Open Access Article
Open Access ArticleResidue-specific identification of phase separation hot spots of Alzheimer's-related protein tau†
Susmitha
Ambadipudi
a,
Jithender G.
Reddy
bc,
Jacek
Biernat
d,
Eckhard
Mandelkow
de and
Markus
Zweckstetter
 *ab
*ab
aDeutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), Von-Siebold-Str. 3a, 37075 Göttingen, Germany. E-mail: mazw@nmr.mpibpc.mpg.de
bMax-Planck-Institut für Biophysikalische Chemie, Am Fassberg 11, 37077 Göttingen, Germany
cNMR & Structural Chemistry Division, CSIR-Indian Institute of Chemical Technology, Hyderabad, 500007, India
dDeutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), Ludwig-Erhard-Allee 2, 53175 Bonn, Germany
eCAESAR Research Center, Bonn, MPI for Metabolism Research, Hamburg Outstation, 22607 Hamburg, Germany
First published on 22nd May 2019
Abstract
Liquid–liquid phase separation (LLPS) of proteins enables the formation of non-membrane-bound organelles in cells and is associated with cancer and neurodegeneration. Little is known however about the structure and dynamics of proteins in LLPS conditions, because of the polymorphic nature of liquid-like protein droplets. Using carbon-detected NMR experiments we here show that the conversion of the aggregation-prone repeat region of the Alzheimer's-related protein tau from the dispersed monomeric state to phase-separated liquid-like droplets involves tau's aggregation-prone hexapeptides and regulatory KXGS motifs. Droplet dissolution in presence of 1,6-hexanediol revealed that chemical shift perturbations in the hexapeptide motifs are temperature driven, while those in KXGS motifs report on phase separation. Residue-specific secondary structure analysis further indicated that tau's repeat region exists in extended conformation in the dispersed state and attains transient β-hairpin propensity upon LLPS. Taken together our work shows that NMR spectroscopy can provide high-resolution insights into LLPS-induced changes in intrinsically disordered proteins.
Introduction
In living cells, biochemical reactions are functionally optimized and occur in both membrane-bound (e.g. mitochondria, Golgi) and non-membrane-bound organelles (e.g. nucleoli, stress granules).1,2 Non-membrane-bound organelles are formed by liquid–liquid phase separation (LLPS) of intrinsically disordered proteins and protein regions and are involved in cell signaling and nucleic acid metabolism.1,3,4 Aberrant phase separation has been linked to protein condensation diseases such as neurodegeneration and cancer.5,6 For example, the proteins Fused in Sarcoma (FUS; associated with amyotrophic lateral sclerosis) and tau protein (associated with Alzheimer's disease and tauopathies) undergo LLPS and form liquid-like droplets in solution.7–12 The composition and biophysical environment in the interior of phase separated protein droplets is distinct from the surrounding aqueous environment.2,13,14 NMR spectroscopy is a versatile tool to study the structural propensities of disordered proteins. Because of the dynamic and polymorphic nature of liquid-like protein droplets, however, the atomistic details and local structural preferences of intrinsically disordered proteins within liquid-like droplets remain largely elusive.Pathogenic aggregation of the microtubule-associated protein tau is the hallmark of Alzheimer's disease and tauopathies.15,16 The 441-residue tau is the longest isoform in the adult human brain and comprises two N-terminal inserts, the proline-rich region and four pseudo-repeats in the C-terminal half.17–20 The four pseudo-repeats are important for microtubule-binding and aggregation.21 A truncated tau version that contains only the four pseudo-repeats (termed K18) has higher aggregation propensity than the full-length protein.22 We and others previously showed that both tau and K18 undergo LLPS in a pH- and temperature-dependent manner, droplet formation is modulated by phosphorylation at KXGS motifs and LLPS promotes tau fibrillization and microtubule polymerization.10–12,23
Here, we used 13C-detected NMR experiments to identify the key residues involved in phase separation of the four-repeat region of tau at high resolution. We show that the local chemical environment at tau's aggregation-prone hexapeptides and the regulatory KXGS motifs is altered upon phase separation and droplet formation. Residue stretches spanning these motifs gain transient β-hairpin propensity, while the protein retains the overall disordered nature upon LLPS.
Results and discussion
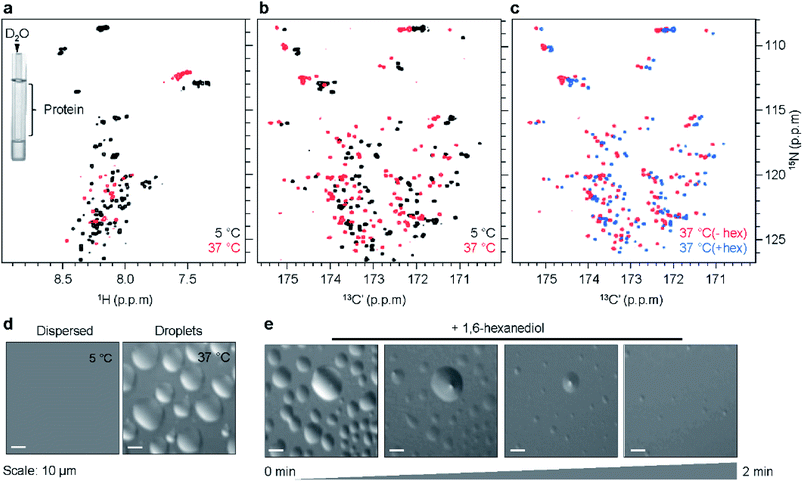
Previous studies showed that K18 undergoes self-coacervation, i.e. phase separation in the absence of co-factors or molecular crowding agents, at 100 μM protein concentration, 37 °C, pH 8.8. K18 droplets form in a temperature-dependent, reversible manner and are liquid-like and dynamic.10 Two-dimensional (2D) 1H/15N NMR correlation spectra of K18 in the monomeric dispersed state at 5 °C and the phase separated state at 37 °C displayed poor signal dispersion (Fig. 1a). In addition, the 1H/15N NMR signals were broadened at 37 °C and pH 8.8 (Fig. 1a). LLPS-induced broadening of tau's NMR signals might be attributed to the conditions of high pH and physiological temperature, which favor rapid exchange of labile amide protons with deuterium atoms of the buffer. To exclude contributions from rapid solvent exchange to NMR signal attenuation, we separated the protein sample from D2O, which is used for field-to-frequency lock during NMR measurements. To this end, a D2O-containing capillary was placed into the NMR sample tube, which contained protein in H2O solvent (Fig. 1a, insert). 1H/15N correlation spectra were then recorded for increasing temperatures from 5 °C to 37 °C (Fig. 1a and S1a†). Droplet formation was confirmed by differential interference contrast (DIC) microscopy (Fig. 1d). Fluorescence microscopy of droplets using Alexa 488 labeled protein showed that the concentration of K18 within the droplets is ∼20–25-fold higher than in the surrounding environment (Fig. S2†).The NMR spectra displayed temperature-dependent chemical shift changes with signal broadening varying across residues. Several of the 1H/15N signals became undetectable above 25 °C (Fig. S1a and b†). The experiments showed that LLPS of K18 induces – independent of rapid solvent exchange – strong NMR signal broadening in 1H/15N correlation spectra. Because little signal broadening was observed in 1H/13C HSQC spectra of phase-separated K18 recorded under identical solution conditions,10 inhomogeneity in magnetic susceptibility anisotropy can be excluded as a major source of broadening. Instead, chemical exchange between different conformations (potentially involving transient hydrogen bonds) might cause line broadening of 1H/15N cross peaks. Consistent with the LLPS-specific nature of NMR signal broadening, other intrinsically disordered proteins showed severe broadening in 1H/15N correlation spectra upon LLPS.7,24
In order to improve spectral resolution and minimize LLPS-induced NMR signal broadening in K18, we recorded 2D 13C-detected NMR experiments.2513CO/15N correlation spectra (CON) are particularly powerful, because the chemical shift dispersion of CO is larger when compared to HN and therefore CON spectra offer better spectral resolution (Fig. 1b and S1c†).26,27 In addition, CON cross-peaks are less affected by solvent exchange at high pH and temperature.25 CON spectra of K18 were recorded in both the monomeric dispersed (5 °C) and phase-separated state (37 °C), keeping sample conditions identical to those used for 1H/15N correlation spectra. Both CON spectra were well-resolved, showed little signal broadening and displayed chemical shift perturbations between 5 °C and 37 °C (Fig. 1b, S1c and d†).
We next asked if the chemical shift changes inherent to temperature changes can be distinguished from those due to LLPS. To this end, we added 1,6-hexanediol, an aliphatic alcohol, to K18 droplets preformed at 37 °C. 1,6-hexanediol dissolves liquid-like droplets in vitro and in cells through disruption of weak interactions between the constituent protein molecules.28 DIC microscopy demonstrated that addition of 3% 1,6-hexanediol dissolved K18 droplets within ∼2 minutes (Fig. 1e). Superposition of CON spectra in the phase-separated (37 °C, without 1,6-hexanediol) and droplet-dissolved states (37 °C, with 1,6-hexanediol) revealed small chemical shift changes (Fig. 1c), suggesting that the two states differ locally in structure and/or dynamics.
The high quality of the 13C-detected NMR experiments at 37 °C suggested that it might be possible to follow the chemical shift perturbations for individual residues, and thus gain residue-specific insights into K18 phase separation. To this end, we assigned the backbone resonances of K18 at 5 °C and pH 8.8, with the help of HNCO,29 HN(C)N,30 HNCACB and HN(CO)CACB31 experiments (Table S1†). The observed chemical shifts were used to assign the peaks (including proline residues) in the CON spectrum at 5 °C. We then recorded a series of CON spectra for increasing temperatures from 5 to 37 °C. High spectral quality was retained for all temperatures (Fig. S1c and d†), in contrast to 1H/15N correlation spectra, where broadening of NMR signals was severe in conditions of LLPS (37 °C) (Fig. S1a and b†). Many of the NMR signals that were broadened beyond detection in the 1H/15N spectra (Fig. S1b†) were from glycine and serine residues (Fig. S1d†). Subsequently, CON spectra at increasing temperatures were sequence-specifically assigned (including prolines) based on the resonance assignment at 5 °C.
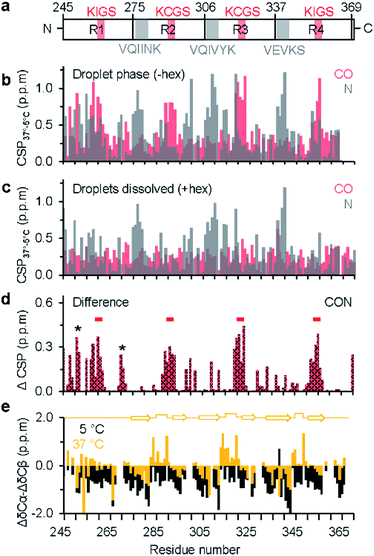
As the dispersed protein proceeded to form droplets when reaching 37 °C, chemical shift perturbation was observed for most K18 residues (Fig. 1b, S1c and d†). Analysis of the perturbations for individual nuclei furthermore showed that CO and N display a distinct sequence behavior. In case of CO, the perturbations are larger in proximity to the KXGS motifs, while for N the perturbation was most pronounced at the hexapeptides (Fig. 2a and b). In presence of 1,6-hexanediol, where the droplets were dissolved, the perturbation (relative to 5 °C) in the hexapeptides was retained, reflecting that these changes are primarily caused by the change in temperature from 5 to 37 °C (Fig. 2c). 1,6-hexanediol showed little or no effect on the chemical shifts of the dispersed protein at 5 °C (Fig. S3†). The combined chemical shift difference plot with and without 1,6-hexanediol (i.e. droplets dissolved vs. K18 droplets at 37 °C) displayed perturbation in the KXGS motifs of K18 (Fig. 2d and S4†). In addition, the proline-rich region and the PGGG motif in repeat R1 were affected by LLPS (Fig. 2d). Consistent with the importance of the KXGS motifs for tau-LLPS, phosphorylation of serine residues in the KXGS motifs by MARK2, a kinase that is involved in regulation of microtubule binding of tau,32 promotes self-coacervation of tau.10,12
We further asked if the changes in chemical shift environment concomitantly induced local structural changes upon LLPS of K18. Our previous study based on amide H and N chemical shift dispersion and far UV circular dichroism of K18 at 37 °C suggested that the protein remains largely disordered within the droplets, in agreement with the intrinsically disordered nature of tau.10 However, these approaches did not allow a residue-specific analysis of the structural content in the phase-separated state. We therefore recorded 2D 13C-detected CBCACO spectra25 of K18 for increasing temperatures from 5 °C (dispersed state) to 37 °C (droplet state) (Fig. S5†). Because CA, CB and CO chemical shifts are sensitive to phi/psi dihedral angles, secondary chemical shifts (Δδ), estimated as the difference between observed and random coil chemical shift values, report on protein secondary structure with positive (negative) deviation of ΔδCA and ΔδCO indicating helical (β-structure) propensity.
First, the CBCACO spectrum at 5 °C was assigned using the assignment obtained at 5 °C by 1H-detected experiments (see above). Next, this assignment was used to follow the temperature-dependent chemical shift changes of CA, CB and CO resonances from 5 °C to 37 °C. Subsequently, temperature-dependent Δδ values of CA, CB and CO were estimated using random coil chemical shifts at pH 8.8 and the given temperature.33 At 5 °C, the secondary structural propensity, calculated as ΔδCA − ΔδCB was negative for almost all residues (Fig. 2e), suggesting an extended K18 conformation. Upon phase separation at 37 °C, V275-H299, S305-H330 and G334-I360, which include the hexapeptides and KXGS motifs, displayed an alternating negative/positive ΔδCA − ΔδCB pattern, as expected for β-hairpin structure (Fig. 2e). A similar trend was observed for CO secondary chemical shifts at 37 °C, in the absence of 1,6-hexanediol (Fig. S6a†). In presence of 1,6-hexanediol where the droplets were dissolved, more negative CO secondary chemical shifts were observed, indicating a higher tendency for extended structure (Fig. S6b†). In the pseudo-repeats R2 and R3, ‘X’ of the KXGS motif corresponds to C291 and C322, respectively. The observed CB chemical shifts of C291 (28.47 ppm) and C322 (28.48 ppm) are characteristic for a reduced thiol group.34 Protein dimerization as a source for the observed changes in secondary structure propensity could therefore be excluded.
A defining property of intrinsically disordered proteins is that they can rapidly exchange between different conformations in solution.35 In this situation, the magnitude of Δδ will be reduced when compared to well-ordered proteins. In conditions of LLPS, the protein also can exchange between the droplet interior and the surrounding environment, potentially resulting in further chemical shift averaging (when the exchange is fast on the NMR time scale). The small changes in secondary structure propensity that were observed for K18 upon LLPS (Fig. 2e) are thus in agreement with the dynamic and polymorphic nature of phase separation of intrinsically disordered proteins. The propensity for β-hairpin formation in repeats 2, 3 and 4 upon LLPS suggests that LLPS favors amyloid promoting structure of tau. Conversion of liquid-like droplets into hydrogels might further stabilize the transient β-hairpin conformation and result in cross-β-structure in amyloid fibrils as previously observed for FUS protein.36
Conclusions
In conclusion, we provided residue-specific insights into liquid–liquid phase separation of the repeat region of tau. We identified hot spots of LLPS and showed that the aggregation-prone hexapeptides and KXGS motifs are important for droplet formation. Our study further demonstrates that 13C-detected NMR spectroscopy is a powerful approach to study liquid–liquid phase separation of intrinsically disordered proteins.Experimental section
Sample preparation
13C/15N-labelled K18 was recombinantly expressed in E. coli BL21(DE3) cells and purified according to published protocols.21,22 Typically, samples contained 100 μM of protein in 50 mM sodium phosphate (pH 8.8), 100 μM of tris(2-carboxyethyl)phosphine and 0.02% sodium azide. For NMR measurements, 50 μM of 4,4-dimethyl-4-silapentane-1-sulfonic acid was added for chemical shift referencing.DIC and fluorescence microscopy
Droplet formation was monitored by DIC microscopy. For quantification, protein was fluorescently labelled with Alexa 488 using Alexa Fluor 488 microscale protein labelling kit (Thermo Fisher Scientific, A30006). Unlabelled and Alexa-488 labelled proteins were mixed in a ratio of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. DIC and fluorescent images were acquired on a DM6000B (Leica) microscope with a 63×-objective (water immersion). Relative protein concentrations in droplets and the surrounding environment were estimated from fluorescence intensities. 15 μl of protein sample was mixed with 7% v/v polyethylene glycol (PEG-3350) and loaded onto a microscope slide with single concavity. For droplet dissolution, 3% of 1,6-hexanediol was added to preformed K18 droplets. Images were acquired every 10 seconds until droplet dissolution was observed. Images were processed using ImageJ.
1. DIC and fluorescent images were acquired on a DM6000B (Leica) microscope with a 63×-objective (water immersion). Relative protein concentrations in droplets and the surrounding environment were estimated from fluorescence intensities. 15 μl of protein sample was mixed with 7% v/v polyethylene glycol (PEG-3350) and loaded onto a microscope slide with single concavity. For droplet dissolution, 3% of 1,6-hexanediol was added to preformed K18 droplets. Images were acquired every 10 seconds until droplet dissolution was observed. Images were processed using ImageJ.
NMR spectroscopy
NMR spectra were recorded on a Bruker 700 MHz spectrometer equipped with a triple resonance cryogenic probe. To avoid solvent exchange effects, protein sample (prepared in H2O solvent) and D2O were separately loaded into a 5 mm Shigemi and a 1.7 mm capillary tube, respectively. The sample was degassed and the tubes were concentrically sealed. For temperature-dependent measurements, the sample was incubated for 20–24 h prior to acquisition at the respective temperature. 1H/15N HSQC spectra were acquired with 200 increments (8 scans per increment) and an inter-scan delay of 1 s. 2D CON and 2D CBCACO experiments were acquired with 1024 and 128 complex points in the direct and indirect dimension, respectively, with 48/128 scans per increment (CON/CBCACO). The inter-scan delay was set to 1 s. NMR spectra were processed using Bruker Topspin 3.5 and analysed in CARA.37 Chemical shift perturbation (CSP) of individual nuclei was plotted as ΔδCO and ΔδN, with ΔδCO = δCO37°C − δCO5°C and ΔδN = δN37°C − δN5°C. Averaged CSPs were calculated as ΔCSP = √0.3 × (ΔδCO)2 + 0.14(ΔδN)2. Secondary chemical shifts were calculated as Δδ = Δδobserved − Δδrandom coil.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
M. Z. was supported by the advanced grant ‘787679 - LLPS-NMR’ of the European Research Council.Notes and references
- D. M. Mitrea and R. W. Kriwacki, Cell Commun. Signaling, 2016, 14, 1 CrossRef PubMed.
- A. A. Hyman, C. A. Weber and F. Julicher, Annu. Rev. Cell Dev. Biol., 2014, 30, 39–58 CrossRef CAS PubMed.
- Y. Lin, D. S. Protter, M. K. Rosen and R. Parker, Mol. Cell, 2015, 60, 208–219 CrossRef CAS PubMed.
- P. A. Chong and J. D. Forman-Kay, Curr. Opin. Struct. Biol., 2016, 41, 180–186 CrossRef CAS PubMed.
- M. Ramaswami, J. P. Taylor and R. Parker, Cell, 2013, 154, 727–736 CrossRef CAS PubMed.
- A. Aguzzi and M. Altmeyer, Trends Cell Biol., 2016, 26, 547–558 CrossRef CAS PubMed.
- K. A. Burke, A. M. Janke, C. L. Rhine and N. L. Fawzi, Mol. Cell, 2015, 60, 231–241 CrossRef CAS PubMed.
- T. Murakami, S. Qamar, J. Q. Lin, G. S. Schierle, E. Rees, A. Miyashita, A. R. Costa, R. B. Dodd, F. T. Chan, C. H. Michel, D. Kronenberg-Versteeg, Y. Li, S. P. Yang, Y. Wakutani, W. Meadows, R. R. Ferry, L. Dong, G. G. Tartaglia, G. Favrin, W. L. Lin, D. W. Dickson, M. Zhen, D. Ron, G. Schmitt-Ulms, P. E. Fraser, N. A. Shneider, C. Holt, M. Vendruscolo, C. F. Kaminski and P. St George-Hyslop, Neuron, 2015, 88, 678–690 CrossRef CAS PubMed.
- A. Patel, H. O. Lee, L. Jawerth, S. Maharana, M. Jahnel, M. Y. Hein, S. Stoynov, J. Mahamid, S. Saha, T. M. Franzmann, A. Pozniakovski, I. Poser, N. Maghelli, L. A. Royer, M. Weigert, E. W. Myers, S. Grill, D. Drechsel, A. A. Hyman and S. Alberti, Cell, 2015, 162, 1066–1077 CrossRef CAS PubMed.
- S. Ambadipudi, J. Biernat, D. Riedel, E. Mandelkow and M. Zweckstetter, Nat. Commun., 2017, 8, 275 CrossRef PubMed.
- X. Zhang, Y. Lin, N. A. Eschmann, H. Zhou, J. N. Rauch, I. Hernandez, E. Guzman, K. S. Kosik and S. Han, PLoS Biol., 2017, 15, e2002183 CrossRef PubMed.
- S. Wegmann, B. Eftekharzadeh, K. Tepper, K. M. Zoltowska, R. E. Bennett, S. Dujardin, P. R. Laskowski, D. MacKenzie, T. Kamath, C. Commins, C. Vanderburg, A. D. Roe, Z. Fan, A. M. Molliex, A. Hernandez-Vega, D. Muller, A. A. Hyman, E. Mandelkow, J. P. Taylor and B. T. Hyman, EMBO J., 2018, 37, e98049 CrossRef PubMed.
- T. J. Nott, T. D. Craggs and A. J. Baldwin, Nat. Chem., 2016, 8, 569–575 CrossRef CAS PubMed.
- A. A. Hyman and C. P. Brangwynne, Dev. Cell, 2011, 21, 14–16 CrossRef CAS PubMed.
- C. Ballatore, V. M. Lee and J. Q. Trojanowski, Nat. Rev. Neurosci., 2007, 8, 663–672 CrossRef CAS PubMed.
- C. Bancher, H. Braak, P. Fischer and K. A. Jellinger, Neurosci. Lett., 1993, 162, 179–182 CrossRef CAS PubMed.
- M. D. Weingarten, A. H. Lockwood, S. Y. Hwo and M. W. Kirschner, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 1858–1862 CrossRef CAS PubMed.
- M. Goedert, M. G. Spillantini, R. Jakes, D. Rutherford and R. A. Crowther, Neuron, 1989, 3, 519–526 CrossRef CAS PubMed.
- K. A. Butner and M. W. Kirschner, J. Cell Biol., 1991, 115, 717–730 CrossRef CAS PubMed.
- N. Gustke, B. Trinczek, J. Biernat, E. M. Mandelkow and E. Mandelkow, Biochemistry, 1994, 33, 9511–9522 CrossRef CAS PubMed.
- M. D. Mukrasch, J. Biernat, M. von Bergen, C. Griesinger, E. Mandelkow and M. Zweckstetter, J. Biol. Chem., 2005, 280, 24978–24986 CrossRef CAS PubMed.
- H. Wille, G. Drewes, J. Biernat, E. M. Mandelkow and E. Mandelkow, J. Cell Biol., 1992, 118, 573–584 CrossRef CAS PubMed.
- A. Hernandez-Vega, M. Braun, L. Scharrel, M. Jahnel, S. Wegmann, B. T. Hyman, S. Alberti, S. Diez and A. A. Hyman, Cell Rep., 2017, 20, 2304–2312 CrossRef CAS PubMed.
- J. P. Brady, P. J. Farber, A. Sekhar, Y. H. Lin, R. Huang, A. Bah, T. J. Nott, H. S. Chan, A. J. Baldwin, J. D. Forman-Kay and L. E. Kay, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E8194–E8203 CrossRef CAS PubMed.
- I. C. Felli, L. Gonnelli and R. Pierattelli, Nat. Protoc., 2014, 9, 2005–2016 CrossRef CAS PubMed.
- L. Duma, S. Hediger, A. Lesage and L. Emsley, J. Magn. Reson., 2003, 164, 187–195 CrossRef CAS PubMed.
- W. Bermel, I. Bertini, L. Duma, I. C. Felli, L. Emsley, R. Pierattelli and P. R. Vasos, Angew. Chem., 2005, 44, 3089–3092 CrossRef CAS PubMed.
- S. Kroschwald, S. Maharana, D. Mateju, L. Malinovska, E. Nuske, I. Poser, D. Richter and S. Alberti, eLife, 2015, 4, e06807 CrossRef PubMed.
- M. J. Blackledge, R. Bruschweiler, C. Griesinger, J. M. Schmidt, P. Xu and R. R. Ernst, Biochemistry, 1993, 32, 10960–10974 CrossRef CAS PubMed.
- S. C. Panchal, N. S. Bhavesh and R. V. Hosur, J. Biomol. NMR, 2001, 20, 135–147 CrossRef CAS PubMed.
- A. Yamazaki, S. Kaya, T. Tsuda, Y. Araki, Y. Hayashi and K. Taniguchi, J. Biochem., 1994, 116, 1360–1369 CrossRef CAS PubMed.
- J. Biernat, N. Gustke, G. Drewes, E. M. Mandelkow and E. Mandelkow, Neuron, 1993, 11, 153–163 CrossRef CAS PubMed.
- M. Kjaergaard and F. M. Poulsen, J. Biomol. NMR, 2011, 50, 157–165 CrossRef CAS PubMed.
- O. A. Martin, M. E. Villegas, J. A. Vila and H. A. Scheraga, J. Biomol. NMR, 2010, 46, 217–225 CrossRef CAS PubMed.
- K. Tamiola, B. Acar and F. A. Mulder, J. Am. Chem. Soc., 2010, 132, 18000–18003 CrossRef CAS PubMed.
- D. T. Murray, M. Kato, Y. Lin, K. R. Thurber, I. Hung, S. L. McKnight and R. Tycko, Cell, 2017, 171, 615–627 CrossRef CAS PubMed.
- R. Keller, 2004, http://www.cara.ethz.ch/Wiki/FrontPage.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc00531e |
| This journal is © The Royal Society of Chemistry 2019 |