Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Magnetically responsive horseradish peroxidase@ZIF-8 for biocatalysis†
Raffaele
Ricco
 *a,
Peter
Wied
*a,
Peter
Wied
 ab,
Bernd
Nidetzky
ab,
Bernd
Nidetzky
 bc,
Heinz
Amenitsch
d and
Paolo
Falcaro
bc,
Heinz
Amenitsch
d and
Paolo
Falcaro
 a
a
aInstitute of Physical and Theoretical Chemistry, Graz University of Technology, 8010 Graz, Austria. E-mail: raffaele.ricco@tugraz.at
bInstitute of Biotechnology and Biochemical Engineering, Graz University of Technology, 8010 Graz, Austria
cAustrian Centre of Industrial Biotechnology (acib), 8010, Graz, Austria
dInstitute of Inorganic Chemistry, Graz University of Technology, 8010 Graz, Austria
First published on 8th April 2020
Abstract
A biocatalytic system based on the zeolitic imidazolate framework-8 (ZIF-8) is obtained in a one-pot process by directly combining the enzyme horseradish peroxidase (HRP), iron oxide magnetic nanoparticles, the ligand and metal ions, in water at room temperature. The resulting system provides a useful platform for the next generation of reusable/repositionable biocatalysts.
Enzyme-coated magnetic particles are widely used for biochemical tests and catalysis.1–3 Typically, bio-conjugation protocols are used to immobilize enzymes on magnetic particles.4 However, immobilization can compromise the bioactivity of the proteins because of conformational changes at the interface,5 and the exposure of grafted and unprotected biomolecules to inhospitable conditions (e.g. organic solvents and high temperature) leads to protein denaturation.6
A method affording immobilization and protection of biomolecules exploits encapsulation in porous metal–organic frameworks (MOFs).7–11 For example, zeolitic imidazolate frameworks (ZIFs) were used to encapsulate enzymes, forming bioactive composites with improved robustness to solvents, temperature, and chaotropic and proteolytic agents.10–14 For ZIF-8, which is composed of 2-methylimidazole (HmIm) and Zn2+, the framework self-assembles spontaneously in water around negatively charged proteins.15 Thus, the synthesis of the ZIF-8-based biocomposite can be conducted using a one-pot method, as the enzyme acts as a seed for the MOF formation.11
Magnetic particles can also act as seeds for MOF crystallization;16 the resulting magnetically active composite (Magnetic Framework Composites, MFCs)17 can be used for dynamic localization,18 triggered heating,19 and catalysis.20,21 As both enzymes and magnetic particles were separately used to spontaneously induce the ZIF-8 formation in aqueous conditions,22 we hypothesized that the simultaneous use of both could lead to the one-step synthesis of magnetically and catalytically active ZIF composites. This would result in a new straightforward procedure for the preparation of recyclable MOF-based biocatalysts.
To date, only a few reports have disclosed the preparation of magnetically active ZIF biocomposites. In the synthesis, some exploited additives such as polyvinylpyrrolidone (PVP)23 or cellulose.24 Alternatively, enzymes were immobilised on the surface of pre-synthesized magnetic MOF core–shell systems.25,26 Thus, the available methods either require the use of compatibilizing agents or multi-step procedures.
Here, we describe a facile one-pot synthesis that yields the spontaneous encapsulation of both magnetic nanoparticles (MNPs) and enzymes in ZIF-8 crystals. The protocol requires the addition of an aqueous solution of enzyme, HmIm and dispersed iron oxide MNPs, to an aqueous solution of Zn2+ ions (Fig. 1).27 The presence of both protein and inorganic nanoparticle triggers the rapid self-assembly of ZIF-8. As model enzyme, we chose horseradish peroxidase (HRP) because: (1) the catalytic activity can be monitored following the degradation of the peroxide (O–O) bond,28 and (2) HRP is considered an effective biomimetic mineralization agent, as its negative zeta potential triggers the formation of a ZIF-8 coating.15 As MNPs, we used Fe3O429 with an approximate diameter of 12 nm, as confirmed by atomic force microscopy (AFM) measurements (Fig. S1, ESI†). Both enzyme and MNPs were studied using 0.33, 0.66, and 1 mg mL−1 concentrations. As crystalline phases of ZIF biocomposites and their properties depend on the synthesis conditions,30 we selected a HmIm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn2+ ratio of 16
Zn2+ ratio of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 that consistently yields protein@ZIF-8 with porous sodalite (sod) topology,31 irrespective of the amounts of protein and MNPs. The invariant properties of the matrix enabled us to inspect the relation between catalytic activity and amounts of protein and MNPs used during the synthesis. The synthesis was conducted at room temperature for ∼18 hours to improve the yield (Table S1 and Fig. S2, S3, ESI†).
1 that consistently yields protein@ZIF-8 with porous sodalite (sod) topology,31 irrespective of the amounts of protein and MNPs. The invariant properties of the matrix enabled us to inspect the relation between catalytic activity and amounts of protein and MNPs used during the synthesis. The synthesis was conducted at room temperature for ∼18 hours to improve the yield (Table S1 and Fig. S2, S3, ESI†).
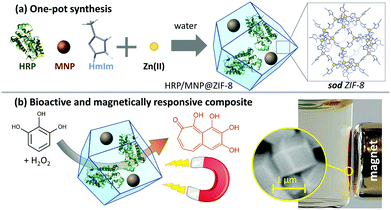
The obtained HRP/MNP@ZIF-8 composite and the related control samples (ZIF-8, MNPs@ZIF-8 and HRP@ZIF-8) were studied with X-ray diffraction methods, and diffraction plots confirmed sod ZIF-8 as the only crystalline phase (Fig. 2a).32 From the SEM images reported in Fig. 2b, all systems clearly show the expected rhombic dodecahedron geometry typical of ZIF-8 in sod phase.33 HRP/MNP@ZIF-8 (1) and the control composite samples HRP@ZIF-8 (2) and MNP@ZIF-8 (3), are found as smaller particles (800 nm in diameter) with respect to the pure ZIF-8 particles (4, diameter of 1.5 μm). The smaller size of the composites with respect to the control (pure ZIF-8) can be explained with the presence of several nucleation seeds (heterogeneous nucleation process) that typically leads to a larger number of smaller crystals.34 To validate this hypothesis, we performed a SAXS/WAXS growth kinetics study at Elettra Synchrotron. Due to the high volume of reagents needed, BSA was chosen as a model protein,35 behaving similarly to HRP in the biomimetic mineralization of ZIFs. The plots (Fig. S4, ESI†) show that both BSA and MNP induced the crystallization of ZIF-8 in 13 seconds. A faster crystal growth is noted in presence of both BSA and MNPs: the formation BSA/MNP@ZIF-8 is detected in 7 seconds. Conversely, for pure ZIF-8, diffraction peaks of sod are observed after ∼20 seconds. Although other crystalline phases are indicated as more thermodynamically stable,36,37 our syntheses yielded sod for all the MOF composites prepared.
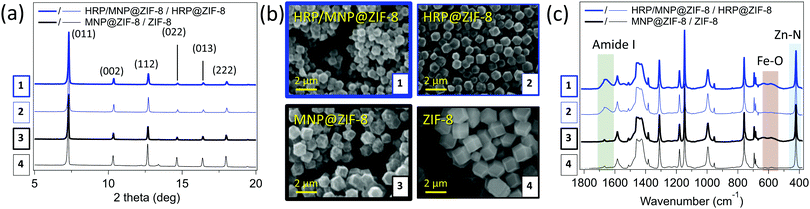 | ||
| Fig. 2 XRD (a), SEM (b), and FTIR (c) analysis of HRP/MNP@ZIF-8 (1, thick blue), and the controls HRP@ZIF-8 (2, thin blue) MNP@ZIF-8 (3, thick black), ZIF-8 (4, think black). WAXS profiles for HRP/MNP@ZIF-8 and HRP@ZIF-8, with different amounts of enzyme and magnetic nanoparticles, along with full FTIR spectra, are reported respectively in Fig. S5 and S6 (ESI†). Data for HRP-FITC/MNP@ZIF-8 and HRP-FITC@ZIF-8 are reported in Fig. S7 (ESI†). | ||
The chemical composition of the biocomposites was studied with FTIR spectroscopy (Fig. 2c). Spectra in the fingerprint region show typical ZIF-8 modes,23,38 such as the band at 421 cm−1 related to the Zn–N stretching. The presence of iron oxide MNPs is confirmed by the band right below 600 cm−1 attributed to the Fe–O stretching,23,39 and the presence of HRP is ascertained by the typical amide bands at about 1545 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching) and 1650 (N–H stretching) cm−1.40
O stretching) and 1650 (N–H stretching) cm−1.40
To study the encapsulation of the protein within the composite, we tagged HRP with fluorescein isothiocyanate (FITC). The composite HRP-FITC/MNP@ZIF-8 and the related control HRP-FITC@ZIF-8 showed identical diffraction patterns and morphology when compared with HRP/MNP@ZIF-8 and HRP@ZIF-8, respectively. We verified the presence of the FITC-tagged HRP enzyme in the composite using confocal scanning microscopy (CSLM, Fig. S8a and b, ESI†). Fluorescein emission at 580 nm (excitation laser: 488 nm, 10 mW) of FITC-HRP confirms the presence of the protein in HRP-FITC/MNP@ZIF-8 as well as in the control sample (HRP-FITC@ZIF-8). Next, to confirm the presence of MNPs in HRP-FITC/MNP@ZIF-8 composite, we collected energy-dispersive X-ray spectroscopy (EDX) maps of HRP-FITC/MNP@ZIF-8. Fig. S8c (ESI†) shows the overlap of Zn and Fe X-ray emissions, associated with ZIF-8 and MNPs, along with the C, O and N signals (Fig. S9, ESI†).
The evaluation of the encapsulation efficiency (EE%) was conducted by Bradford assay before and after reaction (calibration curve in Fig. S10, ESI†). Fig. 3a shows an average EE% of ∼82% in absence of MNPs, increasing from ∼86% to ∼94% when adding 0.33 and 1.00 mg mL−1 of MNPs, respectively. Surprisingly, the presence of MNPs influenced the HRP loading (Table S2, ESI†). A loading of 6.9 mg g−1 was measured for HRP = 0.33 mg mL−1 without MNPs, decreasing to 5.1 mg g−1 for MNP = 0.33 mg mL−1, and then increasing to 9.1 mg g−1 for MNP = 1.00 mg mL−1. The same trend occurred with HRP concentrations of 0.66 and 1.00 mg mL−1, and it can be justified by the competition between HRP and MNPs as heterogeneous nucleation agents. The enzymatic activity of HRP/MNP@ZIF-8 was tested with the reaction: 2 pyrogallol + 3 H2O2 → purpurogallin + CO2 + 5 H2O, utilizing a previously reported enzymatic assay.41,42 However, this assay is generally performed in phosphate buffer, a condition that can decompose ZIF-8 into amorphous zinc phosphate byproducts.43 After confirming that the results are fully comparable with the assay performed in phosphate buffer (Fig. S11 and S12, ESI†), we used pure water to run the assay. As controls, we tested the activity MNP, ZIF-8, and MNP@ZIF-8, finding that they did not interfere with the assay (Fig. S13, ESI†).
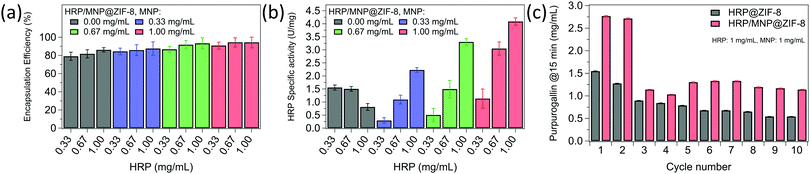 | ||
| Fig. 3 (a) Encapsulation efficiency (EE%) and (b) enzymatic activity (U mg−1), for HRP@ZIF-8 (grey) and HRP/MNP@ZIF-8 composites containing different amounts of MNPs used during the synthesis (blue: 0.33 mg mL−1; green: 0.67 mg mL−1; red: 1.00 mg mL−1). (c) Product obtained after 10 cycles performed on HRP@ZIF-8 (grey) and HRP/MNP@ZIF-8 composite (red). The reaction is: 2 pyrogallol + 3 H2O2 → purpurogallin + CO2 + 5 H2O. All data values are reported in Table S2 (ESI†). All concentrations (mg mL−1) refers to the reaction mixture. | ||
Comparing HRP/MNP@ZIF-8 with HRP@ZIF-8 (Fig. 3b) the presence of MNPs enhanced the specific activity from 1.1 U mg−1 to 3.0 U mg−1 (∼270% increase), when the concentration of MNPs increases from 0.33 mg mL−1 to 1.00 mg mL−1. We noted that previous studies, using nanoparticles for the preparation of MOF composites44 led to improved properties for hydrogen storage,45 catalysis,46,47 and photothermal conversion.48,49 In case of HRP/MNP@ZIF-8, we hypothesize that magnetic nanoparticles, acting as large nucleation seeds, could enhance structural defects in the MOF matrix.50 In this condition, the system would have preferential diffusion pathways facilitating the diffusion of the substrate within the composite.51 Although HRP was immobilized in different MOFs (Table S3, ESI†), the lack of (1) a standard procedures for the material preparation (e.g. same enzymatic loading), (2) available data (e.g. SA of the free enzyme) and, (3) standard activity evaluation procedures (e.g. the specific type of assay used) prevents a precise comparison of the catalytic performance.
As industrial application of enzymes is often hampered by their difficult recovery,52 we focused our attention on the reusability of HRP/MNP@ZIF-8. By testing the activity over 10 cycles (Fig. 3c), the composite showed superior product conversion over the HRP@ZIF-8 control (about 4 times higher). This was justified considering two relevant aspects: higher EE% of HRP/MNP@ZIF-8, and presence of crystal defects (e.g. grain boundaries of polycrystalline particles, Fig. S3b and e, ESI†) that could facilitate mass transfer. However, after 2 cycles we observed a drop of the enzymatic activity. We investigated the leaching of enzymes from the HRP/MNP@ZIF-8 system (3% after 10 cycles, Fig. S14, ESI†), which was not sufficient to justify the performance drop. The loss of enzymatic activity can be attributed to different pathways:53 (1) decomposition induced by excess of H2O2, (2) occlusion of pores in the carrier, (3) degradation of heme center due to interaction with substrate. The importance of each contribution is strongly related to the material properties. Analogous rapid activity losses were reported for other immobilized enzymes.54
In summary, we prepared a MOF composite based on ZIF-8 simultaneously encapsulating HRP and MNPs (HRP/MNP@ZIF-8). The one-pot synthesis was conducted at room temperature in water, overcoming previously reported limitations, such as multistep synthesis,25 need for polymeric additives, organic solvents,23,24 or surface functionalization processes.26 HRP/MNP@ZIF-8 was characterized by SEM, XRD, FTIR, CSLM, and AFM to ascertain morphology, crystallinity, and composition. For the first time, we studied the kinetic of crystallization with in situ SAXS/WAXS, demonstrating that the co-presence of MNPs and enzymes can induce a faster crystallization when compared with HRP@ZIF-8 and MNP@ZIF-8. More importantly, we proved that the presence of MNPs can significantly enhance the enzymatic activity of the ZIF-8 biocomposite. Indeed, the activity of HRP/MNP@ZIF-8 was 5 times higher than HRP@ZIF-8. By using 1 mg mL−1 of both enzyme and MNPs as precursors, it was possible to prepare a magnetic porous carrier with an enzymatic loading of about 5.6 mg g−1. The combination of enzyme, magnetic nanoparticles, and porous metal–organic frameworks, along with its dynamic localization properties,18,21,23 holds much promise for the progress of MOF biocomposites for application to biocatalysts.
This work was supported by the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement #748649 (project “MNEMONIC”), and under the European Union's Horizon 2020 Programme (FP/2014–2020)/ERC Grant Agreement no. 771834 – POPCRYSTAL. The authors acknowledge support from the European Union's Horizon 2020 FETOPEN-1-2016–2017 research and innovation program under grant agreement 801464, and the lead project LP-03 of Graz University of Technology. Marcello Solomon and Mercedes Linares-Moreau are acknowledged for their support in the acquisition of XRD and AFM data. Helmar Wiltsche is particularly acknowledged for the ICP-OES/MS analysis. We gratefully thank Varta Microtechnologies GmbH for the use of SEM and EDX instruments, and CERIC-ERIC Consortium for the travel support and access to the SAXS facility.
Conflicts of interest
There are no conflicts to declare.References
- W. L. Shelver, L. M. Kamp, J. L. Church and F. M. Rubio, J. Agric. Food Chem., 2007, 55, 3758–3763 CrossRef CAS PubMed.
- C. S. Hottenstein, F. M. Rubio, D. P. Herzog, J. R. Fleeker and T. S. Lawruk, J. Agric. Food Chem., 1996, 44, 3576–3581 CrossRef CAS.
- T. S. Lawruk, C. S. Hottenstein, D. P. Herzog and F. M. Rubio, Bull. Environ. Contam. Toxicol., 1992, 48, 643–650 CrossRef CAS PubMed.
- L. H. Reddy, J. L. Arias, J. Nicolas and P. Couvreur, Chem. Rev., 2012, 112, 5818–5878 CrossRef CAS PubMed.
- J. Xu, J. Sun, Y. Wang, J. Sheng, F. Wang and M. Sun, Molecules, 2014, 19, 11465–11486 CrossRef PubMed.
- D. Shortle, FASEB J., 1996, 10, 27–34 CrossRef CAS PubMed.
- R. Ricco, C. Pfeiffer, K. Sumida, C. J. Sumby, P. Falcaro, S. Furukawa, N. R. Champness and C. J. Doonan, CrystEngComm, 2016, 18, 6532–6542 RSC.
- W. Liang, H. Xu, F. Carraro, N. K. Maddigan, Q. Li, S. G. Bell, D. M. Huang, A. Tarzia, M. B. Solomon, H. Amenitsch, L. Vaccari, C. J. Sumby, P. Falcaro and C. J. Doonan, J. Am. Chem. Soc., 2019, 141, 2348–2355 CrossRef CAS PubMed.
- F.-S. Liao, W.-S. Lo, Y.-S. Hsu, C.-C. Wu, S.-C. Wang, F.-K. Shieh, J. V. Morabito, L.-Y. Chou, K. C.-W. Wu and C.-K. Tsung, J. Am. Chem. Soc., 2017, 139, 6530–6533 CrossRef CAS PubMed.
- G. Lu, S. Li, Z. Guo, O. K. Farha, B. G. Hauser, X. Qi, Y. Wang, X. Wang, S. Han, X. Liu, J. S. DuChene, H. Zhang, Q. Zhang, X. Chen, J. Ma, S. C. J. Loo, W. D. Wei, Y. Yang, J. T. Hupp and F. Huo, Nat. Chem., 2012, 4, 310–316 CrossRef CAS PubMed.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed.
- Y. Feng, H. Wang, S. Zhang, Y. Zhao, J. Gao, Y. Zheng, P. Zhao, Z. Zhang, M. J. Zaworotko, P. Cheng, S. Ma and Y. Chen, Adv. Mater., 2019, 31, 1805148 CrossRef PubMed.
- E. Astria, M. Thonhofer, R. Ricco, W. Liang, A. Chemelli, A. Tarzia, K. Alt, C. E. Hagemeyer, J. Rattenberger, H. Schroettner, T. Wrodnigg, H. Amenitsch, D. M. Huang, C. J. Doonan and P. Falcaro, Mater. Horiz., 2019, 6, 969–977 RSC.
- S. Li, M. Dharmarwardana, R. P. Welch, Y. Ren, C. M. Thompson, R. A. Smaldone and J. J. Gassensmith, Angew. Chem., 2016, 128, 10849–10854 CrossRef.
- N. K. Maddigan, A. Tarzia, D. M. Huang, C. J. Sumby, S. G. Bell, P. Falcaro and C. J. Doonan, Chem. Sci., 2018, 9, 4217–4223 RSC.
- T. Zhang, X. Zhang, X. Yan, L. Kong, G. Zhang, H. Liu, J. Qiu and K. L. Yeung, Chem. Eng. J., 2013, 228, 398–404 CrossRef CAS.
- R. Ricco, L. Malfatti, M. Takahashi, A. J. Hill and P. Falcaro, J. Mater. Chem. A, 2013, 1, 13033–13045 RSC.
- P. Falcaro, F. Normandin, M. Takahashi, P. Scopece, H. Amenitsch, S. Costacurta, C. M. Doherty, J. S. Laird, M. D. H. Lay, F. Lisi, A. J. Hill and D. Buso, Adv. Mater., 2011, 23, 3901–3906 CrossRef CAS PubMed.
- M. R. Lohe, K. Gedrich, T. Freudenberg, E. Kockrick, T. Dellmann and S. Kaskel, Chem. Commun., 2011, 47, 3075–3077 RSC.
- M. Faustini, J. Kim, G.-Y. Jeong, J. Y. Kim, H. R. Moon, W.-S. Ahn and D.-P. Kim, J. Am. Chem. Soc., 2013, 135, 14619–14626 CrossRef CAS PubMed.
- T. Toyao, M. J. Styles, T. Yago, M. M. Sadiq, R. Ricco, K. Suzuki, Y. Horiuchi, M. Takahashi, M. Matsuoka and P. Falcaro, CrystEngComm, 2017, 19, 4201–4210 RSC.
- F. Pang, M. He and J. Ge, Chem. – Eur. J., 2015, 21, 6879–6887 CrossRef CAS PubMed.
- C. Hou, Y. Wang, Q. Ding, L. Jiang, M. Li, W. Zhu, D. Pan, H. Zhu and M. Liu, Nanoscale, 2015, 7, 18770–18779 RSC.
- S.-L. Cao, H. Xu, L.-H. Lai, W.-M. Gu, P. Xu, J. Xiong, H. Yin, X.-H. Li, Y.-Z. Ma, J. Zhou, M.-H. Zong and W.-Y. Lou, Bioresour. Bioprocess., 2017, 4, 56 CrossRef.
- M. Zhao, X. Zhang and C. Deng, Chem. Commun., 2015, 51, 8116–8119 RSC.
- A. Samui, A. R. Chowdhuri, T. K. Mahto and S. K. Sahu, RSC Adv., 2016, 6, 66385–66393 RSC.
- K. S. Park, Z. Ni, A. P. Côté, J. Y. Choi, R. Huang, F. J. Uribe-Romo, H. K. Chae, M. O’Keeffe and O. M. Yaghi, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10186–10191 CrossRef CAS PubMed.
- N. C. Veitch, Phytochemistry, 2004, 65, 249–259 CrossRef CAS PubMed.
- A. Schejn, T. Mazet, V. Falk, L. Balan, L. Aranda, G. Medjahdi and R. Schneider, Dalton Trans., 2015, 44, 10136–10140 RSC.
- F. Carraro, M. de, J. Velásquez-Hernández, E. Astria, W. Liang, L. Twight, C. Parise, M. Ge, Z. Huang, R. Ricco, X. Zou, L. Villanova, C. O. Kappe, C. Doonan and P. Falcaro, Chem. Sci., 2020, 11, 3397–3404 RSC.
- W. Liang, R. Ricco, N. K. Maddigan, R. P. Dickinson, H. Xu, Q. Li, C. J. Sumby, S. G. Bell, P. Falcaro and C. J. Doonan, Chem. Mater., 2018, 30, 1069–1077 CrossRef CAS.
- W. Morris, C. J. Stevens, R. E. Taylor, C. Dybowski, O. M. Yaghi and M. A. Garcia-Garibay, J. Phys. Chem. C, 2012, 116, 13307–13312 CrossRef CAS.
- C. Avci, J. Ariñez-Soriano, A. Carné-Sánchez, V. Guillerm, C. Carbonell, I. Imaz and D. Maspoch, Angew. Chem., Int. Ed., 2015, 54, 14417–14421 CrossRef CAS PubMed.
- S. Yang, Y. Hou, B. Zhang, X. H. Yang, H. Zhang, H. J. Zhao and H. G. Yang, CrystEngComm, 2014, 16, 7502–7506 RSC.
- A. S. Determan, B. G. Trewyn, V. S.-Y. Lin, M. Nilsen-Hamilton and B. Narasimhan, J. Controlled Release, 2004, 100, 97–109 CrossRef CAS PubMed.
- R. Galvelis, B. Slater, A. K. Cheetham and C. Mellot-Draznieks, CrystEngComm, 2011, 14, 374–378 RSC.
- Z. Akimbekov, A. D. Katsenis, G. P. Nagabhushana, G. Ayoub, M. Arhangelskis, A. J. Morris, T. Friščić and A. Navrotsky, J. Am. Chem. Soc., 2017, 139, 7952–7957 CrossRef CAS PubMed.
- Y. Hu, H. Kazemian, S. Rohani, Y. Huang and Y. Song, Chem. Commun., 2011, 47, 12694 RSC.
- T. J. Daou, J. M. Grenèche, G. Pourroy, S. Buathong, A. Derory, C. Ulhaq-Bouillet, B. Donnio, D. Guillon and S. Begin-Colin, Chem. Mater., 2008, 20, 5869–5875 CrossRef CAS.
- A. Barth and C. Zscherp, Q. Rev. Biophys., 2002, 35, 369–430 CrossRef CAS.
- B. Chance and A. C. Maehly, Methods Enzymol., 1955, 2, 764–775 Search PubMed.
- L. M. Shannon, E. Kay and J. Y. Lew, J. Biol. Chem., 1966, 241, 2166–2172 CAS.
- M. de, J. Velásquez-Hernández, R. Ricco, F. Carraro, F. T. Limpoco, M. Linares-Moreau, E. Leitner, H. Wiltsche, J. Rattenberger, H. Schröttner, P. Frühwirt, E. M. Stadler, G. Gescheidt, H. Amenitsch, C. J. Doonan and P. Falcaro, CrystEngComm, 2019, 21, 4538–4544 RSC.
- Q. Yang, Q. Xu and H.-L. Jiang, Chem. Soc. Rev., 2017, 46, 4774–4808 RSC.
- G. Li, H. Kobayashi, J. M. Taylor, R. Ikeda, Y. Kubota, K. Kato, M. Takata, T. Yamamoto, S. Toh, S. Matsumura and H. Kitagawa, Nat. Mater., 2014, 13, 802–806 CrossRef CAS PubMed.
- Y. C. Tan and H. C. Zeng, Nat. Commun., 2018, 9, 4326 CrossRef PubMed.
- Z. Li and H. C. Zeng, Chem. Mater., 2013, 25, 1761–1768 CrossRef CAS.
- Y.-Z. Chen, Z. U. Wang, H. Wang, J. Lu, S.-H. Yu and H.-L. Jiang, J. Am. Chem. Soc., 2017, 139, 2035–2044 CrossRef CAS PubMed.
- Q. Yang, Q. Xu, S.-H. Yu and H.-L. Jiang, Angew. Chem., Int. Ed., 2016, 55, 3685–3689 CrossRef CAS PubMed.
- Z. Fang, B. Bueken, D. E. De Vos and R. A. Fischer, Angew. Chem., Int. Ed., 2015, 54, 7234–7254 CrossRef CAS PubMed.
- N. A. Khan, Z. Hasan and S. H. Jhung, Coord. Chem. Rev., 2018, 376, 20–45 CrossRef CAS.
- R. A. Sheldon and S. van Pelt, Chem. Soc. Rev., 2013, 42, 6223–6235 RSC.
- L. Mao, S. Luo, Q. Huang and J. Lu, Sci. Rep., 2013, 3, 1–7 Search PubMed.
- J. P. Henley and A. Sadana, Biotechnol. Bioeng., 1986, 28, 1277–1285 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, AFM, FTIR, WAXS, EDX, and UV-Vis data. See DOI: 10.1039/c9cc09358c |
| This journal is © The Royal Society of Chemistry 2020 |