Generating linear oxygen gradients across 3D cell cultures with block-layered oxygen controlled chips (BLOCCs)†
Matthew W.
Boyce‡
a,
William C.
Simke‡
a,
Rachael M.
Kenney
a and
Matthew R.
Lockett
 *ab
*ab
aDepartment of Chemistry, University of North Carolina at Chapel Hill, Kenan and Caudill Laboratories, 125 South Road, Chapel Hill, NC 27599-3290, USA. E-mail: mlockett@unc.edu
bLineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, 450 West Drive, Chapel Hill, NC 27599-7295, USA
First published on 26th November 2019
Abstract
Oxygen is a transcriptional regulator responsible for tissue homeostasis and maintenance. Studies relating cellular phenotype with oxygen tension often use hypoxia chambers, which expose cells to a single, static oxygen tension. Despite their ease of use, these chambers are unable to replicate the oxygen gradients found in healthy and diseased tissues. Microfabricated devices capable of imposing an oxygen gradient across tissue-like structures are a promising tool for these studies, as they can provide a high density of information in a single experimental setup. We describe the fabrication and characterization of a modular device, which leverages the gas-permeability of silicone to impose gradients of oxygen across cell-containing regions, assembled by layering sheets of laser cut acrylic and silicone rubber. The silicone also acts as a barrier, separating the flowing gases from the cell culture medium, preventing evaporation or bubble formation in experiments that require prolonged periods of incubation. The acrylic components provide a rigid framework to provide a sterile culture environment. Using oxygen-sensing films, we show the device can support gradients of different ranges and steepness by simply changing the composition of the gases flowing through the silicone components of the BLOCC. Using a cell-based reporter assay, we demonstrate that cellular responses to hypoxia are proportional to oxygen tension.
Introduction
Oxygen plays a vital role in maintaining cellular homeostasis, acting as both a metabolite and a transcriptional regulator. Low physiological oxygen tensions can result in hypoxia, a condition in which cells undergo the metabolic reprogramming needed for continued survival.1–3 In solid tumors, hypoxia is correlated with cancer aggressiveness and patient mortality.4,5 Cellular responses to inadequate oxygen supplies are coordinated by hypoxia-inducible factors (HIFs). These transcription factors rely on two constitutively expressed subunits: HIF-α and HIF-β. Under physiologically normal oxygen tensions (physoxia, 15–100 mmHg), the HIF-α subunits are hydroxylated and degraded via the ubiquitin proteasome pathway.6 Under hypoxic conditions (<15 mmHg O2), the stabilized HIF-α subunits translocate to the nucleus and dimerize with a HIF-β subunit. This functional transcriptional complex regulates metabolism, alters the cell cycle, and promotes both angiogenesis and movement.Hypoxia chambers, cobalt chloride, and prolyl hydroxylase inhibitors such as dimethyloxaloylglycine (DMOG) are commonly used to study HIF activation and transcriptional regulation.7,8 Unlike inhibitors that target a specific protein or pathway, hypoxia chambers ensure a systemic cellular response to the loss of oxygen, including mitochondrial stress-induced pathways. While easy to use, hypoxia chambers impose a single, uniform tension across the culture. A previous study quantifying HIF-α proteins across five oxygen tensions ranging from 9–150 mmHg found that 15 mmHg O2 was required for HIF stabilization and nuclear accumulation of six different cancer cell lines.9 While a fundamentally important study in oxygen regulation, the procedure illustrates one of the most significant limitations of using hypoxia chambers, the need to perform experiments serially, focused on a single oxygen tension per setup.
Exposing cells to a spatially defined yet temporally static oxygen gradient has both advantages and disadvantages. An experimental advantage is the increased density of information derived from a single setup. The information-rich datasets pose experimental challenges, as bulk analyses of the average cellular response from a single cell lysate are not possible. Instead, the cells must be evaluated individually in a manner that maintains spatial integrity. The second advantage of an imposed gradient is quantitative maps of HIF-α stabilization and transcriptional regulation in physiologically representative oxygen environments. All tissues contain oxygen gradients, which extend radially from blood vessels.10,11 These gradients become exaggerated in ischemic tissues and solid tumors, where cellular proliferation outpaces angiogenesis.
Microfabricated devices capable of imposing defined oxygen gradients across 2D and 3D cell structures have investigated invasion, HIF-1α and -2α stabilization, and the production of reactive oxygen species.12–19 Despite the multitude of device architectures capable of precisely controlling extracellular gradients,20–23 their widespread adoption is limited by the need for specialized equipment and expertise for fabrication.24 The maker movement and ready access to 3D printers and laser cutters have empowered many laboratories to generate customizable devices and pieces of equipment.25,26 This accessibility is lowering the barrier to incorporate culture-compatible devices into laboratories focused on cellular hypoxia and redox biology. In particular, they provide a means of readily prototyping and modifying device designs without the need of preparing new masters, performing soft lithography, or casting polymers such as PDMS.
In this paper, we prepared and characterized a Block-Layered Oxygen-Controlled Chip (BLOCC, Fig. 1) capable of imposing oxygen gradients across 3D cultures of cells suspended in a hydrogel. To simultaneously characterize the cellular responses to different oxygen tensions, we incorporated oxygen-sensing films as well as a cell-based reporter system that expressed green fluorescent protein upon HIF transactivation. We demonstrate the ability of the BLOCCs to support sustained oxygen gradients over a 48 h period with the oxygen sensing films and show a correlation of HIF activation with oxygen tensions below 15 mmHg with the cell-based reporter system.
Device design, preparation, and assembly
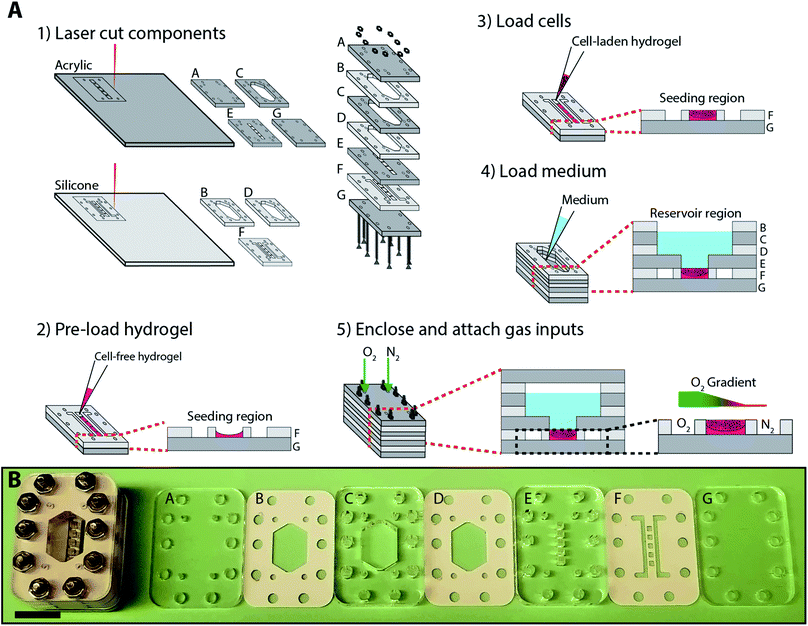
Fig. 1 outlines the fabrication and assembly of a BLOCC device, which consisted of seven alternating layers of clear cast acrylic and silicone rubber. Components A and G enclosed the device, isolating cells and culture medium from the ambient environment. Component F contained five 1.5 × 1.5 × 0.8 mm cell-containing regions, flanked by two gas flow channels. Component E connected the cell-containing regions to the 1 mL culture medium reservoir formed by components B–D.Each component was pre-cut with patterns of interest and are provided in laser cutter-ready Adobe Illustrator files in the ESI.† We chose this approach for several reasons. First, cast acrylic provides structural rigidity to the device and has a low oxygen permeability. It is also optically transparent and allowed us to image cells in the device in real-time with fluorescence microscopy. Second, silicone is highly compressible and an ideal gasket material to provide watertight seals between the acrylic pieces. It also is gas permeable, allowing us to impose oxygen gradients across the cell-containing regions of the device. Third, cutting patterns through the materials avoided variations in channel depth, roughness, and shape that can occur with laser etching.27,28
The BLOCCs were assembled in the following order. First, components F and G were stacked together. Next, 1 μL of cell-free Matrigel (Corning) was loaded into each of the seeding regions in component F and allowed to gelate for 1 min at room temperature. Once gelated, 1 μL of a 2000 cells per μL Matrigel suspension was added. The initial addition of Matrigel ensured the cells were evenly distributed throughout the culture region, as the liquid form was prone to wicking along the silicone gasket and resulted in a concave structure. We note that the addition of cells to the BLOCCs is as straightforward as loading a well plate, making the incorporation of 3D cultures more accessible to labs with little experience with microfabricated devices.
Next, components B–E were assembled and 1 mL of culture medium was added to the reservoir. Finally, component A was placed on top of the stack and the entire device held together with six 25.4 mm stainless steel screws with 4–40 threading and matching nuts. Components A–F of the assembled device formed integrated gas ports, through which we flowed different gas mixtures into the parallel channels in component F.
Results and discussion
BLOCCs are compatible with prolonged cell culture
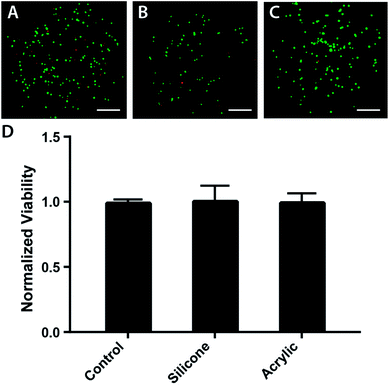
Acrylic and food-grade silicone are biocompatible and have been used to prepare microfabricated devices for cell culture previously.29–31 To ensure the laser-cut components could support prolonged cell culture, we incubated them in contact with parental M231 cells suspended in Matrigel at a final density of 2000 cells per μL. This cell density allowed for easy post-experiment microscopic analyses of the BLOCCs with a widefield fluorescence microscope.First, the cell suspension was loaded into a clear bottom 96 well plate and incubated for 24 hours. Next, a 30 mg piece of laser-cut acrylic or silicone was added to the wells and incubated for 48 h. Lastly, the cells were stained with a mixture of fluorescein diacetate (FDA) and propidium iodide (PI). The percentage of viable cells in the acrylic- and silicone-containing wells were normalized to control wells, exposed to culture medium only. Fig. 2 contains representative images of the live-dead stains for each culture condition. Fig. 2d shows there were no significant differences in viability in any of the wells, confirming the suitability of both components for the HIF transactivation studies described below.
Oxygen gradients are reproducible, tunable, and persistent across 3D cell cultures
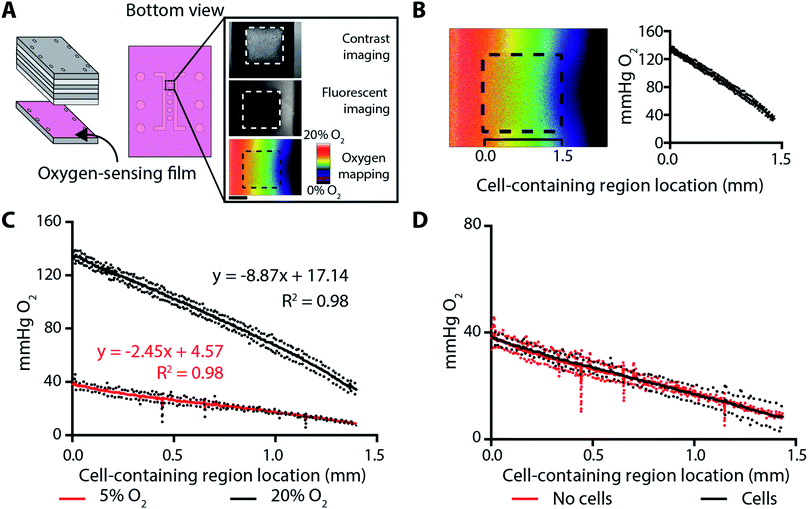
Oxygen gradients were generated across the cell-containing regions by flowing oxygenated and deoxygenated gas mixtures through two parallel channels in component F of the assembled BLOCC (Fig. 1A). We quantified the range and steepness of these gradients with an oxygen-sensitive film coated onto component G of the device (Fig. 3A). The preparation of these films, as well as the conversion of its luminescence intensity to oxygen tension with an experimentally determined Stern–Volmer relationship, was detailed previously.32 These films have a linear response between 0–150 mmHg, with the highest sensitivity in physiologically relevant ranges for epithelial tissues and tumor environments (0–35 mmHg).The combination of a deoxygenated gas mixture (95% N2, 5% CO2) and a high-oxygen gas mixture (75% N2, 20% O2, 5% CO2) resulted in a gradient extending from 132–35 mmHg, with a slope of −67.5 ± 0.2 mmHg O2 per mm (Fig. 3B). The combination of the deoxygenated gas mixture and a low-oxygen gas mixture (90% N2, 5% O2, 5% CO2) resulted in a gradient that was relevant for cellular hypoxia, extending from 35–7 mmHg, with a slope of −18.6 ± 0.2 mmHg O2 per mm (Fig. 3C).
To ensure the oxygen gradients were static during an experiment, we measured them throughout a 12 h incubation in a BLOCC whose seeding regions contained either Matrigel-only or 2000 parental MDA-MB-231 cells suspended in Matrigel. The oxygen gradient was stable throughout this period, with no significant change in slope or range. There also were no differences in the slope or range of the oxygen gradient when cells were added to the BLOCC (Fig. 3D).
Oxygen gradients and HIF activation in MDA-MB-231 cells correlate in BLOCCs
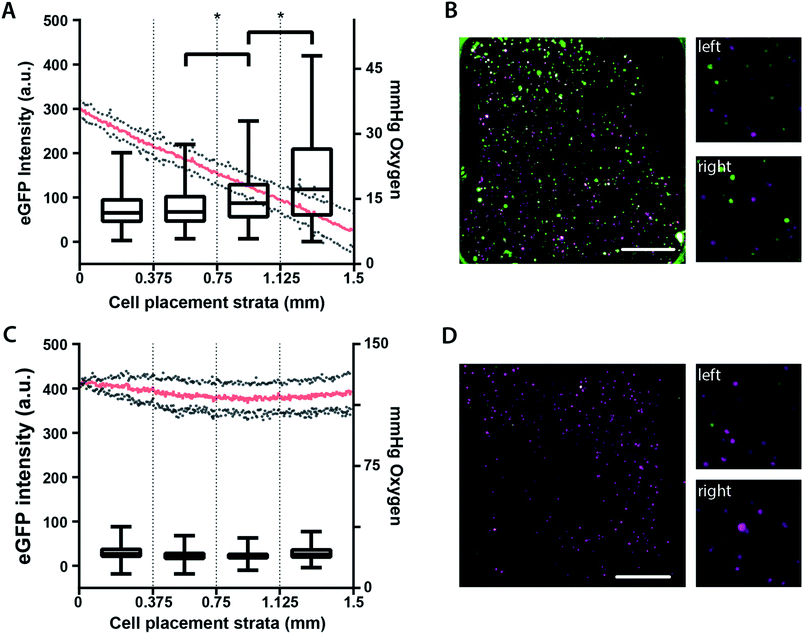
To quantify cellular responses along an imposed oxygen gradient, we prepared BLOCCs whose seeding areas contained 2000 M231-HRE cells suspended in a microliter of Matrigel. Cells in one device were exposed to ambient conditions. Cells in the other device were exposed to a gradient generated by the combination of the deoxygenated and low-oxygen gas mixtures. The M231-HRE cells expressed the mCherry protein constitutively and eGFP upon activation of HIFs. Using z-stack composite fluorescence images, we reconstructed 3D representations of each cell from their mCherry fluorescence intensities. To estimate the average cellular response to hypoxia, we measured the eGFP fluorescence intensity at the center pixel of each cell body. This method accounted for point spread along the z-axis, which can be overestimated from the non-perfect alignment and stitching of the widefield images, as well as non-uniform amounts of background signal.Scatterplots of mCherry and eGFP intensity as a function of cell position in both setups (Fig. S1†) show there was no correlation between mCherry intensity and position in the presence or absence of an oxygen gradient. These data suggest global protein expression was not affected in either setup, and that changes in eGFP were related to response to HIF regulation. Only the oxygen gradient-containing BLOCC had a significant correlation between eGFP fluorescence intensity and position in the device.
To more easily visualize the relationship between eGFP expression and local oxygen tension (Fig. 4), we stratified the cell-containing region into four equal-sized segments corresponding to: 34–27 mmHg O2, 27–21 mmHg O2, 21–14 mmHg O2, or 14–8 mmHg O2. While each segment spans a broad range of oxygen tensions, there is a discernible and inverse relationship between oxygen tension and eGFP expression. Segments 2–4 have statistically significant differences in eGFP expression (Fig. 4A), with cells experiencing the greatest oxygen stress yielding the highest hypoxic response. In the absence of an oxygen gradient, eGFP expression was not significantly different (Fig. 4C). These results are further highlighted with the representative micrographs of the M231-HRE cells in the oxygen gradient-containing (Fig. 4B) and ambient condition (Fig. 4D) BLOCCs.
Conclusion
BLOCCs are a modular platform capable of supporting 3D cell cultures exposed to defined oxygen gradients. The alternating layers of acrylic and silicone components can be laser cut with the pattern of interest, incorporating culture regions of different sizes and shapes beyond those demonstrated here. While gas tanks are needed to impose oxygen gradients across the device, the use of silicone barriers between the gas- and cell-containing regions prevents medium evaporation or the introduction of bubbles. By changing the composition of the gas mixtures, we were able to alter the range and steepness of the oxygen gradient in the same device. An alternate and equally viable means of modulating the oxygen gradients is to laser cut new components with larger cell-containing regions or different gas channel configurations.The incorporation of an oxygen-sensing film and an eGFP-based reporter assay allowed us to map the oxygen gradient and cellular responses simultaneously and in real-time. These data suggest that cellular responses to hypoxia are proportional to oxygen stress. The stratification of these datasets provided further insight into this relationship, narrowing down the oxygen ranges for future studies. BLOCCs will support these investigations, mapping temporal and spatial responses of HIF stabilization in defined oxygen gradients.
Materials and methods
Materials for BLOCCs
All BLOCC components were purchased from McMaster Carr. Components A and G were 3.175 mm thick cast acrylic; components C and E were 6.35 mm thick cast acrylic, and; components B, D, and F were 0.794 mm thick 50A food-grade silicone. Each component was laser cut on a Universal Laser Systems VLS6.60 instrument, with settings of 60 W, 25.4 mm s−1 speed, 1000 dpi resolution, and a 50 mm focusing distance. Debris from the laser-cut silicone components was removed by washing with a 70% (v/v) ethanol solution. Prior to usage, all materials were sterilized under UV light for at least 1 hour.Cell lines and culture reagents
The breast adenocarcinoma MDA-MB-231 cell line (American Type Culture Collection) was engineered to express mCherry fluorescent protein constitutively and enhanced green fluorescent protein under hypoxic conditions. The mCherry gene was incorporated with LPP-MCHR-Lv105-025 lentiviral particles (GeneCopoeia), according to the manufacturer's protocol. The 5HRE/eGFP gene construct (Addgene, plasmid #46926)33 was transfected into the cells with TurboFect (Life Technologies), according to the manufacturer's protocol. We detailed the preparation and characterization of the M231-HRE cells previously.34 The cells were maintained as monolayers at 37 °C and 5% CO2 in RPMI 1640 medium supplemented with 5% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin. Culture medium was exchanged every 48 h, and the cells passaged at 80% confluency. All cell culture medium and supplements were obtained from Gibco (Life Technologies) other than fetal bovine serum (VWR).Oxygen gradient generation and quantification
Deoxygenated and oxygenated gases were interfaced with component A of the assembled BLOCCs, using a wall-plug design.35 The gases were flowed through the channels at a rate of 35 cm3 min−1. The deoxygenated (95% N2, 5% CO2), high-oxygen (75% N2, 20% O2, 5% CO2), and low-oxygen (90% N2, 5% O2, 5% CO2) gas mixtures were prepared and validated by Airgas.Oxygen sensing films were coated directly onto component G. The preparation and characterization of these films was detailed previously.32 Briefly, a 1 mM solution of palladium(II) tetrakis(pentafluorophenyl)porphyrin (PdTFPP) in an 18.75% (w/w) polystyrene–toluene solution was spin coated directly onto the acrylic component. The oxygen sensing films were imaged on an Axiovert 40 CFL (Zeiss) inverted widefield fluorescence microscope equipped with a LED light source (wLS-LG-MB, QImaging) and monochrome 12 bit camera (QIC-F-12 C, QImaging) with a 546/12 nm excitation filter and 590 nm longpass filter, with a 1400 ms exposure.
Cellular viability
Live-dead stains of parental MDA-MB-231 cells used a combination of 5 mg mL−1 FDA and 2 mg mL−1 PI. Once stained, the cells were imaged on an Axiovert 40 CFL. Fluorescein-labeled cells were imaged with a 470/20 nm excitation filter and 540/40 nm emission filter, with a 500 ms exposure. PI-labeled cells were imaged with a 546/12 nm excitation filter and a 590 nm long-pass filter, with a 600 ms exposure. Phase-contrast images were also taken and used to identify the boundaries of all the individual cells in a single image with the Trainable Weka Segmentation plugin in FIJI.36,37 The percentage of viable cells was determined by dividing the number of fluorescein positive cells by the total number of cells measured in a single image. Cells that stained positively for both fluorescein and PI were not included in the viable cell count.Image acquisition and analysis of hypoxia responses
Images of M231-HRE cells in the assembled BLOCCs were obtained on an Olympus IX70 inverted widefield fluorescence microscope equipped with a Hamamatsu Flash 4.0 V2+ sCMOS camera and a 10×/0.30 Ph1 UPlanFL objective. The mCherry protein was imaged with a 555/25 nm excitation and a 605/52 nm emission filter, with 400 ms exposure. eGFP was imaged with a 490/20 nm excitation and a 525/36 nm emission filter, with 200 ms exposure. Z-stack composite images were constructed in the Imaris imaging software package by stitching 10 μm increments together using a 10% overlap. Outlines of the mCherry-expressing cells were identified with the following settings: XY diameter of 20 μm, Z diameter of 40 μm, background subtraction, quality above 10.5, region growing type was set to local contrast. To ensure that we did not overestimate the expression of eGFP from poor image stitching and light scatter, we used the intensity value of the center of each cell for quantitation. Maximum Z-projections for representative images were produced using ImageJ's Imaris BigDataViewer.Statistical analyses
Unless otherwise stated, all reported values are the average and standard deviation (SD) of at least three replicate samples. Datasets were analyzed with GraphPad Prism® v.7.01 and compared with a Brown–Forsythe and Welch ANOVA test. Pearson correlation coefficients were used to define correlations within a single dataset. A p-value of <0.05 was considered significant.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported with funds provided by Eli Lilly and Company's Young Investigator Award as well as National Institute of General Medical Science through Grant Award Number R35GM128697. MWB would like to thank the Graduate School at UNC for support through a Dissertation Completion Fellowship. A portion of this work was performed in the Chapel Hill Analytical and Nanofabrication Laboratory (CHANL), the UNC BeAM Makerspace, and the UNC Microscopy Services Laboratory. CHANL is a member of the North Carolina Research Triangle Network, which is supported by the National Science Foundation Grant Number ECCS-1542015 as part of the National Nanotechnology Coordinated Infrastructure program. We would like to thank Mr Thomas DiProspero for helpful discussions and for reading over this manuscript.References
- R. J. DeBerardinis, J. J. Lum, G. Hatzivassiliou and C. B. Thompson, Cell Metab., 2008, 7, 11–20 CrossRef CAS PubMed.
- J. Chiche, M. C. Brahimi-Horn and J. Pouyssegur, J. Cell. Mol. Med., 2010, 14, 771–794 CrossRef CAS PubMed.
- G. L. Semenza, Physiology, 2009, 24, 97–106 CrossRef CAS PubMed.
- D. M. Gilkes and G. L. Semenza, Future Oncol., 2013, 9, 1623–1636 CrossRef CAS PubMed.
- G. L. Semenza, Biochim. Biophys. Acta, Mol. Cell Res., 2016, 1863, 382–391 CrossRef CAS PubMed.
- R. K. Bruick and S. L. McKnight, Science, 2001, 294, 1337–1340 CrossRef CAS PubMed.
- D. Wu and P. Yotnda, J. Visualized Exp., 2011, e2899 Search PubMed.
- T. L. Yeh, T. M. Leissing, M. I. Abboud, C. C. Thinnes, O. Atasoylu, J. P. Holt-Martyn, D. Zhang, A. Tumber, K. Lippl, C. T. Lohans, I. K. H. Leung, H. Morcrette, I. J. Clifton, T. D. W. Claridge, A. Kawamura, E. Flashman, X. Lu, P. J. Ratcliffe, R. Chowdhury, C. W. Pugh and C. J. Schofield, Chem. Sci., 2017, 8, 7651–7669 RSC.
- C. P. Bracken, A. O. Fedele, S. Linke, W. Balrak, K. Lisy, M. L. Whitelaw and D. J. Peet, J. Biol. Chem., 2006, 281, 22575–22585 CrossRef CAS PubMed.
- F. A. Auget, L. Gibot and D. Lacroix, Ann. Biomed. Eng., 2013, 15, 177–200 CrossRef PubMed.
- A. G. Tsai, P. C. Johnson and M. Intaglietta, Physiol. Rev., 2003, 83, 933–963 CrossRef CAS.
- L. Wang, W. M. Liu, Y. L. Wang, J. C. Wang, Q. Yu, R. Liu and J. Y. Wang, Lab Chip, 2013, 13, 695–705 RSC.
- C. W. Chang, Y. J. Cheng, M. Tu, Y. H. Chen, C. C. Peng, W. H. Liao and Y. C. Tung, Lab Chip, 2014, 14, 3762–3772 RSC.
- M. A. Acosta, X. Jiang, P. K. Huang, K. B. Cutler, C. S. Grant, G. M. Walker and M. P. Gamcsik, Biomicrofluidics, 2014, 8, 054117 CrossRef.
- M. L. Rexius-Hall, J. Rehman and D. T. Eddington, Integr. Biol., 2017, 9, 742–750 CrossRef CAS PubMed.
- M. B. Byrne, M. T. Leslie, H. S. Patel, H. R. Gaskins and P. J. A. Kenis, Biomicrofluidics, 2017, 11, 054116 CrossRef PubMed.
- J. F. Lo, E. Sinkala and D. T. Eddington, Lab Chip, 2010, 10, 2394–2401 RSC.
- B. Mosadegh, M. R. Lockett, K. T. Minn, K. A. Simon, K. Gilbert, S. Hillier, D. Newsome, H. Li, A. B. Hall, D. M. Boucher, B. K. Eustace and G. M. Whitesides, Biomaterials, 2015, 52, 262–271 CrossRef CAS.
- S. M. Cramer, T. S. Larson and M. R. Lockett, Anal. Chem., 2019, 91, 10916–10926 CrossRef CAS.
- M. D. Brennan, M. L. Rexius-Hall, L. J. Elgass and D. T. Eddington, Lab Chip, 2014, 14, 4305–4318 RSC.
- X. Wang, Z. Liu and Y. Pang, RSC Adv., 2017, 7, 29966–29984 RSC.
- Y. Li, L. Li, X. Liu, M. Ding, G. Luo and Q. Liang, Microfluid. Nanofluid., 2016, 20, 97 CrossRef.
- H. C. Shih, T. A. Lee, H. M. Wu, P. L. Ko, W. H. Liao and Y. C. Tung, Sci. Rep., 2019, 9, 8234 CrossRef.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS.
- B. Gross, S. Y. Lockwood and D. M. Spence, Anal. Chem., 2017, 89, 57–60 CrossRef CAS.
- C. P. Chen, B. T. Mehl, A. S. Munshi, A. D. Townsend, D. M. Spence and R. S. Martin, Anal. Methods, 2016, 8, 6005–6012 RSC.
- H. Klank, J. P. Kutter and O. Geschke, Lab Chip, 2002, 2, 242–246 RSC.
- T. F. Hong, W. J. Ju, M. C. Wu, C. H. Tai, C. H. Tsai and L. M. Fu, Microfluid. Nanofluid., 2010, 9, 1125–1133 CrossRef CAS.
- C. W. Tsao, Micromachines, 2016, 7, E225 CrossRef.
- F. Deiss, A. Mazzeo, E. Hong, D. E. Ingber, R. Derda and G. M. Whitesides, Anal. Chem., 2013, 85, 8085–8094 CrossRef CAS.
- N. A. Whitman, Z. W. Lin, T. J. DiProspero, J. C. McIntosh and M. R. Lockett, Anal. Chem., 2018, 90, 11981–11988 CrossRef CAS.
- M. W. Boyce, R. M. Kenney, A. S. Truong and M. R. Lockett, Anal. Bioanal. Chem., 2016, 408, 2985–2992 CrossRef CAS.
- D. Vordermark, T. Shibata and J. M. Brown, Neoplasia, 2001, 3, 527–534 CrossRef CAS.
- A. S. Truong and M. R. Lockett, Analyst, 2016, 141, 3874–3882 RSC.
- L. Capretto, S. Mazzitelli, S. Focaroli and C. Nastruzzi, Chips and Tips, 2010, 2017 Search PubMed , http://blogs.rsc.org/chipsandtips/2010/07/23/wall-plug-inspired-connectors-for-macro-to-microfluidic-interfacing/.
- I. Arganda-Carreras, V. Kaynig, C. Rueden, K. W. Eliceiri, J. Schindelin, A. Cardona and H. S. Seung, Bioinformatics, 2017, 93, 2424–2426 CrossRef.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ay01690b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |