Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Supramolecular fluorogenic peptide sensor array based on graphene oxide for the differential sensing of ebola virus†
Meng-Qi
Fu‡
a,
Xu-Chen
Wang‡
b,
Wei-Tao
Dou‡
a,
Guo-Rong
Chen
a,
Tony D.
James
 c,
Dong-Ming
Zhou
*bd and
Xiao-Peng
He
c,
Dong-Ming
Zhou
*bd and
Xiao-Peng
He
 *ae
*ae
aKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd., Shanghai 200237, China. E-mail: xphe@ecust.edu.cn
bVaccine Research Center, Institut Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai, 200031, China
cDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK
dDepartment of Pathogen Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, 300070, China. E-mail: zhoudongming@tmu.edu.cn
eFrontiers Center for Materiobiology and Dynamic Chemistry, 130 Meilong Rd., Shanghai 200237, China
First published on 15th April 2020
Abstract
We report on a supramolecular sensor array using fluorogenic peptide probes and graphene oxide that can target glycoproteins on a viral caspid, facilitating the differentiation of ebola virus from marburg virus and receptor-extensive vesicular stomatitis virus using principal component analysis.
Ebola virus (EBOV) is an acutely infectious pathogen towards humans. Infection with EBOV can result in a high death rate of 50–90%. The prevalence of EBOV in Africa during 2013–2016 resulted in the infection of more than twenty thousand people, and led to the death of more than ten thousand.1,2 The mechanism of EBOV invasion and the viral persistence are still not well characterized.3 For the effective prevention and control of the spread of EBOV, an effective diagnosis of the viral infection is a high priority in EBOV research.4 However, currently used biochemical methods for EBOV detection are complex and time-consuming.5,6 As a consequence, the development of new chemical tools for the effective sensing of EBOV remains challenging.
Glycoproteins (GPs) exist on the capsid of a number of viral species. They are pivotal targets for the development of virus inhibitors and vaccines.7 The development of anti-GP antibodies is a promising strategy to prevent EBOV infection through neutralization of GP-mediated membrane fusion with host cells.8 A recent report demonstrated that peptide fragments derived from the Fab-region of an anti-GP antibody had a GP-binding affinity comparable to that of the full-length antibody.9 This enables the development of peptide-based biochemical assays for the detection of EBOV.
Over the past decade, rapid development of fluorescent chemosensors for a variety of applications ranging from ion detection in environmental water to imaging of disease-related biomarkers in cells and in vivo has been achieved.10,11 While the majority of these studies use the “lock-and-key” concept, in which a ligand molecule is developed to specifically bind to its target receptor, Anslyn has developed the concept of “array sensing” in order to remove the need for specific ligand/receptor recognition.12 This approach, employs an array of different receptor molecules, which may only have a moderate affinity and selectivity towards the analytes. Resulting in a “fingerprint” response, when combined with an appropriate analytical method, and giving rise to characteristic patterns for each analyte. This differential sensing strategy has been successfully used for detection of ions, small molecules, proteins, cells as well as pathogens.13–15
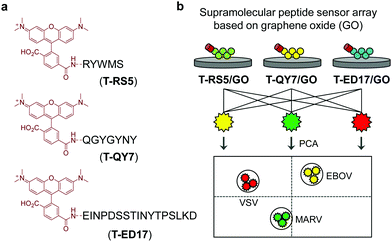
Guided by this elegant strategy, we developed a supramolecular sensor array consisting of graphene oxide (GO)-based fluorogenic peptide probes for the differential sensing of EBOV. GO is a prototypical two-dimensional (2D) material, which has found applications in a number of biomedical applications.16 With its strong fluorescence quenching capacity, GO-based fluorogenic sensors have been extensively developed.17 On the basis of our research on 2D material-based fluorogenic peptide probes,18 we have synthesized three 5-carboxytetramethylrhodamine (5-TAMRA)-labelled peptide probes for self-assembly with GO. Three peptide fragments, RS5, QY7 and ED17 (Fig. 1), derived from a patented anti-GP antibody,19 were used. Marburg virus (MARV), which has similar capsid protein components to EBOV,20 and receptor-extensive vesicular stomatitis virus (VSV), which is a common control virus for RNA-based viral investigations, were also used. For safety reasons, pseudoviruses that only contain the capsid proteins, skeletal genes and reporter genes without the ability to self-replicate were used for the tests.21
The supramolecular peptide probes were constructed by self-assembly between GO and the three fluorescent peptide ligands (T-RS5, T-QY7 and T-ED17) (Fig. 1a). Each peptide ligand was mixed with GO, followed by gently shaking for 15 min, producing the composites (T-RS5/GO, T-QY7/GO and T-ED17/GO). To characterize the association of the peptides with GO, transmission electron microscopy (TEM) was used. As shown in Fig. 2a, small aggregates were observed in the representative TEM images of the peptides alone; similar aggregates were seen on the surface of GO flakes after the self-assembly (Fig. 2b). This is in accordance with our previous observations that fluorescent peptide probes form aggregated species on the surface of 2D materials.22
 | ||
| Fig. 2 Transmission electron microscopic images of (a) T-RS5, T-QY7 and T-ED17 (10 μM), and (b) T-RS5/GO, T-QY7/GO and T-ED17/GO (peptide/GO = 10 μM/40 μg mL−1). | ||
In the UV-vis spectra, we observed a red-shifted absorption peak of TAMRA (from ca. 550 nm to 558–562 nm) after assembly with GO (Fig. S1, ESI†), suggesting a strengthened conjugated system probably due to π–π stacking between the dye and GO.23 Raman spectroscopy was used to characterize the self-assembly. The characteristic D band and G band centered at 1350 cm−1 and 1580 cm−1 were observed for GO, respectively. An increased intensity ratio of D band to G band (ID/IG), an increase in the disorder of carbon sp2-hybridization in the conjugated plane of GO.24 After self-assembly with the peptide probes, we observed an increased ID/IG for T-RS5/GO (1.38), T-QY7/GO (1.27) and T-ED17/GO (1.22) with respect to GO alone (0.98) (Fig. S2, ESI†). This suggests that the adsorption of peptide molecules onto the surface of GO increases the disorder in the carbon sp2-hybridization of the composite system.
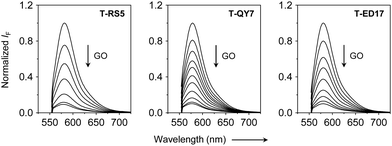
Next, fluorescence spectroscopy was used to evaluate the fluorescence change of the peptide probes after assembly with GO. We observed a concentration-dependent fluorescence quenching of all the peptide probes with increasing GO (Fig. 3), which agrees with the quenching ability of the material for adsorbed photoluminescent species,25 and suggests assembly between the peptide probes and GO. Subsequently, we added the three pseudoviruses to the assembled GO-based fluorogenic peptide probes. We observed a concentration-dependent fluorescence increase with increasing concentrations of all the viruses (Fig. 4). This suggests the removal of the peptide probes from the GO surface probably by forming peptide/virus complexes results in the fluorescence recovery of TAMRA.
Similar phenomena were observed in our previous studies.26 However, these results clearly suggest the inability of the probes to selectively differentiate EBOV from the other two viruses by just referring to the fluorescence recovery rate. We ascribe this observation to the week specificity of the short peptides for GP in the presence of the structurally complex viruses. This is a common problem with peptide-based sensing systems for complex biological samples.27 However, we also observed that while all the GO-based peptide probes showed sensitivity to the three viruses, the extent to which the fluorescence of each peptide probe recovered varied in terms of the different viral species added. This prompted us to construct a supramolecular sensor array for differential sensing of EBOV.
As shown in Fig. 5a, we used the Principal Components Analysis (PCA) for differential sensing of the viruses. PCA is a commonly used method that relies on multivariate statistical dimensional orthogonal linear transformation for the extraction of feature signals by reducing the dimensionality of the data from multivariable problems.28 The advantage of PCA is that it retains the characteristics of the data whilst reducing the dimensions, and provides more intuitive output for subsequent analysis, which promotes PCA as a method of choice for the statistical analysis of complex data.29 By analyzing the different fluorescence recovery rates of the peptide sensor array in the presence of each virus, we determined that the signals for each virus could be differentiated using the 2 top principal components (PCs) of 67% and 31%, respectively (Fig. 5a). Increasing the experimental variables for the sensor array, i.e. concentration of the peptide probes, we were able to determine a more clustered distribution for the three viruses (Fig. 5b); with the 2 top principal components (PCs) being 78% and 21%, respectively. These results suggest that, using the “sensor array” concept, it is possible to differentiate between complex viruses that express different capsid proteins using weakly specific peptide probes. Since sensing stability under varying conditions is crucial for assuring the accuracy in analyte detection, we carried out stability tests for the sensor array. The results suggest good photostability and pH stability of the GO-based fluorogenic peptide sensors (Fig. S4, ESI†). However, we note that non-specific adsorption of viruses to the exposed surface of GO could not be precluded. Therefore, in our future studies, blocking agents will be used to address this issue.
 | ||
| Fig. 5 Differential sensing of EBOV, MARV and VSV through transformed score of the fluorogenic response of (a) T-RS5/GO, T-QY7/GO and T-ED17/GO at a single composite concentration (T-RS5/GO = 0.5 μM/10 μg mL−1, T-QY7/GO = 0.5 μM/40 μg mL−1 and T-ED17/GO = 0.5 μM/24 μg mL−1) and (b) T-RS5/GO (peptide/GO = 0.25 μM/5 μg mL−1, 0.5 μM/10 μg mL−1 and 0.1 μM/20 μg mL−1), T-QY7/GO (peptide/GO = 0.25 μM/20 μg mL−1, 0.5 μM/40 μg mL−1 and 0.1 μM/80 μg mL−1) and T-ED17/GO (peptide/GO = 0.25 μM/12 μg mL−1, 0.5 μM/24 μg mL−1 and 0.1 μM/36 μg mL−1) determined by PCA. The fluorogenic response expressed as histograms determined by a microplate reader of Fig. 5 is shown in Fig. S3 (ESI†). | ||
In summary, we have developed a supramolecular GO-based fluorogenic peptide sensor array for the differential sensing of viruses. Using this method, we were able to differentiate EBOV from two other viruses, MARV and VSV. We believe that our method could become a simple and general strategy for the quick differentiation of viruses of different origins without the need for specific peptide ligands.
The authors thank the National Natural Science Foundation of China (No. 21788102, 91853201, 21722801 and 21776078), The National Key Sci-Tech Special Projects of Infection Diseases of China (2018ZX10732202), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03), the International Cooperation Program of Shanghai Science and Technology Committee (No. 17520750100) and the Fundamental Research Funds for the Central Universities (222201717003) for financial support. WTD wishes to thank the National Postdoctoral Program for Innovative Talents (BX20190115). TDJ wishes to thank the Royal Society for a Wolfson Research Merit Award.
Conflicts of interest
No conflicts of interest.Notes and references
- H. Sweiti, O. Ekwunife, T. Jaschinski and S. T. Lhachimi, Curr. Ther. Res., 2017, 84, 10–21 CrossRef CAS PubMed.
- P. Shears and T. J. D. O'Dempsey, J. Hosp. Infect., 2015, 90, 1–9 CrossRef CAS PubMed.
- M. L. Pleet, C. DeMarino, S. W. Stonier, J. M. Dye, S. Jacobson, M. J. Aman and F. Kashanchi, Viruses, 2019, 11, 410 CrossRef CAS PubMed.
- D. S. Chertow, C. Kleine, J. K. Edwards, R. Scaini, R. Giuliani and A. Sprecher, N. Engl. J. Med., 2014, 371, 2054–2057 CrossRef PubMed.
- S. K. Gire, A. Goba, K. G. Andersen, R. S. G. Sealfon, D. J. Park, L. Kanneh, S. Jalloh, M. Momoh, M. Fullah, G. Dudas, S. Wohl, L. M. Moses, N. L. Yozwiak, S. Winnicki, C. B. Matranga, C. M. Malboeuf, J. Qu, A. D. Gladden, S. F. Schaffner, X. Yang, P.-P. Jiang, M. Nekoui, A. Colubri, M. R. Coomber, M. Fonnie, A. Moigboi, M. Gbakie, F. K. Kamara, V. Tucker, E. Konuwa, S. Saffa, J. Sellu, A. A. Jalloh, A. Kovoma, J. Koninga, I. Mustapha, K. Kargbo, M. Foday, M. Yillah, F. Kanneh, W. Robert, J. L. B. Massally, S. B. Chapman, J. Bochicchio, C. Murphy, C. Nusbaum, S. Young, B. W. Birren, D. S. Grant, J. S. Scheiffelin, E. S. Lander, C. Happi, S. M. Gevao, A. Gnirke, A. Rambaut, R. F. Garry, S. H. Khan and P. C. Sabeti, Science, 2014, 345, 1369–1372 CrossRef CAS PubMed.
- B. Le Guenn, P. Formenty, P. Gounon, F. Walker and C. Boesch, Lancet, 1995, 345, 1271–1274 CrossRef.
- E. K. Nyakatura, J. C. Frei and J. R. Lai, ACS Infect. Dis., 2015, 1, 42–52 CrossRef CAS PubMed.
- C.-H. Tung, Pept. Sci., 2004, 76, 391–403 CrossRef CAS PubMed.
- L. M. Rodríguez-Martínez, A. R. Marquez-Ipiña, F. López-Pacheco, R. Pérez-Chavarría, J. C. González-Vázquez, E. González-González, G. Trujillo-de Santiago, C. A. Ponce-Ponce de León, Y. S. Zhang, M. R. Dokmeci, A. Khademhosseini and M. M. Alvarez, PLoS One, 2015, 10, 1–17 CrossRef PubMed.
- D. Wu, A. C. Sedgwick, T. Gunnlaugsson, E. U. Akkaya, J. Yoon and T. D. James, Chem. Soc. Rev., 2017, 46, 7105–7123 RSC.
- M. H. Lee, J. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 4185–4191 RSC.
- A. T. Wrighta and E. V. Anslyn, Chem. Soc. Rev., 2006, 35, 14–28 RSC.
- K. L. Diehla and E. V. Anslyn, Chem. Soc. Rev., 2013, 42, 8596–8611 RSC.
- J. R. Askim, M. Mahmoudiab and K. S. Suslick, Chem. Soc. Rev., 2013, 42, 8649–8682 RSC.
- S. Stewart, M. A. Ivyb and E. V. Anslyn, Chem. Soc. Rev., 2014, 43, 70–84 RSC.
- Y. Chen, C. Tan, H. Zhang and L. Wang, Chem. Soc. Rev., 2015, 44, 2681–2701 RSC.
- Z. Wang, J. Qiu, X. Wang, Z. Zhang, Y. Chen, X. Huang and W. Huang, Chem. Soc. Rev., 2018, 47, 6128–6174 RSC.
- Y.-H. Ma, W.-T. Dou, Y.-F. Pan, L.-W. Dong, Y.-X. Tan, X.-P. He, H. Tian and H.-Y. Wang, Adv. Mater., 2017, 29, 1604253 CrossRef PubMed.
- M. Hart and J. Wilson. US Pat., 20040053865, 2004 Search PubMed.
- A. Takada, C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt and Y. Kawaoka, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 14764–14769 CrossRef CAS PubMed.
- H. Li, F. Yu, S. Xia, Y. Yu, Q. Wang, M. Lv, Y. Wang, S. Jiang and L. Lu, Antimicrob. Agents Chemother., 2017, 61, e02168–16 CAS.
- W.-T. Dou, Y. Kong, X.-P. He, G.-R. Chen, Y. Zang, J. Li and H. Tian, ACS Appl. Mater. Interfaces, 2017, 9, 14709–14715 CrossRef CAS PubMed.
- Z. Wang, P. Huang, A. Bhirde., A. Jin, Y. Ma, G. Niu, N. Neamati and X. Chen, Chem. Commun., 2012, 48, 9768–9770 RSC.
- C. M. Girish, A. Sasidharan, G. S. Gowd, S. Nair and M. Koyakutty, Adv. Healthcare Mater., 2013, 2, 1489–1500 CrossRef CAS PubMed.
- Y.-X. Liu, X.-C. Dong and P. Chen, Chem. Soc. Rev., 2012, 41, 2283–2307 RSC.
- H.-L. Zhang, X.-L. Wei, Y. Zang, J.-Y. Cao, S. Liu, X.-P. He, Q. Chen, Y.-T. Long, J. Li, G.-R. Chen and K. Chen, Adv. Mater., 2013, 25, 4097–4101 CrossRef CAS PubMed.
- P. Zhang, Y. Cui, C. F. Anderson, C. Zhang, Y. Li, R. Wang and H. Cui, Chem. Soc. Rev., 2018, 47, 3490–3529 RSC.
- N. A. Rakow, A. Sen, M. C. Janzen, J. B. Ponder and K. S. Suslick, Angew. Chem., Int. Ed., 2005, 44, 4528–4532 CrossRef CAS PubMed.
- J. R. Askim, M. Mahmoudiab and K. S. Suslick, Chem. Soc. Rev., 2013, 42, 8649–8682 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details associated with this research. See DOI: 10.1039/c9cc09981f |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2020 |