
- T. A. Wynn and K. M. Vannella, Immunity, 2016, 44, 450–462 CrossRef CAS PubMed.
- F.-X. Yu and K.-L. Guan, Genes Dev., 2013, 27, 355–371 CrossRef CAS PubMed.
- A. Lee, B. Fakler, L. K. Kaczmarek and L. L. Isom, J. Neurosci., 2014, 34, 15159–15169 CrossRef PubMed.
- S. Asuthkar, P. A. Elustondo, L. Demirkhanyan, X. Sun, P. Baskaran, K. K. Velpula, B. Thyagarajan, E. V. Pavlov and E. Zakharian, J. Biol. Chem., 2015, 290, 2659–2669 CrossRef CAS PubMed.
- K. Takahashi, Y. Matsuda and K. Naruse, AIMS Biophys., 2016, 3, 63–74 CAS.
- F. Di Virgilio, A. C. Sarti and F. Grassi, Curr. Opin. Immunol., 2018, 52, 51–59 CrossRef CAS PubMed.
- I. Hafner-Bratkovič and P. Pelegrín, Curr. Opin. Immunol., 2018, 52, 8–17 CrossRef.
- L. Stokes, A. B. MacKenzie and R. Sluyter, Front. Immunol., 2016, 7, 48 Search PubMed.
- G. A. Ramirez, L. A. Coletto, C. Sciorati, E. P. Bozzolo, P. Manunta, P. Rovere-Querini and A. A. Manfredi, Cells, 2018, 7, 70 CrossRef PubMed.
- V. Schatz, P. Neubert, A. Schröder, K. Binger, M. Gebhard, D. N. Müller, F. C. Luft, J. Titze and J. Jantsch, Pediatr. Nephrol., 2017, 32, 201–210 CrossRef.
- K. Salao, L. Jiang, H. Li, V. W.-W. Tsai, Y. Husaini, P. M. Curmi, L. J. Brown, D. A. Brown and S. N. Breit, Biol. Open, 2016, 5, 620–630 CrossRef CAS.
- M. Vaeth and S. Feske, Curr. Opin. Immunol., 2018, 52, 39–50 CrossRef CAS PubMed.
- B. Davenport, Y. Li, J. W. Heizer, C. Schmitz and A.-L. Perraud, Front. Immunol., 2015, 6, 375 Search PubMed.
- L. L. Nohara, S. R. Stanwood, K. D. Omilusik and W. A. Jefferies, Front. Immunol., 2015, 6, 234 Search PubMed.
- S. Mortadza, S. Alawieyah, L. Wang, D. Li and L.-H. Jiang, Front. Immunol., 2015, 6, 407 Search PubMed.
- N. Liu, Y. Zhuang, Z. Zhou, J. Zhao, Q. Chen and J. Zheng, Neurosci. Lett., 2017, 651, 1–8 CrossRef CAS PubMed.
- Y. Mizoguchi, T. A. Kato, Y. Seki, M. Ohgidani, N. Sagata, H. Horikawa, Y. Yamauchi, M. Sato-Kasai, K. Hayakawa and R. Inoue, J. Biol. Chem., 2014, 289, 18549–18555 CrossRef CAS PubMed.
- M. J. Stebbing, J. M. Cottee and I. Rana, Front. Immunol., 2015, 6, 497 Search PubMed.
- R. A. North, Physiol. Rev., 2002, 82, 1013–1067 CrossRef CAS PubMed.
- F. Di Virgilio, G. Schmalzing and F. Markwardt, Trends Cell Biol., 2018, 28, 392–404 CrossRef CAS PubMed.
- R. Ferreira, R. Wong and L. C. Schlichter, Front. Immunol., 2015, 6, 153 Search PubMed.
- M. A. Retamal, M. V. Bennett, P. Pelegrin and R. Fernandez, Mediators Inflammation, 2016, 2016, 6245731, DOI:10.1155/2016/6245731.
- Y. Wu, L. Chen, P. G. Scott and E. E. Tredget, Stem Cells, 2007, 25, 2648–2659 CrossRef CAS PubMed.
- S. Mukherjee, S. Darzi, K. Paul, J. A. Werkmeister and C. E. Gargett, Interface Focus, 2019, 9, 20180089 CrossRef PubMed.
- T. S. Cheung and F. Dazzi, Semin. Immunol., 2018, 35, 59–68 CrossRef CAS PubMed.
- K. Le Blanc and L. C. Davies, Immunol. Lett., 2015, 168, 140–146 CrossRef CAS PubMed.
- E. Eggenhofer and M. J. Hoogduijn, Transplant. Res., 2012, 1, 12 CrossRef CAS PubMed.
- D. Kyurkchiev, I. Bochev, E. Ivanova-Todorova, M. Mourdjeva, T. Oreshkova, K. Belemezova and S. Kyurkchiev, World J. Stem Cells, 2014, 6, 552–570 CrossRef.
- H. Lei, K. Schmidt-Bleek, A. Dienelt, P. Reinke and H. D. Volk, Front. Pharmacol., 2015, 6, 184 Search PubMed.
- L. C. Davies and P. R. Taylor, Immunology, 2015, 144, 541–548 CrossRef CAS PubMed.
- H. Li, S. Shen, H. Fu, Z. Wang, X. Li, X. Sui, M. Yuan, S. Liu, G. Wang and Q. Guo, Stem Cells Int., 2019, 2019, 9671206 Search PubMed.
- J. D. Humphrey, E. R. Dufresne and M. A. Schwartz, Nat. Rev. Mol. Cell Biol., 2014, 15, 802–812 CrossRef CAS PubMed.
- C. Bonnans, J. Chou and Z. Werb, Nat. Rev. Mol. Cell Biol., 2014, 15, 786–801 CrossRef CAS PubMed.
- L. Li, J. Eyckmans and C. S. Chen, Nat. Mater., 2017, 16, 1164–1168 CrossRef CAS PubMed.
- J. Eyckmans, T. Boudou, X. Yu and C. S. Chen, Dev. Cell, 2011, 21, 35–47 CrossRef CAS PubMed.
- F. Martino, A. R. Perestrelo, V. Vinarsky, S. Pagliari and G. Forte, Front. Physiol., 2018, 9, 824 CrossRef PubMed.
- N. Wang, J. Phys. D: Appl. Phys., 2017, 50, 233002 CrossRef.
- R. J. McMurray, M. J. Dalby and P. M. Tsimbouri, J. Tissue Eng. Regener. Med., 2015, 9, 528–539 CrossRef PubMed.
- Y. Zhang, K. Liao, C. Li, A. C. K. Lai, J. J. Foo and V. Chan, Bioengineering, 2017, 4, 72 CrossRef PubMed.
- T. Iskratsch, H. Wolfenson and M. P. Sheetz, Nat. Rev. Mol. Cell Biol., 2014, 15, 825–833 CrossRef CAS PubMed.
- J. L. Alonso and W. H. Goldmann, AIMS Biophys., 2016, 3, 50–62 CAS.
- E. A. Cavalcanti-Adam, T. Volberg, A. Micoulet, H. Kessler, B. Geiger and J. P. Spatz, Biophys. J., 2007, 92, 2964–2974 CrossRef CAS PubMed.
- H. B. Schiller and R. Fassler, EMBO Rep., 2013, 14, 509–519 CrossRef CAS.
- Z. J. Chen, W. P. Wang, Y. C. Chen, J. Y. Wang, W. H. Lin, L. A. Tai, G. G. Liou, C. S. Yang and Y. H. Chi, J. Cell Sci., 2014, 127, 1792–1804 CrossRef CAS PubMed.
- N. Strohmeyer, M. Bharadwaj, M. Costell, R. Fässler and D. J. Müller, Nat. Mater., 2017, 16, 1262 CrossRef CAS PubMed.
- S. Seetharaman and S. Etienne-Manneville, Biol. Cell., 2018, 110, 49–64 CrossRef CAS.
- A. Elosegui-Artola, E. Bazellieres, M. D. Allen, I. Andreu, R. Oria, R. Sunyer, J. J. Gomm, J. F. Marshall, J. L. Jones, X. Trepat and P. Roca-Cusachs, Nat. Mater., 2014, 13, 631–637 CrossRef CAS PubMed.
- K. D. Webster, W. P. Ng and D. A. Fletcher, Biophys. J., 2014, 107, 146–155 CrossRef CAS PubMed.
- T. J. Chen, C. C. Wu, M. J. Tang, J. S. Huang and F. C. Su, PLoS One, 2010, 5, e14392 CrossRef CAS PubMed.
- P. Naumanen, P. Lappalainen and P. Hotulainen, J. Microsc., 2008, 231, 446–454 CrossRef CAS PubMed.
- P. Weiss, J. Exp. Zool., 1945, 100, 353–386 CrossRef CAS PubMed.
- K. Metavarayuth, P. Sitasuwan, X. Zhao, Y. Lin and Q. Wang, ACS Biomater. Sci. Eng., 2016, 2, 142–151 CrossRef CAS.
- N. Gui, W. Xu, D. E. Myers, R. Shukla, H. P. Tang and M. Qian, Biomater. Sci., 2018, 6, 250–264 RSC.
- J. Park, D. H. Kim and A. Levchenko, Biophys. J., 2018, 114, 1257–1263 CrossRef CAS PubMed.
- A. M. Ross, Z. Jiang, M. Bastmeyer and J. Lahann, Small, 2012, 8, 336–355 CrossRef CAS PubMed.
- A. G. Harvey, E. W. Hill and A. Bayat, Expert Rev. Med. Devices, 2013, 10, 257–267 CrossRef CAS PubMed.
- C. Rianna, M. Ventre, S. Cavalli, M. Radmacher and P. A. Netti, ACS Appl. Mater. Interfaces, 2015, 7, 21503–21510 CrossRef CAS PubMed.
- F. F. B. Hulshof, B. Papenburg, A. Vasilevich, M. Hulsman, Y. Zhao, M. Levers, N. Fekete, M. de Boer, H. Yuan, S. Singh, N. Beijer, M. A. Bray, D. J. Logan, M. Reinders, A. E. Carpenter, C. van Blitterswijk, D. Stamatialis and J. de Boer, Biomaterials, 2017, 137, 49–60 CrossRef CAS PubMed.
- F. Hulshof, C. Schophuizen, M. Mihajlovic, C. van Blitterswijk, R. Masereeuw, J. de Boer and D. Stamatialis, J. Tissue Eng. Regener. Med., 2018, 12, e817–e827 CAS.
- D. S. Hernandez, E. T. Ritschdorff, J. L. Connell and J. B. Shear, J. Am. Chem. Soc., 2018, 140, 14064–14068 CrossRef CAS PubMed.
- A. T. Nguyen, S. R. Sathe and E. K. Yim, J. Phys.: Condens. Matter, 2016, 28, 183001 CrossRef PubMed.
- C. L. Gilchrist, D. S. Ruch, D. Little and F. Guilak, Biomaterials, 2014, 35, 10015–10024 CrossRef CAS PubMed.
- B. K. Teo, S. T. Wong, C. K. Lim, T. Y. Kung, C. H. Yap, Y. Ramagopal, L. H. Romer and E. K. Yim, ACS Nano, 2013, 7, 4785–4798 CrossRef CAS PubMed.
- S. Wang, J. Li, Z. Zhou, S. Zhou and Z. Hu, Molecules, 2018, 24, 75 CrossRef PubMed.
- J. Padmanabhan, M. J. Augelli, B. Cheung, E. R. Kinser, B. Cleary, P. Kumar, R. Wang, A. J. Sawyer, R. Li, U. D. Schwarz, J. Schroers and T. R. Kyriakides, Sci. Rep., 2016, 6, 33277 CrossRef CAS PubMed.
- T. Khampieng, V. Yamassatien, P. Ekabutr, P. Pavasant and P. Supaphol, Adv. Polym. Technol., 2018, 37, 2030–2042 CrossRef CAS.
- I. Firkowska-Boden, X. Zhang and K. D. Jandt, Adv. Healthcare Mater., 2018, 7, 1700995 CrossRef PubMed.
- X. Wang, R. Berger, J. I. Ramos, T. Wang, K. Koynov, G. Liu, H.-J. Butt and S. Wu, RSC Adv., 2014, 4, 45059–45064 RSC.
- X. J. Liu, Y. Xie, S. J. Shi, Q. L. Feng, A. Bachhuka, X. D. Guo, Z. D. She, R. W. Tan, Q. Cai and K. Vasilev, Appl. Surf. Sci., 2019, 473, 838–847 CrossRef CAS.
- M. S. Lord, M. Foss and F. Besenbacher, Nano Today, 2010, 5, 66–78 CrossRef CAS.
- C. Rianna, L. Rossano, R. H. Kollarigowda, F. Formiggini, S. Cavalli, M. Ventre and P. A. Netti, Adv. Funct. Mater., 2016, 26, 7572–7580 CrossRef CAS.
- G. M. de Peppo, H. Agheli, C. Karlsson, K. Ekstrom, H. Brisby, M. Lenneras, S. Gustafsson, P. Sjovall, A. Johansson, E. Olsson, J. Lausmaa, P. Thomsen and S. Petronis, Int. J. Nanomed., 2014, 9, 2499–2515 CrossRef PubMed.
- H. M. Khanlou, B. C. Ang, M. M. Barzani, M. Silakhori and S. Talebian, Neural Comput. Appl., 2015, 26, 1751–1761 CrossRef.
- H. Chen, X. Huang, M. Zhang, F. Damanik, M. B. Baker, A. Leferink, H. Yuan, R. Truckenmuller, C. van Blitterswijk and L. Moroni, Acta Biomater., 2017, 59, 82–93 CrossRef CAS PubMed.
- M. F. Sola-Ruiz, C. Perez-Martinez, J. J. Martin-del-Llano, C. Carda-Batalla and C. Labaig-Rueda, Med. Oral Patol. Oral Cir. Bucal., 2015, 20, e88–e93 CrossRef PubMed.
- S. Barr, E. W. Hill and A. Bayat, J. Mech. Behav. Biomed. Mater., 2017, 75, 75–81 CrossRef CAS PubMed.
- K. Anselme and M. Bigerelle, Scanning, 2014, 36, 11–20 CrossRef CAS PubMed.
- V. Belaud, T. Petithory, A. Ponche, C. Mauclair, C. Donnet, L. Pieuchot, S. Benayoun and K. Anselme, Biointerphases, 2018, 13, 06D408 CrossRef CAS PubMed.
- O. Raimbault, S. Benayoun, K. Anselme, C. Mauclair, T. Bourgade, A. M. Kietzig, P. L. Girard-Lauriault, S. Valette and C. Donnet, Mater. Sci. Eng., C, 2016, 69, 311–320 CrossRef CAS PubMed.
- A. B. Faia-Torres, S. Guimond-Lischer, M. Rottmar, M. Charnley, T. Goren, K. Maniura-Weber, N. D. Spencer, R. L. Reis, M. Textor and N. M. Neves, Biomaterials, 2014, 35, 9023–9032 CrossRef CAS PubMed.
- P. Mazón, D. García-Bernal, L. Meseguer-Olmo, F. Cragnolini and N. Piedad, Ceram. Int., 2015, 41, 6631–6644 CrossRef.
- R. Olivares-Navarrete, S. L. Hyzy, M. E. Berg, J. M. Schneider, K. Hotchkiss, Z. Schwartz and B. D. Boyan, Ann. Biomed. Eng., 2014, 42, 2551–2561 CrossRef PubMed.
- R. Xing, S. P. Lyngstadaas, J. E. Ellingsen, S. Taxt-Lamolle and H. J. Haugen, Clin. Oral. Implants Res., 2015, 26, 649–656 CrossRef PubMed.
- S. F. Lamolle, M. Monjo, S. P. Lyngstadaas, J. E. Ellingsen and H. J. Haugen, J. Biomed. Mater. Res., Part A, 2009, 88, 581–588 CrossRef PubMed.
- F. S. L. Bobbert and A. A. Zadpoor, J. Mater. Chem. B, 2017, 5, 6175–6192 RSC.
- F. G. Torres, O. P. Troncoso, O. Gamucci, S. Corvaglia, V. Brunetti and G. Bardi, Int. J. Biol. Macromol., 2015, 75, 460–466 CrossRef CAS PubMed.
- C. Jin, G. Lee, C. Oh, H. J. Kim and H. M. Kim, J. Periodontal Implant Sci., 2017, 47, 116–131 CrossRef CAS PubMed.
- E. M. Lee, K. Smith, K. Gall, B. D. Boyan and Z. Schwartz, Biomaterials, 2016, 110, 34–44 CrossRef CAS PubMed.
- O. Andrukhov, R. Huber, B. Shi, S. Berner, X. Rausch-Fan, A. Moritz, N. D. Spencer and A. Schedle, Dent. Mater., 2016, 32, 1374–1384 CrossRef CAS PubMed.
- K. Yusa, O. Yamamoto, H. Takano, M. Fukuda and M. Iino, Sci. Rep., 2016, 6, 29462 CrossRef CAS PubMed.
- Y. Zhang, S. E. Chen, J. Shao and J. van den Beucken, ACS Appl. Mater. Interfaces, 2018, 10, 36652–36663 CrossRef CAS PubMed.
- D. O. Costa, P. D. Prowse, T. Chrones, S. M. Sims, D. W. Hamilton, A. S. Rizkalla and S. J. Dixon, Biomaterials, 2013, 34, 7215–7226 CrossRef CAS.
- L. Saldana, L. Crespo, F. Bensiamar, M. Arruebo and N. Vilaboa, J. Biomed. Mater. Res., Part A, 2014, 102, 128–140 CrossRef PubMed.
- S. Giljean, M. Bigerelle and K. Anselme, Scanning, 2014, 36, 2–10 CrossRef CAS PubMed.
- B. Zhou, X. Gao, C. Wang, Z. Ye, Y. Gao, J. Xie, X. Wu and W. Wen, ACS Appl. Mater. Interfaces, 2015, 7, 17181–17187 CrossRef CAS.
- B. S. Moon, S. Kim, H. E. Kim and T. S. Jang, Mater. Sci. Eng., C, 2017, 73, 90–98 CrossRef CAS PubMed.
- W. Wan, K. K. Bjorkman, E. S. Choi, A. L. Panepento, K. S. Anseth and L. A. Leinwand, bioRxiv, 2019, 682930 Search PubMed.
- S. Mei, L. Yang, Y. Pan, D. Wang, X. Wang, T. Tang and J. Wei, Colloids Surf., B, 2019, 174, 207–215 CrossRef CAS PubMed.
- N. Fukuda, M. Kanazawa, K. Tsuru, A. Tsuchiya, J. Sunarso, R. Toita, Y. Mori, Y. Nakashima and K. Ishikawa, Sci. Rep., 2018, 8, 16887 CrossRef PubMed.
- H. Begam, B. Kundu, A. Chanda and S. K. Nandi, Ceram. Int., 2017, 43, 3752–3760 CrossRef CAS.
- S. F. Lamolle, M. Monjo, M. Rubert, H. J. Haugen, S. P. Lyngstadaas and J. E. Ellingsen, Biomaterials, 2009, 30, 736–742 CrossRef CAS PubMed.
- H. Jeon, H. Lee and G. Kim, Tissue Eng., Part C, 2014, 20, 951–963 CrossRef CAS PubMed.
- C. Ribeiro, V. Sencadas, A. C. Areias, F. M. Gama and S. Lanceros-Mendez, J. Biomed. Mater. Res., Part A, 2015, 103, 2260–2268 CrossRef CAS PubMed.
- P. Slepicka, I. Michaljanicova, S. Rimpelova and V. Svorcik, Mater. Sci. Eng., C, 2017, 76, 818–826 CrossRef CAS PubMed.
- S. Prauzner-Bechcicki, J. Raczkowska, J. Rysz, J. Wiltowska-Zuber, J. Pabijan, M. Marzec, A. Budkowski and M. Lekka, Appl. Surf. Sci., 2018, 457, 881–890 CrossRef CAS.
- M. Strickstrock, H. Rothe, S. Grohmann, G. Hildebrand, I. M. Zylla and K. Liefeith, BioNanoMaterials, 2017, 18(1–2), 20160013 Search PubMed.
- Y. Deng, X. Liu, A. Xu, L. Wang, Z. Luo, Y. Zheng, F. Deng, J. Wei, Z. Tang and S. Wei, Int. J. Nanomed., 2015, 10, 1425–1447 CAS.
- G. C. Machado, E. Garcia-Tunon, R. V. Bell, M. Alini, E. Saiz and M. Peroglio, J. Eur. Ceram. Soc., 2018, 38, 949–961 CrossRef.
- C. Wu, M. Chen, T. Zheng and X. Yang, Biomed. Mater. Eng., 2015, 26(Suppl 1), S155–S164 Search PubMed.
- X. Li, Q. Huang, T. A. Elkhooly, Y. Liu, H. Wu, Q. Feng, L. Liu, Y. Fang, W. Zhu and T. Hu, Biomed. Mater., 2018, 13, 045013 CrossRef PubMed.
- S. Migita and K. Araki, AIMS Bioeng., 2017, 4, 162–170 Search PubMed.
- K. H. Vining and D. J. Mooney, Nat. Rev. Mol. Cell Biol., 2017, 18, 728–742 CrossRef CAS PubMed.
- R. Olivares-Navarrete, E. M. Lee, K. Smith, S. L. Hyzy, M. Doroudi, J. K. Williams, K. Gall, B. D. Boyan and Z. Schwartz, PLoS One, 2017, 12, e0170312 CrossRef PubMed.
- P. Moshayedi, G. Ng, J. C. Kwok, G. S. Yeo, C. E. Bryant, J. W. Fawcett, K. Franze and J. Guck, Biomaterials, 2014, 35, 3919–3925 CrossRef CAS PubMed.
- S. van Helvert, C. Storm and P. Friedl, Nat. Cell Biol., 2018, 20, 8–20 CrossRef CAS PubMed.
- A. D. Doyle, N. Carvajal, A. Jin, K. Matsumoto and K. M. Yamada, Nat. Commun., 2015, 6, 8720 CrossRef CAS PubMed.
- C. D. Hartman, B. C. Isenberg, S. G. Chua and J. Y. Wong, Exp. Cell Res., 2017, 359, 361–366 CrossRef CAS.
- J. Li, D. Han and Y. P. Zhao, Sci. Rep., 2014, 4, 3910 CrossRef.
- M. Nii, J. H. Lai, M. Keeney, L. H. Han, A. Behn, G. Imanbayev and F. Yang, Acta Biomater., 2013, 9, 5475–5483 CrossRef CAS PubMed.
- L. M. Wang, H. Chang, H. Zhang, K. F. Ren, H. Li, M. Hu, B. C. Li, M. C. L. Martins, M. A. Barbosa and J. Ji, J. Mater. Chem. B, 2015, 3, 7546–7553 RSC.
- L. G. Vincent, Y. S. Choi, B. Alonso-Latorre, J. C. del Alamo and A. J. Engler, Biotechnol. J., 2013, 8, 472–484 CrossRef CAS PubMed.
- P. Tseng and D. Di Carlo, Adv. Mater., 2014, 26, 1242–1247 CrossRef CAS PubMed.
- E. B. Evans, S. W. Brady, A. Tripathi and D. Hoffman-Kim, Biomater. Res., 2018, 22, 14 CrossRef PubMed.
- W. J. Hadden, J. L. Young, A. W. Holle, M. L. McFetridge, D. Y. Kim, P. Wijesinghe, H. Taylor-Weiner, J. H. Wen, A. R. Lee, K. Bieback, B. N. Vo, D. D. Sampson, B. F. Kennedy, J. P. Spatz, A. J. Engler and Y. S. Choi, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5647–5652 CrossRef CAS PubMed.
- Y. Wu, Z. Yang, J. B. Law, A. Y. He, A. A. Abbas, V. Denslin, T. Kamarul, J. H. Hui and E. H. Lee, Tissue Eng., Part A, 2017, 23, 43–54 CrossRef CAS.
- J. H. Seo, K. Sakai and N. Yui, Acta Biomater., 2013, 9, 5493–5501 CrossRef CAS.
- M. Bao, J. Xie, N. Katoele, X. Hu, B. Wang, A. Piruska and W. T. Huck, ACS Appl. Mater. Interfaces, 2018, 11, 1754–1759 CrossRef PubMed.
- Y. Zhang, Y. Xing, J. Li, Z. Zhang, H. Luan, Z. Chu, H. Gong and Y. Fan, BioMed Res. Int., 2018, 2018, 4025083 Search PubMed.
- J. You, S. A. Park, D. S. Shin, D. Patel, V. K. Raghunathan, M. Kim, C. J. Murphy, G. Tae and A. Revzin, Tissue Eng., Part A, 2013, 19, 2655–2663 CrossRef CAS PubMed.
- Z. Li, Y. Gong, S. Sun, Y. Du, D. Lu, X. Liu and M. Long, Biomaterials, 2013, 34, 7616–7625 CrossRef CAS PubMed.
- A. S. Mao, J. W. Shin and D. J. Mooney, Biomaterials, 2016, 98, 184–191 CrossRef CAS PubMed.
- J. Xu, M. Sun, Y. Tan, H. Wang, H. Wang, P. Li, Z. Xu, Y. Xia, L. Li and Y. Li, Differentiation, 2017, 96, 30–39 CrossRef CAS PubMed.
- D. A. Foyt, D. K. Taheem, S. A. Ferreira, M. D. A. Norman, J. Petzold, G. Jell, A. E. Grigoriadis and E. Gentleman, Acta Biomater., 2019, 89, 73–83 CrossRef CAS PubMed.
- S. H. Medina, B. Bush, M. Cam, E. Sevcik, F. W. DelRio, K. Nandy and J. P. Schneider, Biomaterials, 2019, 202, 1–11 CrossRef CAS.
- I. Hopp, A. Michelmore, L. E. Smith, D. E. Robinson, A. Bachhuka, A. Mierczynska and K. Vasilev, Biomaterials, 2013, 34, 5070–5077 CrossRef CAS PubMed.
- S. Sur, C. J. Newcomb, M. J. Webber and S. I. Stupp, Biomaterials, 2013, 34, 4749–4757 CrossRef CAS PubMed.
- M. Guvendiren, M. Perepelyuk, R. G. Wells and J. A. Burdick, J. Mech. Behav. Biomed. Mater., 2014, 38, 198–208 CrossRef CAS PubMed.
- L. S. Wang, C. Du, W. S. Toh, A. C. Wan, S. J. Gao and M. Kurisawa, Biomaterials, 2014, 35, 2207–2217 CrossRef CAS PubMed.
- K. Ye, X. Wang, L. Cao, S. Li, Z. Li, L. Yu and J. Ding, Nano Lett., 2015, 15, 4720–4729 CrossRef CAS PubMed.
- J. M. Banks, L. C. Mozdzen, B. A. Harley and R. C. Bailey, Biomaterials, 2014, 35, 8951–8959 CrossRef CAS PubMed.
- J. C. Courtenay, C. Deneke, E. M. Lanzoni, C. A. Costa, Y. Bae, J. L. Scott and R. I. Sharma, Cellulose, 2018, 25, 925–940 CrossRef CAS PubMed.
- X. Lu, Z. Ding, F. Xu, Q. Lu and D. L. Kaplan, ACS Appl. Bio Mater., 2019, 2(7), 3108–3119 CrossRef CAS.
- K. E. Coombs, A. T. Leonard, M. N. Rush, D. A. Santistevan and E. L. Hedberg-Dirk, J. Biomed. Mater. Res., Part A, 2017, 105, 51–61 CrossRef CAS.
- M. Lin, H. Wang, C. Ruan, J. Xing, J. Wang, Y. Li, Y. Wang and Y. Luo, Biomacromolecules, 2015, 16, 973–984 CrossRef CAS.
- C. Pinese, S. Jebors, P. E. Stoebner, V. Humblot, P. Verdie, L. Causse, X. Garric, H. Taillades, J. Martinez, A. Mehdi and G. Subra, Mater. Today Chem., 2017, 4, 73–83 CrossRef.
- R. M. d. Nascimento, S. M. Ramos, I. H. Bechtold and A. C. Hernandes, ACS Biomater. Sci. Eng., 2018, 4, 2784–2793 CrossRef CAS.
- L. N. Chen, C. Yan and Z. J. Zheng, Mater. Today, 2018, 21, 38–59 CrossRef CAS.
- S. Sakata, Y. Inoue and K. Ishihara, Langmuir, 2014, 30, 2745–2751 CrossRef CAS PubMed.
- S. Kumar, S. Raj, E. Kolanthai, A. K. Sood, S. Sampath and K. Chatterjee, ACS Appl. Mater. Interfaces, 2015, 7, 3237–3252 CrossRef CAS PubMed.
- S. Zhao, J. Zhang, M. Zhu, Y. Zhang, Z. Liu, Y. Ma, Y. Zhu and C. Zhang, J. Mater. Chem. B, 2015, 3, 1612–1623 RSC.
- J. J. Li, N. Kawazoe and G. Chen, Biomaterials, 2015, 54, 226–236 CrossRef CAS PubMed.
- H. Amani, H. Arzaghi, M. Bayandori, A. S. Dezfuli, H. Pazoki-Toroudi, A. Shafiee and L. Moradi, Adv. Mater. Interfaces, 2019, 6, 1900572 CrossRef.
- H. C. Bygd, K. D. Forsmark and K. M. Bratlie, Biomaterials, 2015, 56, 187–197 CrossRef CAS PubMed.
- S. J. Jenkins, D. Ruckerl, P. C. Cook, L. H. Jones, F. D. Finkelman, N. van Rooijen, A. S. MacDonald and J. E. Allen, Science, 2011, 332, 1284–1288 CrossRef CAS.
- Y. Shen, M. Gao, Y. Ma, H. Yu, F. Z. Cui, H. Gregersen, Q. Yu, G. Wang and X. Liu, Colloids Surf., B, 2015, 126, 188–197 CrossRef CAS PubMed.
- A. Rashad, K. Mustafa, E. B. Heggset and K. Syverud, Biomacromolecules, 2017, 18, 1238–1248 CrossRef CAS PubMed.
- Y. Zhang, H. Pan, P. Zhang, N. Gao, Y. Lin, Z. Luo, P. Li, C. Wang, L. Liu, D. Pang, L. Cai and Y. Ma, Nanoscale, 2013, 5, 5919–5929 RSC.
- L. Wang, M. He, T. Gong, X. Zhang, L. Zhang, T. Liu, W. Ye, C. Pan and C. Zhao, Biomater. Sci., 2017, 5, 2416–2426 RSC.
- X. Liu, S. Shi, Q. Feng, A. Bachhuka, W. He, Q. Huang, R. Zhang, X. Yang and K. Vasilev, ACS Appl. Mater. Interfaces, 2015, 7, 18473–18482 CrossRef CAS PubMed.
- S. Shahabi, L. Treccani, R. Dringen and K. Rezwan, ACS Appl. Mater. Interfaces, 2015, 7, 13821–13833 CrossRef CAS PubMed.
- A. Hasan, S. K. Pattanayek and L. M. Pandey, ACS Biomater. Sci. Eng., 2018, 4, 3224–3233 CrossRef CAS.
- F. Meder, C. Brandes, L. Treccani and K. Rezwan, Acta Biomater., 2013, 9, 5780–5787 CrossRef CAS PubMed.
- S. N. Christo, A. Bachhuka, K. R. Diener, A. Mierczynska, J. D. Hayball and K. Vasilev, Adv. Healthcare Mater., 2016, 5, 956–965 CrossRef CAS PubMed.
- Z. Chen, A. Bachhuka, S. Han, F. Wei, S. Lu, R. M. Visalakshan, K. Vasilev and Y. Xiao, ACS Nano, 2017, 11, 4494–4506 CrossRef CAS PubMed.
- Y. Yang, P. Qi, Y. Ding, M. F. Maitz, Z. Yang, Q. Tu, K. Xiong, Y. Leng and N. Huang, J. Mater. Chem. B, 2015, 3, 72–81 RSC.
- D. Zeng, X. Zhang, X. Wang, Q. Huang, J. Wen, X. Miao, L. Peng, Y. Li and X. Jiang, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 1425–1435 CrossRef CAS PubMed.
- Q. Li, B. Zhang, N. Kasoju, J. Ma, A. Yang, Z. Cui, H. Wang and H. Ye, Int. J. Mol. Sci., 2018, 19, 2344 CrossRef PubMed.
- Y. Zhang, J. P. Luan, S. X. Jiang, X. Zhou and M. M. Li, Composites, Part B, 2019, 172, 397–405 CrossRef CAS.
- G. Aziz, N. De Geyter, H. Declercq, R. Cornelissen and R. Morent, Surf. Coat. Technol., 2015, 271, 39–47 CrossRef CAS.
- C. Insomphun, J.-A. Chuah, S. Kobayashi, T. Fujiki and K. Numata, ACS Biomater. Sci. Eng., 2016, 3, 3064–3075 CrossRef.
- J. P. Yapor, A. Lutzke, A. Pegalajar-Jurado, B. H. Neufeld, V. B. Damodaran and M. M. Reynolds, J. Mater. Chem. B, 2015, 3, 9233–9241 RSC.
- Y. Y. Zheng, L. H. Liu, Y. Ma, L. Xiao and Y. Liu, Ind. Eng. Chem. Res., 2018, 57, 10403–10410 CrossRef CAS.
- M. G. L. Olthof, M. A. Tryfonidou, X. Liu, B. Pouran, B. P. Meij, W. J. A. Dhert, M. J. Yaszemski, L. Lu, J. Alblas and D. H. R. Kempen, Tissue Eng., Part A, 2018, 24, 819–829 CrossRef CAS PubMed.
- Y. Bai, H. Xing, P. Wu, X. Feng, K. Hwang, J. M. Lee, X. Y. Phang, Y. Lu and S. C. Zimmerman, ACS Nano, 2015, 9, 10227–10236 CrossRef CAS PubMed.
- Z. Shen, J. Wu, Y. Yu, S. Liu, W. Jiang, H. Nurmamat and B. Wu, Sci. Rep., 2019, 9, 7557 CrossRef PubMed.
- A. K. Fuchs, T. Syrovets, K. A. Haas, C. Loos, A. Musyanovych, V. Mailander, K. Landfester and T. Simmet, Biomaterials, 2016, 85, 78–87 CrossRef CAS PubMed.
- J. Zhang, L. Li, Y. Peng, Y. Chen, X. Lv, S. Li, X. Qin, H. Yang, C. Wu and Y. Liu, Biochim. Biophys. Acta, Mol. Cell Res., 2018, 1865, 172–185 CrossRef CAS PubMed.
- C. Morke, H. Rebl, B. Finke, M. Dubs, P. Nestler, A. Airoudj, V. Roucoules, M. Schnabelrauch, A. Kortge, K. Anselme, C. A. Helm and J. B. Nebe, ACS Appl. Mater. Interfaces, 2017, 9, 10461–10471 CrossRef.
- T. Chen, L. Li, G. Xu, X. Wang, J. Wang, Y. Chen, W. Jiang, Z. Yang and G. Lin, Front. Pharmacol., 2018, 9, 763 CrossRef PubMed.
- Y. Y. Zheng, L. H. Liu, C. D. Xiong and L. F. Zhang, Mater. Lett., 2018, 213, 84–87 CrossRef CAS.
- Y. Zheng, C. Xiong, S. Zhang, X. Li and L. Zhang, Mater. Sci. Eng., C, 2015, 55, 512–523 CrossRef CAS PubMed.
- B. Cao, Y. Peng, X. Liu and J. Ding, ACS Appl. Mater. Interfaces, 2017, 9, 23574–23585 CrossRef CAS PubMed.
- T. T. Yu, F. Z. Cui, Q. Y. Meng, J. Wang, D. C. Wu, J. Zhang, X. X. Kou, R. L. Yang, Y. Liu, Y. S. Zhang, F. Yang and Y. H. Zhou, ACS Biomater. Sci. Eng., 2017, 3, 1119–1128 CrossRef CAS.
- R. Ion, S. Vizireanu, C. E. Stancu, C. Luculescu, A. Cimpean and G. Dinescu, Mater. Sci. Eng., C, 2015, 48, 118–125 CrossRef CAS PubMed.
- X. Lin, K. Fukazawa and K. Ishihara, ACS Appl. Mater. Interfaces, 2015, 7, 17489–17498 CrossRef CAS PubMed.
- M. Vetten and M. Gulumian, Toxicol. Appl. Pharmacol., 2019, 363, 131–141 CrossRef CAS PubMed.
- T. Lu, Y. Qiao and X. Liu, Interface Focus, 2012, 2, 325–336 CrossRef PubMed.
- E. M. McCarthy, H. Floyd, O. Addison, Z. J. Zhang, P. G. Oppenheimer and L. M. Grover, ACS Omega, 2018, 3, 10129–10138 CrossRef CAS PubMed.
- G. Socrates, Infrared and Raman characteristic group frequencies: tables and charts, John Wiley & Sons, 2004 Search PubMed.
- S. Staehlke, H. Rebl, B. Finke, P. Mueller, M. Gruening and J. B. Nebe, Cell Biosci., 2018, 8, 22 CrossRef PubMed.
- Y. Zhao, K. W. K. Yeung and P. K. Chu, Appl. Surf. Sci., 2014, 310, 11–18 CrossRef CAS.
- W. H. Jin and P. K. Chu, Surf. Coat. Technol., 2018, 336, 2–8 CrossRef CAS.
- C. H. Yang, Y. C. Li, W. F. Tsai, C. F. Ai and H. H. Huang, Clin. Oral. Implants Res., 2015, 26, 166–175 CrossRef PubMed.
- W. H. Jin, G. S. Wu, H. Q. Feng, W. H. Wang, X. M. Zhang and P. K. Chu, Corros. Sci., 2015, 94, 142–155 CrossRef CAS.
- H. Qin, H. Cao, Y. Zhao, G. Jin, M. Cheng, J. Wang, Y. Jiang, Z. An, X. Zhang and X. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 10785–10794 CrossRef CAS.
- R. I. Iziumov, A. Y. Beliaev, I. V. Kondyurina, I. N. Shardakov, A. V. Kondyurin, M. M. Bilek and D. R. McKenzie, Mater. Sci. Eng., 2016, 123(1), 012003 Search PubMed.
- C. R. Mariappan and N. Ranga, Mater. Sci. Forum, 2016, 860, 25–28 Search PubMed.
- G. G. Fuentes, Micromanufacturing Engineering and Technology, 2010, pp. 459–486 Search PubMed.
- J. Jagielski, A. Piatkowska, P. Aubert, L. Thome, A. Turos and A. A. Kader, Surf. Coat. Technol., 2006, 200, 6355–6361 CrossRef CAS.
- C. Park, Y. J. Seong, I. G. Kang, E. H. Song, H. Lee, J. Kim, H. D. Jung, H. E. Kim and T. S. Jang, ACS Appl. Mater. Interfaces, 2019, 11, 10492–10504 CrossRef CAS.
- B. Li, H. Cao, Y. Zhao, M. Cheng, H. Qin, T. Cheng, Y. Hu, X. Zhang and X. Liu, Sci. Rep., 2017, 7, 42707 CrossRef CAS.
- Y. Yu, G. Jin, Y. Xue, D. Wang, X. Liu and J. Sun, Acta Biomater., 2017, 49, 590–603 CrossRef CAS PubMed.
- T. Lu, S. Qian, F. Meng, C. Ning and X. Liu, Colloids Surf., B, 2016, 142, 192–198 CrossRef CAS PubMed.
- K. Gan, H. Liu, L. Jiang, X. Liu, X. Song, D. Niu, T. Chen and C. Liu, Dent. Mater., 2016, 32, e263–e274 CrossRef CAS PubMed.
- E. A. Wakelin, G. C. Yeo, D. R. McKenzie, M. M. M. Bilek and A. S. Weiss, APL Bioeng., 2018, 2, 026109 CrossRef PubMed.
- U. Walschus, A. Hoene, M. Patrzyk, S. Lucke, B. Finke, M. Polak, G. Lukowski, R. Bader, C. Zietz, A. Podbielski, J. B. Nebe and M. Schlosser, J. Funct. Biomater., 2017, 8, 30 CrossRef PubMed.
- D. K. Shiau, C. H. Yang, Y. S. Sun, M. F. Wu, H. B. Pan and H. H. Huang, Surf. Coat. Technol., 2019, 365, 173–178 CrossRef CAS.
- Y. S. Sun, L. Zhang, H. Q. Zhu, W. E. Yang, M. Y. Lan, S. W. Lee and H. H. Huang, J. Vac. Sci. Technol., A, 2016, 34, 041402 CrossRef.
- H. H. Huang, D. K. Shiau, C. S. Chen, J. H. Chang, S. Wang, H. B. Pan and M. F. Wu, Surf. Coat. Technol., 2019, 365, 179–188 CrossRef CAS.
- S. Won, Y. H. Huh, L. R. Cho, H. S. Lee, E. S. Byon and C. J. Park, Tissue Eng. Regener. Med., 2017, 14, 123–131 CrossRef CAS PubMed.
- C. Xia, D. S. Cai, J. Tan, K. Q. Li, Y. Q. Qiao and X. Y. Liu, ACS Biomater. Sci. Eng., 2018, 4, 3185–3193 CrossRef CAS.
- S. Qiao, H. Cao, X. Zhao, H. Lo, L. Zhuang, Y. Gu, J. Shi, X. Liu and H. Lai, Int. J. Nanomed., 2015, 10, 653–664 Search PubMed.
- S. Viswanathan, L. Mohan, M. Chakraborty, C. Mandal, P. Bera, S. T. Aruna and C. Anandan, Mater. Manuf. Processes, 2018, 33, 1121–1127 CrossRef CAS.
- X. Cheng, J. Fei, A. Kondyurin, K. Fu, L. Ye, M. M. M. Bilek and S. Bao, Mater. Sci. Eng., C, 2019, 99, 863–874 CrossRef CAS PubMed.
- V. Chudinov, I. Kondyurina, V. Terpugov and A. Kondyurin, Nucl. Instrum. Methods Phys. Res., Sect. B, 2019, 440, 163–174 CrossRef CAS.
- E. Kosobrodova, W. J. Gan, A. Kondyurin, P. Thorn and M. M. M. Bilek, ACS Appl. Mater. Interfaces, 2018, 10, 227–237 CrossRef CAS PubMed.
- T. S. Jang, J. H. Lee, S. Kim, C. Park, J. Song, H. J. Jae, H. E. Kim, J. W. Chung and H. D. Jung, Biomaterials, 2019, 223, 119461 CrossRef CAS PubMed.
- L. M. de Andrade, C. Paternoster, V. Montaño-Machado, G. Barucca, M. Sikora-Jasinska, R. Tolouei, S. Turgeon and D. Mantovani, MRS Commun., 2018, 8, 1404–1412 CrossRef.
- K. Schrock, J. Lutz, S. Mandl, M. C. Hacker, M. Kamprad and M. Schulz-Siegmund, J. Orthop. Res., 2015, 33, 325–333 CrossRef PubMed.
- M. A. Shafique, G. Murtaza, S. Saadat, M. K. H. Uddin and R. Ahmad, Radiat. Eff. Defects Solids, 2017, 172, 590–599 CrossRef CAS.
- C. Park, S. W. Lee, J. Kim, E. H. Song, H. D. Jung, J. U. Park, H. E. Kim, S. Kim and T. S. Jang, Biomater. Sci., 2019, 7, 2907–2919 RSC.
- L. Tong, W. Zhou, Y. Zhao, X. Yu, H. Wang and P. K. Chu, Colloids Surf., B, 2016, 148, 139–146 CrossRef CAS PubMed.
- M. A. Wronska, I. B. O'Connor, M. A. Tilbury, A. Srivastava and J. G. Wall, Adv. Mater., 2016, 28, 5485–5508 CrossRef CAS PubMed.
- M. W. Tibbitt, C. B. Rodell, J. A. Burdick and K. S. Anseth, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 14444–14451 CrossRef CAS PubMed.
- A. V. Bryksin, A. C. Brown, M. M. Baksh, M. G. Finn and T. H. Barker, Acta Biomater., 2014, 10, 1761–1769 CrossRef CAS PubMed.
- A. Srivastava, L. B. O'Connor, A. Pandit and J. G. Wall, Prog. Polym. Sci., 2014, 39, 308–329 CrossRef CAS.
- T. Garg, G. Rath and A. K. Goyal, J. Drug Targeting, 2015, 23, 202–221 CrossRef CAS PubMed.
- Q. Wei, T. Becherer, S. Angioletti-Uberti, J. Dzubiella, C. Wischke, A. T. Neffe, A. Lendlein, M. Ballauff and R. Haag, Angew. Chem., Int. Ed., 2014, 53, 8004–8031 CrossRef CAS PubMed.
- M. Aizawa, T. Matsuura and Z. Zhuang, Biol. Pharm. Bull., 2013, 36, 1654–1661 CrossRef CAS PubMed.
- T. Sarkar, Y. Gao and A. Mulchandani, Appl. Biochem. Biotechnol., 2013, 170, 1011–1025 CrossRef CAS PubMed.
- C. A. Custodio, C. M. Alves, R. L. Reis and J. F. Mano, J. Tissue Eng. Regener. Med., 2010, 4, 316–323 CrossRef CAS PubMed.
- S. L. Hirsh, D. R. McKenzie, N. J. Nosworthy, J. A. Denman, O. U. Sezerman and M. M. Bilek, Colloids Surf., B, 2013, 103, 395–404 CrossRef CAS PubMed.
- P. Zucca and E. Sanjust, Molecules, 2014, 19, 14139–14194 CrossRef PubMed.
- B. R. Coad, M. Jasieniak, S. S. Griesser and H. J. Griesser, Surf. Coat. Technol., 2013, 233, 169–177 CrossRef CAS.
- B. R. Coad, K. Vasilev, K. R. Diener, J. D. Hayball, R. D. Short and H. J. Griesser, Langmuir, 2012, 28, 2710–2717 CrossRef CAS PubMed.
- E. A. Wakelin, A. Fathi, M. Kracica, G. C. Yeo, S. G. Wise, A. S. Weiss, D. G. McCulloch, F. Dehghani, D. R. McKenzie and M. M. Bilek, ACS Appl. Mater. Interfaces, 2015, 7, 23029–23040 CrossRef CAS PubMed.
- H. Main, J. Radenkovic, E. Kosobrodova, D. McKenzie, M. Bilek and U. Lendahl, Cell. Mol. Life Sci., 2014, 71, 3841–3857 CrossRef CAS PubMed.
- R. P. Tan, A. H. Chan, S. Wei, M. Santos, B. S. Lee, E. C. Filipe, B. Akhavan, M. M. Bilek, M. K. Ng and Y. Xiao, JACC: Basic to Translational Science, 2019, 4, 56–71 Search PubMed.
- H. Lee, N. F. Scherer and P. B. Messersmith, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12999–13003 CrossRef CAS PubMed.
- C. D. Spicer and B. G. Davis, Nat. Commun., 2014, 5, 4740 CrossRef CAS PubMed.
- M. P. Deonarain, G. Yahioglu, I. Stamati and J. Marklew, Expert Opin. Drug Discovery, 2015, 10, 463–481 CrossRef CAS PubMed.
- A. Srivastava, C. Cunningham, A. Pandit and J. G. Wall, Macromol. Biosci., 2015, 15, 682–690 CrossRef CAS PubMed.
- S. Cao, M. N. Barcellona, F. Pfeiffer and M. T. Bernards, J. Appl. Polym. Sci., 2016, 133(40), 43985 CrossRef.
- X. X. Yang, X. Y. Wang, F. Yu, L. L. Ma, X. H. Pan, G. J. Luo, S. Lin, X. M. Mo, C. L. He and H. S. Wang, RSC Adv., 2016, 6, 99720–99728 RSC.
- K. A. Totaro, X. Liao, K. Bhattacharya, J. I. Finneman, J. B. Sperry, M. A. Massa, J. Thorn, S. V. Ho and B. L. Pentelute, Bioconjugate Chem., 2016, 27, 994–1004 CrossRef CAS PubMed.
- E. J. Park, T. N. Gevrek, R. Sanyal and A. Sanyal, Bioconjugate Chem., 2014, 25, 2004–2011 CrossRef CAS PubMed.
- C. A. Custodio and J. F. Mano, ChemNanoMat, 2016, 2, 376–384 CrossRef CAS.
- P. Nacharaju, F. N. Boctor, B. N. Manjula and S. A. Acharya, Transfusion, 2005, 45, 374–383 CrossRef CAS.
- M. E. Smith, F. F. Schumacher, C. P. Ryan, L. M. Tedaldi, D. Papaioannou, G. Waksman, S. Caddick and J. R. Baker, J. Am. Chem. Soc., 2010, 132, 1960–1965 CrossRef CAS PubMed.
- Z. Y. Zhang, N. Vanparijs, S. Vandewalle, F. E. Du Prez, L. Nuhn and B. G. De Geest, Polym. Chem., 2016, 7, 7242–7248 RSC.
- N. Vanparijs, R. De Coen, D. Laplace, B. Louage, S. Maji, L. Lybaert, R. Hoogenboom and B. G. De Geest, Chem. Commun., 2015, 51, 13972–13975 RSC.
- H. Kakwere, C. K. Chun, K. A. Jolliffe, R. J. Payne and S. Perrier, Chem. Commun., 2010, 46, 2188–2190 RSC.
- Y. Zou, L. Zhang, L. Yang, F. Zhu, M. Ding, F. Lin, Z. Wang and Y. Li, J. Controlled Release, 2018, 273, 160–179 CrossRef CAS PubMed.
- J. Gopinathan and I. Noh, Tissue Eng. Regener. Med., 2018, 15, 531–546 CrossRef CAS PubMed.
- K. Nagahama, Y. Kimura and A. Takemoto, Nat. Commun., 2018, 9, 2195 CrossRef PubMed.
- B. K. Lee, J. H. Noh, J. H. Park, S. H. Park, J. H. Kim, S. H. Oh and M. S. Kim, Tissue Eng. Regener. Med., 2018, 15, 393–402 CrossRef CAS PubMed.
- G. Yi, J. Son, J. Yoo, C. Park and H. Koo, Biomater. Res., 2018, 22, 13 CrossRef PubMed.
- S. L. Liu, M. J. Dong, Z. H. Zhang and G. D. Fu, Polym. Adv. Technol., 2017, 28, 1065–1070 CrossRef CAS.
- S. Fu, H. Dong, X. Deng, R. Zhuo and Z. Zhong, Carbohydr. Polym., 2017, 169, 332–340 CrossRef CAS PubMed.
- S. Li, L. Wang, X. Yu, C. Wang and Z. Wang, Mater. Sci. Eng., C, 2018, 82, 299–309 CrossRef CAS PubMed.
- Y. L. Li, Y. Tan, K. Xu, C. G. Lu and P. X. Wang, Polym. Degrad. Stab., 2017, 137, 75–82 CrossRef CAS.
- J. Collins, Z. Y. Xiao, M. Mullner and L. A. Connal, Polym. Chem., 2016, 7, 3812–3826 RSC.
- L. J. Macdougall, K. L. Wiley, A. M. Kloxin and A. P. Dove, Biomaterials, 2018, 178, 435–447 CrossRef CAS PubMed.
- J. Gopinathan and I. Noh, Biomater. Res., 2018, 22, 11 CrossRef PubMed.
- C. S. Jung, B. K. Kim, J. Lee, B. H. Min and S. H. Park, Tissue Eng. Regener. Med., 2018, 15, 155–162 CrossRef CAS PubMed.
- W. Tang and M. L. Becker, Chem. Soc. Rev., 2014, 43, 7013–7039 RSC.
- M. Lotfi, M. Nejib and M. Naceur, Adv. Biomater. Sci. Biomed. Appl., 2013, 8, 208–240 Search PubMed.
- M. M. Gentleman and E. Gentleman, Int. Mater. Rev., 2014, 59, 417–429 CrossRef CAS.
- T. Razafiarison, C. N. Holenstein, T. Stauber, M. Jovic, E. Vertudes, M. Loparic, M. Kawecki, L. Bernard, U. Silvan and J. G. Snedeker, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4631–4636 CrossRef CAS PubMed.
- M. M. Gentleman and J. A. Ruud, Langmuir, 2010, 26, 1408–1411 CrossRef CAS PubMed.
- R. J. Good, J. Adhes. Sci. Technol., 1992, 6, 1269–1302 CrossRef CAS.
- C. Della Volpe and S. Siboni, J. Colloid Interface Sci., 1997, 195, 121–136 CrossRef CAS PubMed.
- C. Della Volpe and S. Siboni, J. Adhes. Sci. Technol., 2000, 14, 235–272 CrossRef CAS.
- C. J. van Oss, W. Wu and R. F. Giese, Lifshitz-van der Waals, Lewis acid–base and electrostatic interactions in adhesion in aqueous media, 1998 Search PubMed.
- C. J. Vanoss, R. J. Good and M. K. Chaudhury, Langmuir, 1988, 4, 884–891 CrossRef CAS.
- A. B. D. Cassie and S. Baxter, Trans. Faraday Soc., 1944, 40, 0546–0550 RSC.
- R. N. Wenzel, Ind. Eng. Chem., 1936, 28, 988–994 CrossRef CAS.
- T. Young, Philos. Trans. R. Soc. London, 1805, 95, 65–87 CrossRef.
- B. Wang, P. Liu, Z. Liu, H. Pan, X. Xu and R. Tang, Biotechnol. Bioeng., 2014, 111, 386–395 CrossRef CAS.
- M. Nakamura, N. Hori, H. Ando, S. Namba, T. Toyama, N. Nishimiya and K. Yamashita, Mater. Sci. Eng., C, 2016, 62, 283–292 CrossRef CAS PubMed.
- O. Savvova, G. Shadrina, O. Babich and O. Fesenko, Chem. Chem. Technol., 2015, 9(3), 349–354 CrossRef CAS.
- R. E. Baier, A. E. Meyer, J. R. Natiella, R. R. Natiella and J. M. Carter, J. Biomed. Mater. Res., 1984, 18, 337–355 CrossRef CAS PubMed.
- T. Groth and G. Altankov, Biomaterials, 1996, 17, 1227–1234 CrossRef CAS PubMed.
- J. M. Schakenraad, H. J. Busscher, C. R. Wildevuur and J. Arends, Cell Biophys., 1988, 13, 75–91 CrossRef CAS PubMed.
- P. Parhi, A. Golas and E. A. Vogler, J. Adhes. Sci. Technol., 2010, 24, 853–888 CrossRef CAS.
- X. Liu, J. Y. Lim, H. J. Donahue, R. Dhurjati, A. M. Mastro and E. A. Vogler, Biomaterials, 2007, 28, 4535–4550 CrossRef CAS PubMed.
- J. Y. Lim, X. Liu, E. A. Vogler and H. J. Donahue, J. Biomed. Mater. Res., Part A, 2004, 68, 504–512 CrossRef PubMed.
- C. Xie, Biosurf. Biotribol., 2019, 5, 83–92 CrossRef.
- R. Sa, Y. Yan, Z. Wei, L. Zhang, W. Wang and M. Tian, ACS Appl. Mater. Interfaces, 2014, 6, 21730–21738 CrossRef CAS PubMed.
- J. H. Liang, R. Song, Q. L. Huang, Y. Yang, L. X. Lin, Y. M. Zhang, P. L. Jiang, H. P. Duan, X. Dong and C. J. Lin, Electrochim. Acta, 2015, 174, 1149–1159 CrossRef CAS.
- M. Nasrollahzadeh, S. M. Sajadi and M. Iqbal, Nanomaterials and Plant Potential, Springer, 2019, pp. 31–70 Search PubMed.
- L. Lunelli, C. Potrich, L. Pasquardini and C. Pederzolli, Nanostructured Functionalized Surfaces, Encyclopedia of Nanotechnology, Springer, Dordrecht, 2012, pp. 1760–1766 Search PubMed.
- R. Rasouli, A. Barhoum and H. Uludag, Biomater. Sci., 2018, 6, 1312–1338 RSC.
- G. S. Wang, Y. Wan, B. Ren, T. Wang and Z. Q. Liu, Adv. Eng. Mater., 2017, 19, 1700299 CrossRef.
- C. Frey, A. Sales, K. Athanasopulu, J. Spatz and R. Kemkemer, Hydrogels with precisely nano-functionalized micro-topography for cell guidance, Conference: 48th DGBMT Annual Conference, 2015, vol. 59. pp. S6–S9 Search PubMed.
- S. Christo, A. Bachhuka, K. R. Diener, K. Vasilev and J. D. Hayball, Sci. Rep., 2016, 6, 26207 CrossRef CAS PubMed.
- M. B. Hovgaard, K. Rechendorff, J. Chevallier, M. Foss and F. Besenbacher, J. Phys. Chem. B, 2008, 112, 8241–8249 CrossRef CAS PubMed.
- J. Huang, S. V. Grater, F. Corbellini, S. Rinck, E. Bock, R. Kemkemer, H. Kessler, J. Ding and J. P. Spatz, Nano Lett., 2009, 9, 1111–1116 CrossRef CAS PubMed.
- V. Marzaioli, C. J. Groß, I. Weichenmeier, C. B. Schmidt-Weber, J. Gutermuth, O. Groß and F. Alessandrini, Nanomaterials, 2017, 7, 355 CrossRef PubMed.
- F. Karimi, T. G. McKenzie, A. J. O'Connor, G. G. Qiao and D. E. Heath, J. Mater. Chem. B, 2017, 5, 5942–5953 RSC.
- V. M. Aguilar and B. D. Cosgrove, Skeletal Muscle Development, Springer, 2017, pp. 75–92 Search PubMed.
- L. Filipponi, P. Livingston, O. Kaspar, V. Tokarova and D. V. Nicolau, Biomed. Microdevices, 2016, 18, 9 CrossRef PubMed.
- S. Arango-Santander, A. Pelaez-Vargas, S. C. Freitas and C. Garcia, J. Nanotechnol., 2018, 2018, 8624735, DOI:10.1155/2018/8624735.
- D. Hamelinck, H. Zhou, L. Li, C. Verweij, D. Dillon, Z. Feng, J. Costa and B. B. Haab, Mol. Cell. Proteomics, 2005, 4, 773–784 CrossRef CAS PubMed.
- M. Woodson and J. Liu, Phys. Chem. Chem. Phys., 2007, 9, 207–225 RSC.
- A. M. M. Brito, E. Belleti, L. R. Menezes, A. J. C. Lanfredi and I. L. Nantes-Cardoso, An. Acad. Bras. Cienc., 2019, 91(4), e20181236 CrossRef.
- G. Oteri, M. Pisanom and M. Cicciu, J. Appl. Spectrosc., 2016, 83, 316–321 CrossRef CAS.
- S. Sen-Britain, D. M. Britain, W. L. Hicks, Jr. and J. A. Gardella, Jr., Biointerphases, 2019, 14, 051003 CrossRef PubMed.
- K. Gajos, A. Budkowski, P. Petrou, V. Pagkali, K. Awsiuk, J. Rysz, A. Bernasik, K. Misiakos, I. Raptis and S. Kakabakos, Appl. Surf. Sci., 2018, 444, 187–196 CrossRef CAS.
- L. Yahia and L. Mireles, Characterization of Polymeric Biomaterials, Elsevier, 2017, pp. 83–97 Search PubMed.
- J. L. Calvo-Guirado, A. B. Montilla, P. N. De Aza, M. Fernandez-Dominguez, S. A. Gehrke, P. Cegarra-Del Pino, L. Mahesh, A. A. Pelegrine, J. M. Aragoneses and J. E. M. S. de Val, Materials, 2019, 12, 380 CrossRef CAS PubMed.
- D. G. Castner, Surf. Interface Anal., 2018, 50, 981–990 CrossRef CAS.
- P. Kingshott, G. Andersson, S. L. McArthur and H. J. Griesser, Curr. Opin. Chem. Biol., 2011, 15, 667–676 CrossRef CAS PubMed.
- R. N. Foster, E. T. Harrison and D. G. Castner, Langmuir, 2016, 32, 3207–3216 CrossRef CAS PubMed.
- S. Cometa, M. A. Bonifacio, A. M. Ferreira, P. Gentile and E. De Giglio, Materials, 2020, 13, 705 CrossRef CAS PubMed.
- M. Stevanović, M. Đošić, A. Janković, V. Kojic, M. Vukasinovic-Sekulic, J. Stojanović, J. Odović, M. Crevar Sakač, K. Y. Rhee and V. Mišković-Stanković, ACS Biomater. Sci. Eng., 2018, 4, 3994–4007 CrossRef.
- N. Metoki, D. Mandler and N. Eliaz, Cryst. Growth Des., 2016, 16, 2756–2764 CrossRef CAS.
- E. T. Harrison, T. Weidner, D. G. Castner and G. Interlandi, Biointerphases, 2017, 12, 02D401 CrossRef PubMed.
- S. Muramoto, D. J. Graham, M. S. Wagner, T. G. Lee, D. W. Moon and D. G. Castner, J. Phys. Chem. C, 2011, 115, 24247–24255 CrossRef CAS PubMed.
- M. S. Wagner, T. A. Horbett and D. G. Castner, Biomaterials, 2003, 24, 1897–1908 CrossRef CAS PubMed.
- N. G. Welch, R. M. T. Madiona, J. A. Scoble, B. W. Muir and P. J. Pigram, Langmuir, 2016, 32, 10824–10834 CrossRef CAS PubMed.
- J. Yu, Y. Zhou, M. Engelhard, Y. Zhang, J. Son, S. Liu, Z. Zhu and X. Y. Yu, Sci. Rep., 2020, 10, 3695 CrossRef CAS PubMed.
- T. Weidner and D. G. Castner, Phys. Chem. Chem. Phys., 2013, 15, 12516–12524 RSC.
- M. Deighan and J. Pfaendtner, Langmuir, 2013, 29, 7999–8009 CrossRef CAS PubMed.
- I. Donderwinkel, J. C. M. van Hest and N. R. Cameron, Polym. Chem., 2017, 8, 4451–4471 RSC.
- S. Derakhshanfar, R. Mbeleck, K. Xu, X. Zhang, W. Zhong and M. Xing, Bioact. Mater., 2018, 3, 144–156 CrossRef PubMed.
- B. Zhang, L. Gao, L. Ma, Y. C. Luo, H. Y. Yang and Z. F. Cui, Engineering, 2019, 5, 777–794 CrossRef CAS.
- H. Lee and D. W. Cho, Lab Chip, 2016, 16, 2618–2625 RSC.
- S. Freeman, R. Ramos, P. Alexis Chando, L. Zhou, K. Reeser, S. Jin, P. Soman and K. Ye, Acta Biomater., 2019, 95, 152–164 CrossRef CAS PubMed.
- W. Lim, B. Kim and Y. L. Moon, Ann. Joint, 2019, 4, 7 CrossRef.
- E. Ning, G. Turnbull, J. Clarke, F. Picard, P. Riches, M. Vendrell, D. Graham, A. W. Wark, K. Faulds and W. Shu, Biofabrication, 2019, 11, 045018 CrossRef CAS PubMed.
- Y. Luo, A. Elumalai, A. Humayun and D. K. Mills, 3D Bioprinting in Medicine, Springer, 2019, pp. 163–189 Search PubMed.
- S. Vijayavenkataraman, N. Vialli, J. Fuh and W. F. Lu, Int. J. Bioprint., 2019, 5, 229 Search PubMed.
- C. Yu, X. Ma, W. Zhu, P. Wang, K. L. Miller, J. Stupin, A. Koroleva-Maharajh, A. Hairabedian and S. Chen, Biomaterials, 2019, 194, 1–13 CrossRef CAS PubMed.
- W. L. Ng, J. M. Lee, M. Zhou and W. Y. Yeong, Rapid Prototyping of Biomaterials, Elsevier, 2020, pp. 183–204 Search PubMed.
- J. Yin, M. Yan, Y. Wang, J. Fu and H. Suo, ACS Appl. Mater. Interfaces, 2018, 10, 6849–6857 CrossRef CAS PubMed.
- S. Massa, M. A. Sakr, J. Seo, P. Bandaru, A. Arneri, S. Bersini, E. Zare-Eelanjegh, E. Jalilian, B. H. Cha, S. Antona, A. Enrico, Y. Gao, S. Hassan, J. P. Acevedo, M. R. Dokmeci, Y. S. Zhang, A. Khademhosseini and S. R. Shin, Biomicrofluidics, 2017, 11, 044109 CrossRef PubMed.
- M. Hospodiuk, M. Dey, D. Sosnoski and I. T. Ozbolat, Biotechnol. Adv., 2017, 35, 217–239 CrossRef CAS PubMed.
- M. Rahmati, J. J. Blaker, S. P. Lyngstadaas, J. F. Mano and H. J. Haugen, Mater. Today Adv., 2020, 5, 100051 CrossRef.
- Y. J. Seol, H. W. Kang, S. J. Lee, A. Atala and J. J. Yoo, Eur. J. Cardiothorac. Surg., 2014, 46, 342–348 CrossRef PubMed.
- H. Gudapati, M. Dey and I. Ozbolat, Biomaterials, 2016, 102, 20–42 CrossRef CAS PubMed.
- S. H. Park, C. S. Jung and B. H. Min, Tissue Eng. Regener. Med., 2016, 13, 622–635 CrossRef CAS PubMed.
- A.-V. Do, B. Khorsand, S. M. Geary and A. K. Salem, Adv. Healthcare Mater., 2015, 4(12), 1742–1762 CrossRef CAS PubMed.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Cell, 2006, 126, 677–689 CrossRef CAS PubMed.
- Y. C. Li, Y. S. Zhang, A. Akpek, S. R. Shin and A. Khademhosseini, Biofabrication, 2016, 9, 012001 CrossRef PubMed.
- X. Kuang, D. J. Roach, J. T. Wu, C. M. Hamel, Z. Ding, T. J. Wang, M. L. Dunn and H. J. Qi, Adv. Funct. Mater., 2019, 29, 1805290 CrossRef.
- W. Li, J. Zhou and Y. Xu, Biomed. Rep., 2015, 3, 617–620 CrossRef CAS PubMed.
- R. Rai, T. Keshavarz, J. A. Roether, A. R. Boccaccini and I. Roy, Mater. Sci. Eng., R, 2011, 72, 29–47 CrossRef.
- S. Sinn, T. Scheuermann, S. Deichelbohrer, G. Ziemer and H. P. Wendel, J. Mater. Sci.: Mater. Med., 2011, 22, 1521–1528 CrossRef CAS PubMed.
- B. Swetha, S. Mathew, B. S. Murthy, N. Shruthi and S. H. Bhandi, Int. Dent. Med. J. Adv. Res., 2015, 1, 1–6 Search PubMed.
- A. K. Jain, D. Singh, K. Dubey, R. Maurya, S. Mittal and A. K. Pandey, In Vitro Toxicology, Elsevier, 2018, pp. 45–65 Search PubMed.
- C. Frumento, Medical Writing, 2017, 26, 39–40 Search PubMed.
- A. Migliore, Expert Rev. Med. Devices, 2017, 14(12), 921–923 CrossRef CAS PubMed.
- S. K. Gupta, J. Young Pharm., 2016, 8, 6–11 CrossRef.
- M. N. Helmus, D. F. Gibbons and D. Cebon, Toxicol. Pathol., 2008, 36, 70–80 CrossRef CAS PubMed.
- M. Grennan and R. J. Town, Penn Wharton Public Policy Initiative. Book 7, 2016 Search PubMed.
- K. S. Jones, Semin. Immunol., 2008, 20, 130–136 CrossRef CAS PubMed.
- A. Przekora, Mater. Sci. Eng., C, 2019, 97, 1036–1051 CrossRef CAS.
- A. Przekora, J. Czechowska, D. Pijocha, A. Ślósarczyk and G. Ginalska, Open Life Sci., 2014, 9, 277–289 CAS.
- K. Klimek, A. Belcarz, R. Pazik, P. Sobierajska, T. Han, R. J. Wiglusz and G. Ginalska, Mater. Sci. Eng., C, 2016, 65, 70–79 CrossRef CAS PubMed.
- W. De Jong, J. Carraway and R. Geertsma, Biocompatibility and performance of medical devices, Elsevier, 2012, pp. 120–158 Search PubMed.
- L. M. Wancket, Vet. Pathol., 2015, 52, 842–850 CrossRef CAS PubMed.
- P. H. Long, Toxicol. Pathol., 2008, 36, 85–91 CrossRef CAS PubMed.
- X. Struillou, H. Boutigny, A. Soueidan and P. Layrolle, Open Dent. J., 2010, 4, 37 CrossRef PubMed.
- D. W. Grainger, Nat. Biotechnol., 2013, 31, 507 CrossRef CAS PubMed.
- H. B. van der Worp, D. W. Howells, E. S. Sena, M. J. Porritt, S. Rewell, V. O'Collins and M. R. Macleod, PLoS Med., 2010, 7, e1000245 CrossRef PubMed.
- I. W. Mak, N. Evaniew and M. Ghert, Am. J. Transl. Res., 2014, 6, 114–118 Search PubMed.
- C. H. Evans, Tissue Eng., Part B, 2011, 17, 437–441 CrossRef PubMed.
- M. I. Martic-Kehl, R. Schibli and P. A. Schubiger, Eur. J. Nucl. Med. Mol. Imaging, 2012, 39, 1492–1496 CrossRef PubMed.
- V. Marx, Nature, 2015, 522, 373–377 CrossRef CAS PubMed.
- M. A. Lancaster and J. A. Knoblich, Science, 2014, 345, 1247125 CrossRef PubMed.
- S. Kim, A. N. Cho, S. Min, S. Kim and S. W. Cho, Adv. Ther., 2019, 2, 1800087 CrossRef.
- A. L. Bredenoord, H. Clevers and J. A. Knoblich, Science, 2017, 355(6322), eaaf9414 CrossRef PubMed.
- K. M. Conlee and A. N. Rowan, Hastings Cent. Rep., 2012,(Suppl), S31–S34 CrossRef PubMed.
- S. Schulze, S. G. Henkel, D. Driesch, R. Guthke and J. Linde, Front. Microbiol., 2015, 6, 65 Search PubMed.
- A. Denchai, D. Tartarini and E. Mele, Front. Bioeng. Biotechnol., 2018, 6, 155 CrossRef PubMed.
- S. S. Shera, S. Sahu and R. M. Banik, Advances in Polymer Sciences and Technology, Springer, 2018, pp. 65–74 Search PubMed.
| This journal is © The Royal Society of Chemistry 2020 |