Open Access Article
Open Access ArticleUltraspecific live imaging of the dynamics of zebrafish neutrophil granules by a histopermeable fluorogenic benzochalcone probe†
Emma
Colucci-Guyon
 *ab,
Ariane S.
Batista‡
*ab,
Ariane S.
Batista‡
 c,
Suellen D. S.
Oliveira‡
c,
Suellen D. S.
Oliveira‡
 d,
Magali
Blaud
d,
Magali
Blaud
 e,
Ismael C.
Bellettini
e,
Ismael C.
Bellettini
 f,
Benoit S.
Marteyn
f,
Benoit S.
Marteyn
 gh,
Karine
Leblanc
i,
Philippe
Herbomel
gh,
Karine
Leblanc
i,
Philippe
Herbomel
 ab and
Romain
Duval
ab and
Romain
Duval
 *j
*j
aInstitut Pasteur, Unité Macrophages et Développement de l'Immunité, Paris, 75015, France. E-mail: emma.colucci@pasteur.fr
bCNRS, UMR 3738, Paris, France
cNanotechnology Engineering Program, Instituto Alberto Luiz Coimbra de Pós-Graduação e Pesquisa de Engenharia – COPPE, Universidade Federal do Rio de Janeiro, Rio de Janeiro, 21941-972, Brazil
dDepartment of Anesthesiology, University of Illinois, Chicago, 60612, USA
eLCRB, CNRS, Université Paris 5, Sorbonne Paris Cité, Paris, 75006, France
fDepartamento de Ciências Exatas e Educaçao, Universidade Federal de Santa Catarina, Blumenau, 89036-256, Brazil
gInstitut Pasteur, Unité de Pathogénie Microbienne Moléculaire, Paris, 75015, France
hINSERM, UMR 786, Paris, France
iBioCIS, CNRS, Université Paris-Sud 11, Châtenay-Malabry, 92290, France
jMERIT, IRD, Université Paris 5, Sorbonne Paris Cité, Paris, 75006, France. E-mail: romain.duval@ird.fr
First published on 14th February 2019
Abstract
Neutrophil granules (NGs) are key components of the innate immune response and mark the development of neutrophilic granulocytes in mammals. However, there has been no specific fluorescent vital stain up to now to monitor their dynamics within a whole live organism. We rationally designed a benzochalcone fluorescent probe (HAB) featuring high tissue permeability and optimal photophysics such as elevated quantum yield, pronounced solvatochromism and target-induced fluorogenesis. Phenotypic screening identified HAB as the first cell- and organelle-specific small-molecule fluorescent tracer of NGs in live zebrafish larvae, with no labeling of other cell types or organelles. HAB staining was independent of the state of neutrophil activation, labeling NGs of both resting and phagocytically active neutrophils with equal specificity. By high-resolution live imaging, we documented the dynamics of HAB-stained NGs during phagocytosis. Upon zymosan injection, labeled NGs were rapidly recruited to the forming phagosomes. Despite being a reversible ligand, HAB could not be displaced by high concentrations of pharmacologically relevant competing chalcones, indicating that this specific labeling was the result of the HAB's precise physicochemical signature rather than a general feature of chalcones. However, one of the competitors was discovered as a promising interstitial fluorescent tracer illuminating zebrafish histology, similarly to BODIPY-ceramide. As a yellow-emitting histopermeable vital stain, HAB functionally and spectrally complements most genetically incorporated fluorescent tags commonly used in live zebrafish biology, holding promise for the study of neutrophil-dependent responses relevant to human physiopathology such as developmental defects, inflammation and infection. Furthermore, HAB intensely labeled isolated live human neutrophils at the level of granulated subcellular structures consistent with human NGs, suggesting that the labeling of NGs by HAB is not restricted to the zebrafish model but also relevant to mammalian systems.
The zebrafish (Danio rerio), a precious vertebrate model in developmental biology, has recently emerged as a powerful non-mammalian model to study the development and the function of the immune system and to address the host–pathogen interaction in the context of an entire live organism. The small size and the natural translucency of the zebrafish embryo and swimming larva make it possible to follow leukocyte deployment, behavior and functions in vivo at high resolution throughout the organism.1,2 Maturation and deployment of myeloid cell lineages have been characterized for more than a decade. Zebrafish possesses a multi-lineage myeloid compartment with two types of granulocytes (heterophils/neutrophils and eosinophils), and monocytes/macrophages, each with characteristic morphological and histochemical features. Macrophages appear during the first day of zebrafish development, followed by neutrophils that arise a day later, both leukocytes representing together a first, efficient immune system for the developing fish.3–5 Zebrafish neutrophils are morphologically and functionally similar to their mammalian counterpart. They are equipped with granules containing microbicidal compounds, they engulf and kill invading microorganisms, and have been extensively studied for their key roles in innate immunity and inflammation. The availability of transgenic fish lines expressing GFP or other fluorescent proteins under the control of specific neutrophil promoters allows the live imaging of neutrophil behavior in the context of the entire organism.6,7 Although several small-molecule tracers of neutrophil granules have been described (e.g., quinacrine, LysoTrackerRed®, fluorescent tyramide conjugates or Sudan Black staining for endogenous peroxidase-containing granules), they are either non-specific in terms of labeled organelles or limited to fixed samples and therefore unusable as vital stains for live animal imaging.5,8,9
In our development of fluorescent organic dyes for biological applications, we searched for small size, non-canonical fluorophores with novel structure–fluorescence relationships (SFRs),10 and identified the chalcone scaffold as such a motif. Chalcones (1,3-diphenyl-2-propen-1-ones) have been investigated as fluorescent chemosensors for several analytical applications,11–13 and as pharmacological probes in a few cases.14–17 However, their development as fluotracers has remained hampered by suboptimal photophysics such as minute to modest quantum yields,11–13 by poor cytopermeabilities limiting biological labeling to cell surfaces14,15 or extracellular deposits,17 by substantial non-specific labeling,16 and by weakly documented SFRs.14–16,18 These constitute severe drawbacks for potential fluorophores that are otherwise compact, easy to synthesize and also spectrally tractable by chemical modulation. They define the challenges to be addressed in the design of an optimal chalcone probe for biological studies.
Results
Probe synthesis and fluorescence structure–activity relationships
Chalcone itself is intrinsically non-fluorescent. Towards emissive chalcones possessing optimal photophysics and high cytopermeability for live-cell and live-animal imaging, we rationally delineated minimal structural modifications of the 1,3-diarylpropenone system (Fig. 1). Organic fluorophores are classically composed of push–pull moieties connected by means of an extended conjugation, allowing efficient excited state intramolecular charge transfers (ICTs).10 Accordingly, an electron donating aromatic ring (B) would be conjugated to the enone system, with the arylcarbonyl (A) acting as the electron-withdrawing moiety in the chalcone system (Fig. 1). The electron donating aryl was chosen as a naphthyl group, known to increase fluorescence intensity while exerting bathochromic effects with respect to the phenyl group.10,19 This shift is useful for microscopic imaging as it diminishes light scattering, cellular auto-fluorescence and phototoxicity.20,21 In addition, the more lipophilic naphthyl nucleus would dramatically increase the ability of a given probe to penetrate cells and organelles. We finally settled on a pivotal primary amino group as the electron donating substituent, having in mind its useful transformation to the azide for fluorogenic photolabeling strategies of cells or proteomes.22 Accordingly, we targeted the 3-aminobenzochalcone core as the optimal fluorophore in this series, featuring the most extended captodative system, and expected to display maximal fluorescence intensity and bathochromism (Fig. 1).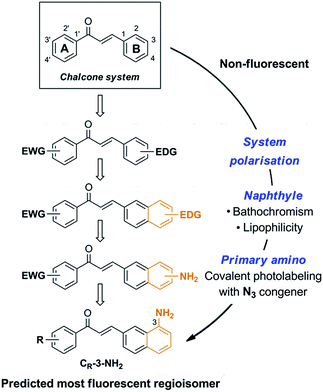 | ||
| Fig. 1 Stepwise design of a minimal cytopermeable chalcone fluorophore (EWG, electron withdrawing group; EDG, electron donating group; R, any substituent). | ||
These 3-aminobenzochalcones were obtained by reduction of the corresponding nitro intermediates, accessed by Claisen–Schmidt condensation of various acetophenones with 8-nitro-2-naphthaldehyde 2. Starting from β-naphthaldehyde 1,23 a small library of benzochalcones 3–8 was thus obtained where the electron density of the acetophenyl part was tuned from severely deficient (R = 4′-CF3) to strongly enriched (R = 4′-OH) (Fig. 2A). Two close analogues were synthesized for the physicochemical standardization of this new series: (i) 5-aminobenzochalcone 12 is a regioisomer of 3-aminobenzochalcone 8, possessing a less extended captodative system as a pseudo-para conjugation; (ii) 2-aminochalcone 14 is the exact chalcone congener of 3-aminobenzochalcone 6, and should permit to quantify the influence of the naphthyl ring both in terms of fluorescence and lipophilicity. Compound 12 was obtained from β-methylnaphthalene 9 upon regioselective nitration24 and then methyl oxidation. The obtained 6-nitro-2-naphthaldehyde 11 was converted to 5-aminobenzochalcone 12 by a “one-pot” condensation–reduction as for aminochalcones 3–8. 2-Aminochalcone 14 was obtained from 2-nitrobenzaldehyde 13 following the same strategy (Fig. 2A) (see ESI† for all synthetic procedures).
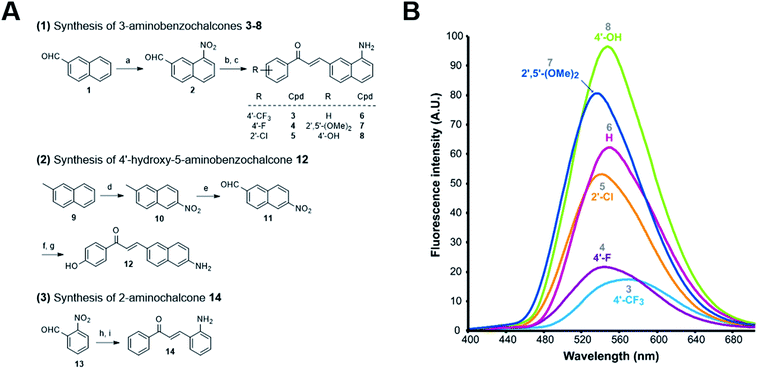 | ||
| Fig. 2 Synthesis of representative compounds in the 3-aminobenzochalcone, 5-aminobenzochalcone and 2-aminochalcone series and SFRs in the 3-aminobenzochalcone series. (A) Reagents and conditions: (a) HNO3 (40 equiv.), H2SO4 (1.5 equiv.), AcOH, r. t., 13% 2; (b) substituted acetophenone (1 equiv.), NaOH (0.75–2 equiv.), EtOH, r. t.; (c) SnCl2 (5–10 equiv.) or cat. Pd–C (10%), H2, AcOH, r. t., 38% 3, 31% 4, 26% 5, 37% 6, 18% 7, 69% 8; (d) HNO3 (1.25 equiv.), Ac2O, r. t., 11% 10; (e) SeO2 (2 equiv.), neat, 150 °C, 28% 11 (45% based on conversion); (f) 4-hydroxyacetophenone (1 equiv.), NaOH (2 equiv.), EtOH, r. t.; (g) Fe (10 equiv.), AcOH, reflux, 56% 12; (h) acetophenone (1 equiv.), NaOH (0.75 equiv.), EtOH, r. t.; (i) Pd–C (10%), H2, AcOH, r. t., 12% 14 (yields of chalcones indicated for two-step, “one-pot” reactions). See ESI† for all synthetic procedures. (B) Semi-quantitative emission spectra of 3-aminobenzochalcones 3–8 (50 μM) in toluene at 20 °C. | ||
3-Aminobenzochalcones were indeed fluorescent compounds, in agreement with our design of this novel fluorophore. However, while electron-withdrawing acetophenyl rings were expected to yield the most fluorescent compounds (Fig. 1), the strongest fluorescence was associated with the electron donating 4′-hydroxy group (cpd 8), while a 4′-trifluoromethyl group (cpd 3) induced a much weaker fluorescence (Fig. 2B). The spectral properties of these fluorophores are given in Table S1 in the ESI† and depicted in Fig. 3. 3-Aminobenzochalcones optimally display high Stokes shifts predicting an absence of internal quenching, with emission in the green-yellow region of the visible spectrum in toluene. As anticipated from Fig. 2B, 4′-hydroxy-3-aminobenzochalcone 8 (HAB), with a 54.0% quantum yield, was significantly more fluorescent than its non-substituted congener 6. This relation is validated by the further weaker quantum yield of 38.2% of the p-trifluoromethyl derivative 3. Importantly, the favourable influence of a strongly electron donating A ring called for revision of fluorophore representation in the chalcone series, with permanent enolization of the carbonyl occurring in an extended conjugation pattern similarly to the prototypical fluorescein or rhodamine fluorophores (Fig. 3). Further, HAB was far more fluorescent than its 5-amino regioisomer 12 with a ca. 20-fold higher quantum yield.
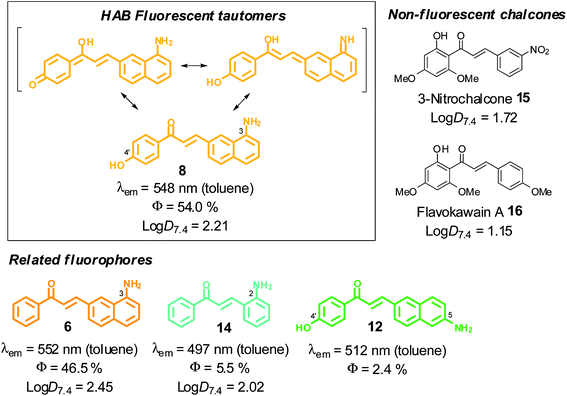 | ||
Fig. 3 SFRs and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 values at 20 °C in the 3-aminobenzochalcone, 5-aminobenzochalcone and 2-aminochalcone series (colours indicative of fluorescence emissions). D7.4 values at 20 °C in the 3-aminobenzochalcone, 5-aminobenzochalcone and 2-aminochalcone series (colours indicative of fluorescence emissions). | ||
This enormous difference in quantum yield between HAB and 12 cannot be explained solely by conjugation patterns: HAB features rotational hindrance of the 3-amino group (having peri relationships with H-2), a phenomenon absent in the 5-amino regioisomer 12.25,26 As a result, HAB should be less susceptible to non-radiative de-excitation than 12, thereby favouring fluorescence transitions. Comparing the isoelectronic compounds 3-aminobenzochalcone 6 and 2-aminochalcone 14 showed as expected that the substitution of the (B) phenyl group for a naphthyl in 6 resulted in much stronger fluorescent emission. Compound 6 thus possesses a quantum yield over eight-fold higher than 14, the latter also displaying cyan emission and a smaller Stokes shift. Noteworthy for future biological studies, the quantum yield of 54.0% for the most fluorescent aminobenzochalcone HAB is higher than those of dansylamides (e.g., DPP)27 and indocyanines (e.g., ICG),28 prototypical fluorescent probes for live-cell and live-animal imaging, respectively, and is also, to our knowledge, the highest described to date in the chalcone series (see Table S1 in the ESI†).
Solvatochromic behavior
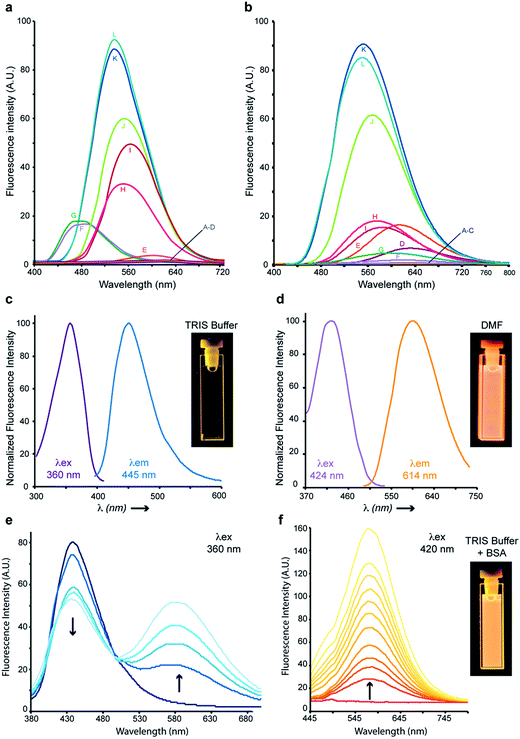
HAB and its non-substituted analogue 6 exhibited pronounced solvatochromism (Δmaxλem ∼ 140 nm), being virtually non-fluorescent in water, biological buffers and alcohols, moderately emissive in polar aprotic solvents (e. g., ethyl acetate) and strongly fluorescent in non-polar solvents (e.g., toluene), with the exception of alcanes. While 6 was significantly fluorescent in n-hexane and n-octane, its 4′-hydroxy derivative HAB proved undetectable in these solvents, and showed only weak fluorescence in n-alkanols used to model biological lipids (e.g., n-heptanol) (Fig. 4a and b). Although the lack of fluorescence of HAB in biological buffers may be perceived as a limitation for imaging studies, it must be realized that the binding of HAB to the amphipathic affinity sites of protein targets should result in a 4–500 fold fluorescence turn-on (FTO) according to Fig. 4b.10 As a proof-of-principle, bovine serum albumin (BSA) quenched HAB's weak blue fluorescence (λex = 360 nm, λem = 445 nm) in a dose–response manner while inducing the formation of a strongly yellow-emitting HAB-protein fluorescent complex (λex = 420 nm, λem = 578 nm) (Fig. 4e and f). A FTO of 21.5 (excitation at 420 nm, at a BSA concentration of 10 mg ml−1) was observed upon binding of HAB to the protein (Fig. 4f), associated with an important bathochromic shift of its fluorescence spectrum (Δλex = 60 nm, Δλem = 133 nm) which was similar to that obtained in the polar aprotic solvent DMF (Fig. 4d–f). This behaviour is opposite to that of chalcones binding the physiological sites of BSA and displaying slight hypsochromic shifts and quenching effects indicative of an apolar environment,18 suggesting that HAB binds non-conventional BSA sites. Overall, HAB should feature a high signal-to-noise ratio and elevated pharmacological sensitivity, showing low or negligible fluorescence in compartments such as the extracellular medium, the cytosol and the membranes in free form, and high fluorescence when target-bound (“in-target” fluorogenesis). Moreover, its absence of fluorescence in water or buffers has the potential to greatly simplify and time-optimize the staining protocols, with suppression of tedious washing steps and diffusion liabilities following dye administration.Cytopermeability evaluation
Lipophilicity is a key physicochemical parameter affecting cellular penetration and subcellular distribution in vitro, as well as pharmacokinetics in vivo.29 We measured the partition coefficients of key benzochalcones compared to reference chalcones using n-octanol and pH 7.4 phosphate buffered saline (PBS), to draw structure–cytopermeability relationships in this novel series (Fig. 3). As expected, the favorable influence of a naphthyl vs. a phenyl ring regarding fluorescence was also exerted in terms of lipophilicity, as 3-aminobenzochalcone 6 and HAB possessed log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 values significantly higher than that of mere 2-aminochalcone 14. In particular, 6 was almost three times more lipophilic than its exact 2-aminochalcone congener 14. On the other hand, the presence of a 4′-hydroxy group on HAB made it 2-fold more hydrophilic than its 4′-deshydroxy congener 6. Further, HAB was three to eleven times more lipophilic than 3-nitrochalcone 15 and natural flavokawain A 16 chosen as representative biologically active chalcones30,31 (Fig. 3). Overall, the neat amphipathicity (log
D7.4 values significantly higher than that of mere 2-aminochalcone 14. In particular, 6 was almost three times more lipophilic than its exact 2-aminochalcone congener 14. On the other hand, the presence of a 4′-hydroxy group on HAB made it 2-fold more hydrophilic than its 4′-deshydroxy congener 6. Further, HAB was three to eleven times more lipophilic than 3-nitrochalcone 15 and natural flavokawain A 16 chosen as representative biologically active chalcones30,31 (Fig. 3). Overall, the neat amphipathicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 between 1.5 and 2.5), and neutrality as well as small molecular weight (289.3 Da) of HAB
D7.4 between 1.5 and 2.5), and neutrality as well as small molecular weight (289.3 Da) of HAB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32 predict an optimal histopermeability for this compound, which thus could behave as a fast-acting fluorescent probe in vivo.
32 predict an optimal histopermeability for this compound, which thus could behave as a fast-acting fluorescent probe in vivo.
HAB labels specific cells in live zebrafish embryos and swimming larvae
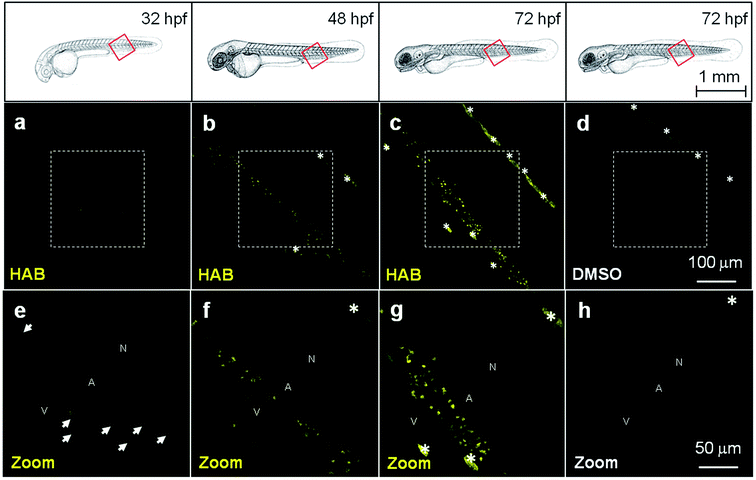
HAB, the most fluorescent, solvatochromic and histopermeable derivative obtained, was tested in representative transparent animal models. We chose the nematode Caenorhabditis elegans and the zebrafish Danio rerio to study the cell and organelle distribution of HAB in complete live organisms and assess its biological specificity. HAB could not be microscopically visualized in C. elegans due to excessive autofluorescence of the nematode (data not shown). On the other hand, HAB displayed a unique labeling pattern in zebrafish that deserved in-depth investigation. When HAB was incubated at 10 μM for 60 min on live zebrafish embryos and swimming larvae ranging from 24 hours post-fertilization (hpf) to 72 hpf, fluorescence emission was detected mainly in the 550–650 nm window following excitation at 488 nm, indicative of a non-aqueous environment (see Fig. 4b for the solvatochromic properties of HAB, and Fig. S2 in the ESI† for the emission spectrum of HABin vivo). Since the fluo-labeling was maximal in the yellow-orange region of the visible spectrum, this suggested that HAB was bound to specific proteins with strong target-induced bathochromic fluorogenesis and FTOs (Fig. 4e and f). This labeling displayed a neat pattern of punctuated areas that first appeared by 32 hpf in the ventral tail of the embryo (Fig. 5a and e). The number and intensity of HAB fluorescent spots progressively increased at 48 hpf and 72 hpf (Fig. 5b, f and c, g) following the known deployment of macrophages and neutrophils during zebrafish development,4,5 suggesting that these cell types could be targeted by HAB. The shape and size of the fluorescent spots within the caudal region of the specimens suggested labeling of some subcellular compartment rather than whole cells.5 It is noteworthy that the already autofluorescent pigment cells present in 72 hpf larvae showed a significant increase of fluorescence in the presence of HAB (Fig. 5c and d) suggesting that the probe also accumulated within these cells due to their high content of aromatic pigments (i.e., pteridine and guanine derivatives).33,34HAB labeling is reversible
HAB labeling was found to be fairly photostable under the conditions used so far, with ca. 50% fluorescence lost after 30 min of continuous illumination at 488 nm. However, since the embryos and larvae were systematically washed and mounted in HAB-free agarose for live imaging, the diffusion of HAB out of its binding sites might be responsible for the loss of labeling overtime. To discriminate between free diffusion and photobleaching as the two possible causes of signal loss, we performed time-lapse imaging experiments in which either of these two phenomena was kept to a minimum. To test the free diffusion hypothesis, 72 hpf zebrafish larvae were treated with 10 μM HAB for 1 h, washed and mounted in tracer-free mounting medium, and then imaged over time with short exposure-acquisition pulses every 10 min. Under these conditions of lower illumination hence limited photobleaching, the detection of HAB-labeled structures was no longer possible after ca. 1 h, with again ca. 50% of fluorescence lost after 30 min exposure (see Fig. S3 in the ESI†). To test the photobleaching hypothesis, we treated the 72 hpf larvae with 10 μM HAB for 1 h and mounted them in agarose and tricaine containing 10 μM HAB. We then imaged the zebrafish larva embedded in HAB 10 μM over time under continuous illumination by the 488 nm laser. Under these conditions of maximum photobleaching and abolished free diffusion, HAB labeling proved highly photostable, as it could be detected for up to 15 h of time-lapse imaging (see Video S4 in the ESI†). This observation indicates that HAB bound in vivo continually exchanges with free non-fluorescent HAB, at a rate faster than the photobleaching of the bound HAB during continuous 488 nm laser illumination. These conditions of constant equilibrium by continuous contact, made possible by the absence of HAB fluorescence in aqueous media, constitute optimal settings for HAB labeling on the zebrafish model and are particularly suitable for long-term experiments.HAB labels neutrophil granules in live zebrafish larvae
To identify the cell type(s) labeled by HAB, which according to their localization and developmental timing of detection could be the first leukocytes of the zebrafish embryo and larva (macrophages and/or neutrophils), we stained zebrafish transgenic 72 hpf larvae harbouring fluorescent macrophages or neutrophils35–37 with HAB. Upon treatment with 10 μM HAB of Tg(mfap4:mCherry-F) larvae possessing red-fluorescent macrophages expressing mCherry, we found that the probe did not co-localize with the red-fluorescent macrophages (Fig. 6a–d). Due to the wide emission range of the mCherry protein (560–760 nm), which overlapped with that of the yellow-orange emitting HAB, mCherry was imaged following excitation at 552 nm and collecting only the “far red” portion of its fluorescence emission (660–750 nm), and HAB was imaged following excitation at 448 nm and collecting the 550–650 nm range of its emission spectrum. Upon treatment with 10 μM HAB of Tg(mpx:GAL4/UAS-E1b:nsfB-mCherry) larvae harbouring neutrophils expressing mCherry and using the same HAB/mCherry acquisition parameters as above, we could show that HAB perfectly colocalized with all neutrophils of the imaged ventral tail region, labeling a specific compartment within them (Fig. 6e–h). Using GFP-labeled neutrophils (Tg(mpx:GFP) strain), and sequentially detecting GFP emission in the 500–520 nm range and HAB using the “red” portion of its emission spectrum (>600 nm) by exciting both fluorophores by the 448 nm laser (Fig. 6i–l) we confirmed that HAB targeted a neutrophil-specific compartment. Not only did HAB stain all neutrophils of the ventral tail region of 72 hpf larvae, where they are known to be particularly abundant,2,4,37 but HAB-labeled neutrophils could also be found around the eye, ear and in the yolk sac of 72 hpf larvae (data not shown), indicating that HAB labels the entire neutrophil population in vivo. Collectively, these observations demonstrated that zebrafish neutrophils are the cell type selectively targeted by HAB, which specifically labels a compartment within these cells. To identify the compartment targeted by HAB within neutrophils, we performed high-resolution confocal imaging of HAB-labeled cells in 72 hpf larvae harbouring red neutrophils, combining the fluorescence detection of HAB and mCherry with DIC microscopy. HAB was found to co-localize with neutrophil granules, which are easily recognizable in transmitted light/DIC imaging5,38 for they are refractile and constantly moving in the cytoplasm (Fig. 6m–p). This colocalization was unambiguously demonstrated by the perfect overlap of fluorescence HAB signals and DIC images during time-lapse acquisitions of live patrolling neutrophils (see high-resolution Videos S5 and S6 in the ESI†). Similar to their mammalian counterpart, the granules of zebrafish neutrophils contain myeloperoxidase.2,38,39 The diazo dye Sudan Black B (SB) is a classical lipid stain that specifically labels the myeloperoxidase-positive granules of zebrafish neutrophils,5,40 as also observed in human neutrophils.41,42 The first SB-stained granules are detected by 32–35 hpf in zebrafish embryos, within the first, still immature embryonic neutrophils; this staining then increases in number and intensity, revealing the deployment of neutrophils during zebrafish development.5 To check that HAB was targeting the same granule population as SB in neutrophils, we tried to combine the two dyes. We first tested if the HAB label was compatible with the fixation protocol classically used to fix zebrafish embryos and larvae, but failed to detect HAB labeling in the larva post-fixation for two hours at room temperature with either 1 or 4% formaldehyde, or when performing HAB labeling before the fixation. This could be due to the loss of HAB pharmacology in fixed samples, or to the reaction of HAB with formaldehyde, leading to fluorophore destruction: even though HAB is both a very weak organic base (predicted pKa value of 3.97 for its amino tautomers, see Fig. S7 in the ESI†) and a weak nucleophile, possessing the electronic features of a vinylogous amide, this possibility cannot be ruled out in the absence of a dedicated study. In any case, it was not possible to combine the SB staining with the HAB labeling.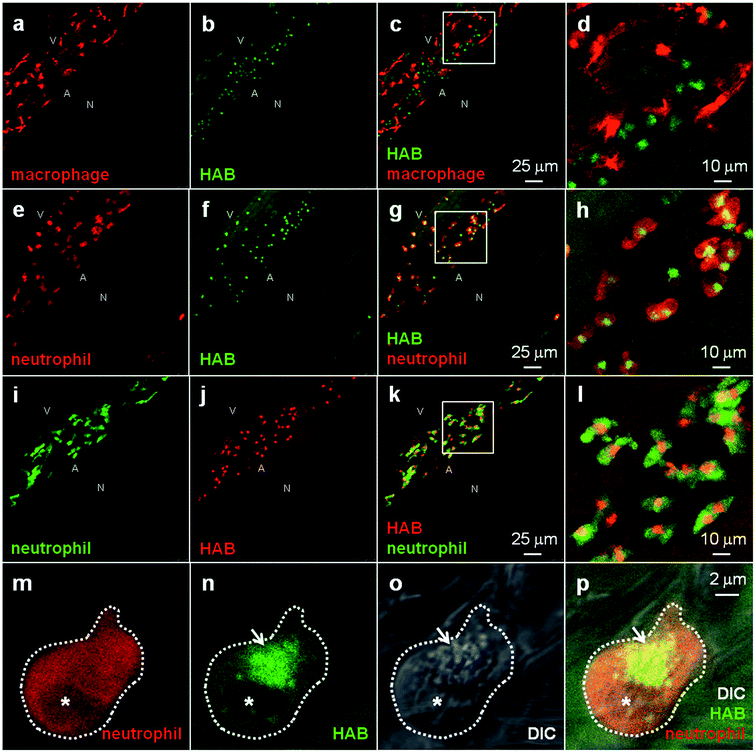 | ||
| Fig. 6 HAB labels zebrafish neutrophil granules. (a–l) Confocal fluorescence imaging of HAB (10 μM) in live transgenic 72 hpf zebrafish larvae following excitation at 448 nm under equilibrium conditions. Detection parameters were as follows: for HAB/mCherry: λex 448 nm, λem 550–650 nm, mCherry: λex 552 nm, λem 660–750 nm using sequential modes of acquisition. For GFP/HAB: GFP λex 448 nm, λem 500–520 nm, HAB: λex 448 nm, λem 550–650 nm using sequential modes of acquisition. Maximum intensity Z-projection images (2 μm serial optical sections) are shown. (m–p) High-resolution DIC and confocal fluorescence imaging of HAB (10 μM) in live 72 hpf zebrafish larvae harbouring red neutrophils. Arrows point to HAB-labeled granules according to the DIC images in neutrophils. A single 0.4 μm optical section is shown. Boxes in (c), (g) and (k) indicate the regions magnified in the insets (d), (h) and (l) respectively. Abbreviations used: A (aorta); N (notochord); V (vein); asterisk = nucleus. See ESI for Videos S5 and S6† related to (m–p). | ||
HAB reveals the behavior of neutrophil granules during phagocytosis in live zebrafish larvae
To circumvent this technical limitation, we sought further evidence for the identity of neutrophil granules as the targets of HAB. One of the most important functions of neutrophils is to eliminate invading microorganisms. Neutrophils are highly phagocytic cells, able to engulf and kill microbes with the arsenal of microbicidal compounds stored in their granules. Once they have engulfed microbes, neutrophils release the granule content (e.g., myeloperoxidase-produced hypochlorous acid and proteases) into the phagosome. We have shown that in vivo, zebrafish neutrophils degranulate into the phagosome following microbe phagocytosis and that the myeloperoxidase activity initially contained in the granules often relocalises to the phagosome, while the SB staining correlatively disappears.43 These steps have been advantageously reproduced with opsonized zymosan particles in cultured mammalian neutrophils, to show that neutrophil granules are recruited to the nascent phagosome in which they deliver their content by exocytosis.8,44 To determine whether HAB would allow the visualization of neutrophil granule dynamics during phagocytosis in vivo, we vitally stained a zebrafish larva with HAB, then subcutaneously injected red-fluorescent Cy5-zymosan, and immediately monitored the interaction between HAB-labeled neutrophil granules and zymosan particles by high resolution confocal imaging (Fig. 7). We found that upon zymosan phagocytosis, HAB-labeled granules are massively recruited to the particle-containing phagosomes (Fig. 7a, b, d and related Video S8† in ESI), similar to the myeloperoxidase containing SB-labeled granules which we found relocalised around the zymosan-containing phagosome in the fixed larva (Fig. 7c). These observations strongly suggest that the myeloperoxidase-containing granules are the targets of HABin vivo. They also represent, to our knowledge, the first documentation of the dynamics of neutrophil degranulation upon phagocytosis in an entire live organism.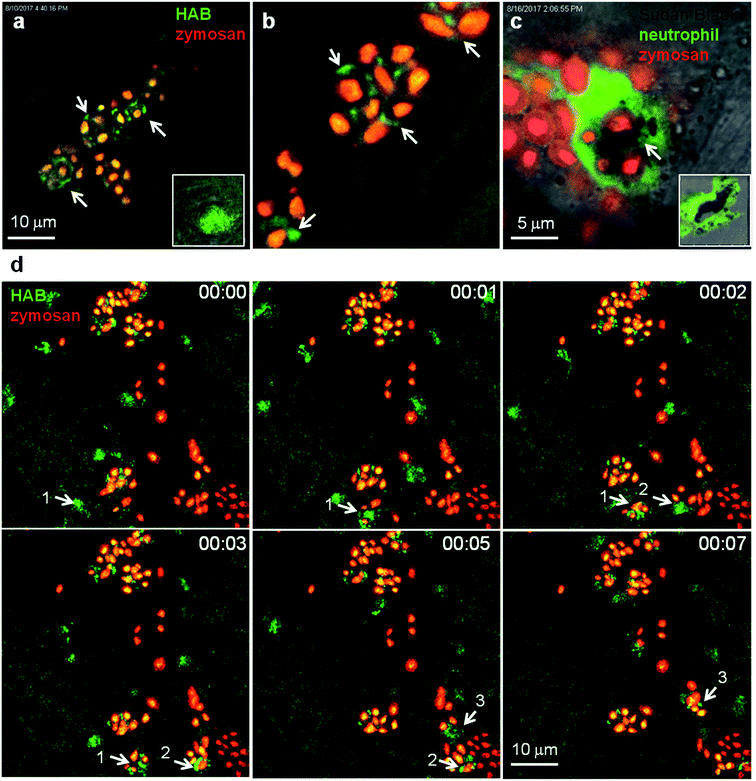 | ||
| Fig. 7 HAB reveals the dynamics of neutrophil granules upon phagocytosis of zymosan particles in live zebrafish. (a and b) Confocal live imaging of HAB-labeled neutrophil granules upon phagocytosis of subcutaneously injected zymosan in a live 72 hpf zebrafish larva under diffusion conditions. HAB is recruited to the forming phagosomes (arrows). Inset: HAB labeling of a resting neutrophil. (c) Sudan Black (SB) staining of myeloperoxidase-containing neutrophil granules showing granule recruitment to the phagosome upon zymosan phagocytosis in fixed zebrafish larvae. Inset: SB staining of a resting neutrophil; a single 1 μm optical section is shown. (d) Frames extracted from an in vivo time-lapse confocal imaging sequence (time step = 1 min). Arrows point to HAB-labeled neutrophil granules that are recruited to the nascent zymosan containing phagosome. Three neutrophils (pointed with number 1 to 3) were tracked during the time lapse sequence. Maximum intensity Z-projection (1 μm serial optical sections). See ESI for Video S8† related to (d). | ||
HAB does not target neutrophil myeloperoxidase, and the staining of neutrophil granules by HAB is not a general feature of chalcones
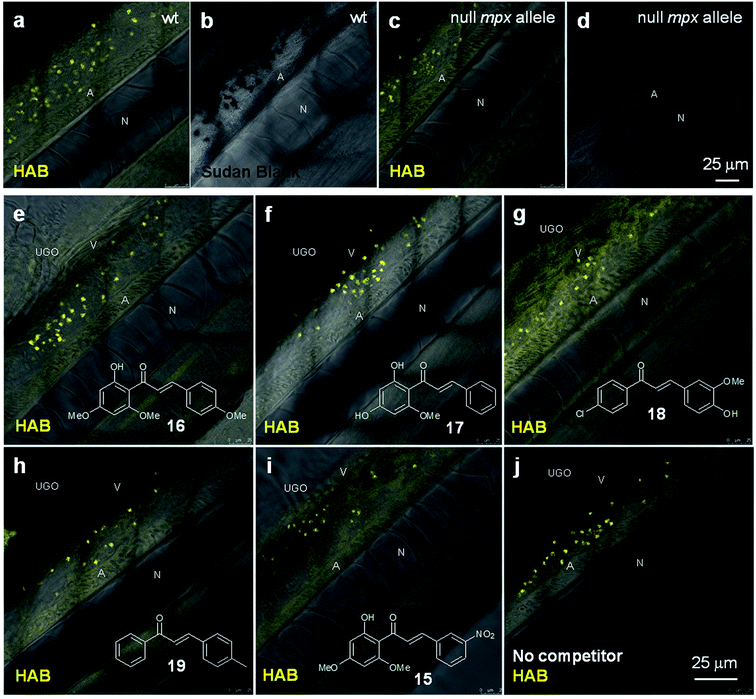
To tentatively identify the specific protein target of HAB in the neutrophil granules, we performed HAB labeling in myeloperoxidase knock-out zebrafish larvae (“spotless” mutant strain)45 under equilibrium conditions. While mutant larvae showed the expected absence of SB staining, they nevertheless still exhibited a fluorescent labeling with HAB qualitatively and quantitatively indistinguishable from wild-type larvae (Fig. 8a–d). This result shows that the myeloperoxidase abundant in neutrophil granules is not the biochemical target of HAB. We also tested a selection of biologically active chalcones as possible HAB competitors, based on their known or putative action on neutrophils such as inhibition of myeloperoxidase activity or repressing effects on pro-inflammatory mediators such as NF-κB, TNF-α, COX-2 and various interleukines.46–48 The natural chalcones flavokawain A 16 and cardamonin 17 are potent anti-inflammatory, proapoptotic and antitumour agents in mouse models, due to their ability to block NF-κB signaling.31,49–51 Synthetic chalcone 18 is an inhibitor of chemokine CXCL12 that blocks its binding to several chemokine receptors, preventing eosinophil infiltration in an asthma mouse model.52 3-Nitrochalcone 15 possesses pronounced antinociceptive properties in mice that are anti-inflammatory in origin.53 Last, 4-methylchalcone 19 was selected due to its homology with mere (unsubstituted) chalcone, reported to exert anti-myeloperoxidase and anti-migratory effects on zebrafish neutrophils.46 Larvae were first incubated with the competitors alone at 100 μM for 1 h to assess their toxicity, as well as possible basal fluorescence in the zebrafish. Under these conditions, all specimens remained alive with no apparent toxicity from the chalcones. Moreover, using the same detection settings as for HAB at 10 μM (λex 488 nm, λem 550–650 nm), none of the competitors showed fluorescence and their labeling was undistinguishable from that of the negative control DMSO (data not shown), except for chalcone 18 (see below and Fig. S9 in the ESI†). When these various potential competitors were co-incubated at 100 μM with HAB at 10 μM under the same conditions, we could observe no extinction or diminution of HAB fluorescence in vivo with any of these competitors relative to the control (Fig. 8e–j). Moreover, fluorimetric association experiments in TRIS buffer or DMF (where HAB shows the same type of yellow-orange fluorescence as in neutrophil granules, see Fig. 4d) demonstrated that none of these chalcones, when incubated in the presence of HAB at the same 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry, induced any significant qualitative or quantitative change in HAB fluorescence. The only exception was chalcone 18 which caused strong emission quenching in both media (data not shown), a feature not observed in zebrafish larvae in vivo (Fig. 8g). The binding of HAB to neutrophil granules thus appears to be the result of the probe's precise physicochemical signature, rather than a general feature of chalcones.
1 stoichiometry, induced any significant qualitative or quantitative change in HAB fluorescence. The only exception was chalcone 18 which caused strong emission quenching in both media (data not shown), a feature not observed in zebrafish larvae in vivo (Fig. 8g). The binding of HAB to neutrophil granules thus appears to be the result of the probe's precise physicochemical signature, rather than a general feature of chalcones.
During the course of this competition study, we discovered that 4-hydroxy-3-methoxy-4′-chlorochalcone (Cpd. 18) at 100 μM was responsible for unusual fluorescence in the presence of HAB (Fig. 8g), and deserved further investigation. When incubated alone at the same concentration on 72 hpf larvae and excited at 488 nm, chalcone 18 exhibited a green-yellow fluorescence that illuminated the anatomy of the specimens. Indeed, confocal imaging showed that the fluorescence of 18 delineated the trunk neuromasts, somite muscle fibers and boundaries, blood vessels, notochord, spinal cord, spinal canal and caudal hematopoietic tissue (Fig. S9 in the ESI†). Detailed examination revealed no detectable intracellular staining, but interstitial histological labeling. This behavior is reminiscent of that of BODIPY-ceramide, a fluorescent dye previously used at similarly high concentration as a histological counterstain for the confocal imaging of live zebrafish embryos.54,55 The fact that chalcone 18 was not cytopermeable even at high concentration further validates the global design of HAB, a benzochalcone congener, as a histo- and cytopermeable tracer for in vivo studies. Interestingly, chalcone 18 was recently evaluated in the zebrafish model as an inhibitor of the chemokine CXCL12a, for its activity against the collective migration of cells of the posterior lateral line primordium. At 10 μM, 18 showed moderate biological effects and was not fluorescently detected by GFP filters.56 The complete study of chalcone 18 regarding the fluorescence detection threshold, photostability, phenotypic effects and long-term toxicity is currently underway, in order to validate this compound as a novel anatomical interstitial live stain in the zebrafish model.
HAB labels granules in live human primary neutrophils
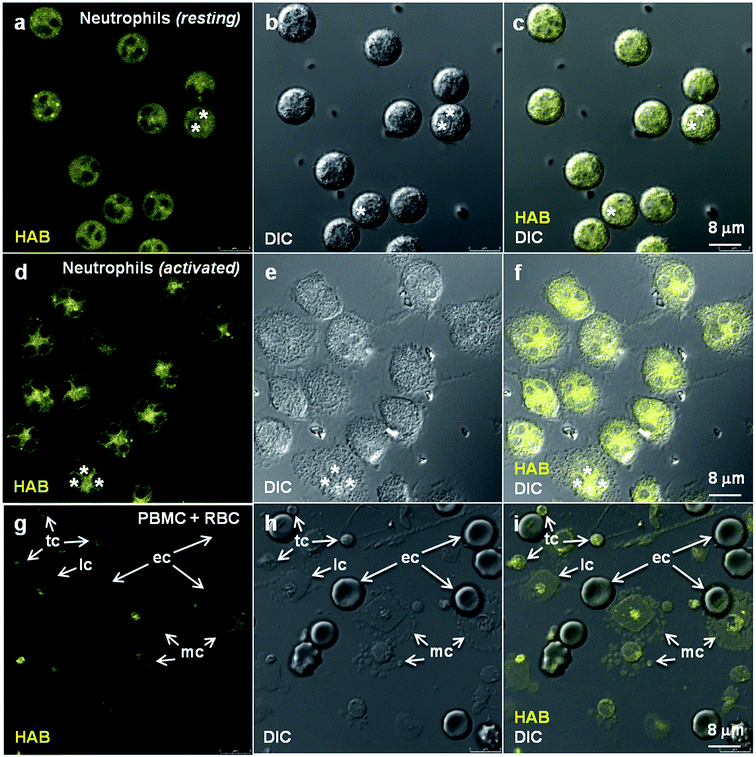
In order to validate HAB as a specific vital stain of neutrophil granules not only in fish but also in mammals, and to assess its relevance for human studies, we attempted the HAB labeling of live primary human neutrophils freshly isolated from total blood. Human neutrophils exist in two morphologically and phenotypically distinct forms depending on their state of activation. Whereas non-activated neutrophils consist of round-shaped non-adhering circulating cells, activated neutrophils are polyhedral cells expressing a number of cytoadhesins and adhering to endothelium walls, glass or plastic surfaces.57,58 When stained with 10 μM HAB, non-activated neutrophils exhibited intense but diffuse fluorescent labeling, presumably due to the difficulty of resolving subcellular structures when imaging non-adherent cells (Fig. 9a–c). An equally intense labeling of cells at the level of subcellular granular structures was detected under the same conditions in activated neutrophils, consistent with the morphology and distribution of human neutrophil granules,59,60 in addition to a strong perinuclear staining possibly corresponding to the nuclear membrane (Fig. 9d–f and S10 in the ESI†). To address the human blood cell selectivity issue, we assessed HAB labeling on other freshly isolated cell populations from whole human blood (peripheral blood mononuclear cells, PBMC) under the same conditions as those used to image purified neutrophils. HAB was found to exhibit an impressive selectivity for neutrophils over lymphocytes, monocytes, erythrocytes and thrombocytes (platelets), which were all weakly to faintly labeled (Fig. 9g–i). This selectivity seems to be the result of the accumulation of HAB in neutrophil granules, consistent with the complete cell and organelle specificity observed in the zebrafish model in vivo. Although these results deserve further investigation regarding the subset of granules labeled in human neutrophils,59 they constitute a strong presumption that the specific labeling of neutrophil granules by HAB in the zebrafish model is also relevant to mammalian systems.Discussion
A novel 3-aminobenzochalcone, HAB, was rationally designed, synthesized and validated as a histopermeable and fluorogenic vital stain for in vivo biological studies. Photophysically, HAB has elevated Stokes shifts, the highest fluorescence quantum yield described to date in this series, elevated signal-to-noise ratios upon protein binding, and unprecedented photostability. This also chemically very stable tracer possesses the minimal architecture to be strongly fluorescent, and stains neutrophils in zebrafish embryos and larvae with extraordinary specificity, with no labeling of other, even related, cell types (e.g., macrophages). Subcellularly, HAB proved to be specific for zebrafish neutrophil granules, with no staining of other organelles or compartments. This behavior was rationalized as target-induced bathochromic fluorogenesis based on various chemical and biochemical model systems. Neutrophil labeling was independent of the state of cell activation, since HAB stained the granules of both resting and phagocytically active neutrophils with equal intensity and specificity. HAB labeling in the zebrafish followed the deployment of neutrophils from 32 hpf embryos to 72 hpf swimming larvae and was even visualised in neutrophil granules in 6 day-old larvae with similar sensitivity despite the increase in tissue density and thickness (see Fig. S11 in the ESI†). Due to its absence of toxicity up to 48 h incubation and outstanding photostability, HAB is ideal for the long-term, high temporal resolution live imaging of developing zebrafish, with the potential of dynamic monitoring of the neutrophil granules in virtually any physiological or physiopathological context. To our knowledge, no other related tracer is endowed with such precious characteristics. Last, HAB intensely labeled freshly purified neutrophils from human blood at the level of granular structures consistent with neutrophil granules. Fluorescent dyes for neutrophil granules are limited, including quinacrine (QA),8 LysoTrackerRed® (LTR),9 dihydrorhodamine (DHR) 123,9 and rhodamine-thiolactones (RTLs),61 which are non-specific by essence. QA and LTR as weak bases constitute general acidotropic stains, sequestrated in their protonated form in various low pH organelles and vesicles (e.g., lysosomes,62–64 apoptotic vesicles,65 mast cell granules66,67), to which mammalian basophil and neutrophil secreted granules also belong.68,69 DHR 123 and RTLs are part of ROS-activated profluorophores used to detect hydrogen peroxide/peroxynitrite and hypochlorous acid, respectively, in activated neutrophils as well as in various other cell systems and organelles.61,70–72 A comparative analysis of the various existing tracers of neutrophil granules is provided in Table 1. In marked contrast, the unique staining of live zebrafish neutrophil granules by HAB over other cell types and acidic compartments in the whole organism, together with HAB very weak basicity (about 105-fold lower than that of acidotropic amines63 with a predicted pKa value of 3.97 for the amino tautomers, see Fig. S7 in the ESI†) supports a specific target-induced labeling of neutrophil granules by HAB. Although we showed that the myeloperoxidase present in the granules is not involved in their specific staining by HAB, these granules also contain abundant neutral serine proteases (e.g., elastase, proteinase 3, and cathepsin G),59 which constitute putative biochemical targets for HAB based on their reactivity towards reversible electrophilic inhibitors.73,74 In this context, HAB has an immediate surrogate for proteomic interrogation in the form of a photoalkylative azido analogue (Fig. 1) easily accessible through Sandmeyer-type modification of the aromatic amino group.22 The covalent photolabeling of live zebrafish larvae or isolated human neutrophils with a fluorogenic 3-azido-4′-hydroxybenzochalcone, FACS purification of labeled cells followed by Western Blot identification of the covalently stained molecular target(s) of HAB, therefore seems within reach. Overall, due to its optimal photophysical and physicochemical properties and unprecedented biological specificity, HAB constitutes a valuable small-molecule addition to the fluorescent proteins toolbox used in the zebrafish model, which it functionally and spectrally complements, and appears suitable for functional studies in live mammalian neutrophils. HAB therefore holds great promise as a vital stain to monitor the dynamics and behaviour of neutrophil granules in various aspects of the neutrophil function in the context of a live organism, with potential relevance to human physiology and physiopathology.| Tracer | Specificity for neutrophil granules | Histopermeability for live animal imaging | Photostability | Fluorogenesis | Ref. |
|---|---|---|---|---|---|
| a FA, formaldehyde; FITC, fluorescein isothiocyanate; FTO, fluorescence turn-on; MeOH, methanol; MPO, myeloperoxidase; NA, non-applicable; RNS, reactive nitrogen species; ROS, reactive oxygen species. | |||||
| Quinacrine | No (acidotropic stain, DNA stain) | Yes | Yes | No | 8, 65 and 75–77 |
| LysoTrackers | No (acidotropic stains) | Yes | No | No | 9, 65 and 77–79 |
| Acridine orange | No (acidotropic stain, DNA/RNA stain) | Yes | Yes | No | 65 and 80–83 |
| Dihydrorhodamine 123 | No (ROS/RNS-dependent - also labels mitochondria) | No (in vitro only) | No | Yes (ROS-activated) | 9 and 84–87 |
| Rhodamine-thiolactones | No (ROS-dependent) | Yes | Yes | Yes (ROS-reactive) | 61 and 88 |
| Sudan Black B | Yes (lipids/MPO-dependent) | No (FA fixation in vivo) | NA (colorimetric stain) | NA (colorimetric stain) | 5, 42 and 89 |
| Luminol | No (ROS-dependent, MPO-amplified) | No (in vitro only) | NA (chemiluminescent stain) | NA (chemiluminescent stain) | 90 |
| Tyramide-FITC/Cy3 | Yes (MPO-reactive) | No (FA fixation in vivo) | No (FITC conjugate) | No | 5 and 91 |
| Yes (Cy3 conjugate) | |||||
| MUB40/RI-MUB40-Cy5 | Yes (lactoferrin ligands) | No (peptides, FA fixation in vitro, in vivo) | Yes | No | 60 |
| NE680 | Yes (elastase substrate) | No (peptide, MeOH fixation ex vivo, intranasal instillation in mouse) | Yes | Yes (enzyme-activated) | 92 |
| Elastase, cathepsin G, proteinases 3 and 4 substrates | Yes (enzyme substrates) | No (peptides, FA fixation in vitro) | Yes | Yes (enzyme-activated) | 93 |
| HAB | Yes (pH, ROS, MPO-independent fluorescence) |
Yes (amphipathic small-molecule, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 = 2.21) D7.4 = 2.21) |
Yes (>15 h under continuous 488 nm irradiation) | Yes (target protein-induced FTO) | Our study |
Methods
Compound synthesis, structural characterization and pKa prediction
Detailed synthetic procedures and analytical identification for compounds 2–8, 10–12 and 14, as well as pKa prediction for HAB, can be found in the ESI.†Zebrafish
Transgenic and mutant stocks of zebrafish were raised and staged according to Westerfield.94 AB wild-type fish and transgenic lines Tg(mpx:GFP)i114, Tg(mpx:GAL4.VP16)i22, Tg(mfap4:mCherry-F) and Tg(UAS-E1b:nfsB.mCherry)c264 were used.35–37 Embryos were reared at 28 °C or 24 °C according to the desired speed of development. All timings in the text refer to the developmental stage at the reference temperature of 28.5 °C.94 Embryos and larvae were anesthetized with 200 μg mL−1 tricaine (Sigma-Aldrich) during live in vivo imaging.HAB labeling of live zebrafish embryos and larvae
Sudan Black staining (Fig. 7 and 8)
Zebrafish larvae (72 hpf) were fixed with 4% methanol-free formaldehyde (Poly-sciences, Warrington, PA) in phosphate-buffered saline (PBS) for 2 hours at room temperature, rinsed in PBS, incubated in Sudan Black (SB; Sigma-Aldrich) for 20 minutes, washed extensively in 70% ethanol in water, and then progressively rehydrated in 0.1% Tween 20 in PBS as previously described.5Zymosan microinjection in zebrafish larvae (Fig. 7)
Zebrafish larvae (72 hpf) were anaesthetized by immersion in buffered tricaine immediately after the HAB labeling. They were injected with 1 nL of 0.4 × 108 mL−1 zymosan-Cy5 particle suspension using pulled borosilicate glass microcapillary (GC100F-15 Harvard apparatus) pipettes under a stereomicroscope (Stemi 2000, Carl Zeiss, Germany) with a mechanical micromanipulator (M-152; Narishige), and a Picospritzer III pneumatic microinjector (Parker Hannifin) set at a pressure of 20 psi and an injection time of 20 ms (subcutaneous injections) as previously described.43Competition experiments (Fig. 8)
72 hpf wild-type AB zebrafish larvae were placed into individual wells as previously described and co-incubated for 1 h at room temperature with HAB (10 μM from a 10 mM stock solution in DMSO) and each competitor (100 μM from a 10 mM stock solution in DMSO) in Volvic water in the dark as previously described. Negative controls consisted of Volvic water containing 1.1% DMSO. In this case, specimens were rapidly rinsed, anaesthetized and mounted in HAB-free media for biological imaging. Practically, homogenous aqueous solutions of HAB ± competitors were obtained by adding the drop of stock DMSO solutions on the inner wall of a plastic tube containing Volvic water at room temperature (20–23 °C), and vortexing without interruption for 20 s.HAB labeling of live human neutrophils (Fig. 9 and S10†)
Peripheral human blood was collected from healthy patients at the ICAReB service of the Pasteur Institute (authorization DC no. 2008-68). All donors gave written informed consent in accordance with the Declaration of Helsinki principles. Blood was collected from the antecubital vein into tubes containing sodium citrate (3.8% final) as the anticoagulant. Human polymorphonuclear neutrophils were purified as described previously.95 Briefly, plasma was removed by centrifugation (450g, 15 min), and blood cells were resuspended in 0.9% NaCl solution supplemented with 0.72% Dextran. After red blood cell sedimentation, white blood cells were pelleted and further separated on a two layer Percoll (GE Healthcare) gradient (51–42%) by centrifugation (240 g, 20 min). Peripheral Blood Mononuclear Cells (PBMC) (top layer) were isolated from polymorphonuclear neutrophils (bottom layer). Red blood cells were removed from the latter fraction using CD235a (glycophorin) microbeads (Miltenyi Biotec). Purified polymorphonuclear neutrophils were resuspended in the autologous plasma until experimental use. For HAB labeling, they were immediately put in RPMI 10 mM Hepes at 2 × 106 cells per mL. 1 mL of neutrophil suspension was distributed in 35 mm glass-bottom dishes and incubated at room temperature for 20 min for neutrophil adhesion. The medium was then replaced by the HAB staining solution (at 1, 5 or 10 μM) in RPMI 10 mM Hepes; after 5–15 min at room temperature, HAB-labeled neutrophils were observed with a SP8 Leica confocal microscope, using a 63× water immersion objective. The 10 μM HAB concentration gave the best results.Live imaging, image processing and analysis
Confocal microscopy to detect HAB single labeling in wild-type zebrafish embryos and larvae was performed at 23–26 °C using a Leica SPE inverted microscope and a 16× oil immersion objective (PL FLUOTAR 16×/0.50 IMM). HAB was excited with a 488 nm laser and the fluorescence emission was collected within the 550–650 nm range unless otherwise indicated. A Leica SP8 confocal microscope with a 20× oil immersion objective (HC PL APO CS2 20×/0.75 IMM) was used to live image transgenic larvae for colocalization studies and wt larvae for competition studies. Regarding HAB/mCherry acquisition, the following settings were applied: HAB (excitation 448 nm; emission 550–650 nm) and mCherry (excitation 552 nm; emission 660–750 nm) using a sequential acquisition mode. For GFP/HAB acquisition the following settings were applied: GFP (excitation 448 nm; emission 500–520 nm) and HAB (excitation 448 nm; emission >600 nm). A 40× water immersion objective (HC PL APO CS2 40×/1.1 water) was also used with the SP8 LEICA confocal microscope, to document at high resolution HAB labeling in neutrophils. For HAB-zymosan acquisition, the following settings were applied: HAB (excitation 488 nm; emission 491–601 nm) and Cy5-zymosan (excitation 638 nm; emission 683–795 nm). A 63× water immersion objective (HC PL APO CS2 63×/1.2 water) was used to image HAB in live human neutrophils, following excitation at 448 nm and collecting the fluorescence emission in the 550–650 nm range. The 3D or 4D files generated by the confocal acquisitions were processed using the LAS-AF Leica software. Acquired Z-stacks (2 μm serial optical sections for images taken using 16× and 20× objectives) were projected using maximum intensity projection and exported as AVI or TIFF files. Z stacks acquired with the 40× or 63× objectives (0.4 μm to 1 μm serial optical sections) were exported as the AVI file. Frames were captured from the AVI files and handled with PowerPoint software to mount figures.Author contributions
E. C.-G. designed, performed and analysed the HAB-zebrafish experiments and contributed to the writing of the manuscript. A. S. B. and S. D. S. O. performed HAB phenotypic screening. M. B. and I. C. B. performed the fluorescence spectroscopy experiments. B. S. M. provided human neutrophils and PBMC. K. L. performed the HRMS analysis. P. H. assisted with helpful discussion and participated in the writing of the manuscript. R. D. designed HAB, performed the synthesis and structural analysis of all compounds, performed log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 measurements, co-analysed all biological experiments, and wrote the manuscript.
D7.4 measurements, co-analysed all biological experiments, and wrote the manuscript.
Conflicts of interest
The authors declare no competing financial interests.Abbreviations
| AcOEt | Ethyl acetate |
| AcOH | Acetic acid |
| BF | Bright field |
| CC | Column chromatography |
| CHCl3 | Chloroform |
| DHR | Dihydrorhodamine |
| DIC | Differential interference contrast |
| DMF | Dimethylformamide |
| DMSO | Dimethylsulfoxide |
| DPP | N-Dansyl-N′-phenylpiperazine |
| FACS | Fluorescence-activated cell sorting |
| F | Fluorescence quantum yield |
| HAB | 4′-Hydroxy-3-aminobenzochalcone |
| HRMS | High-resolution mass spectra |
| ICG | Indocyanine green |
| LTR | LysoTrackerRed |
| MeCN | Acetonitrile |
| MPO | Myeloperoxidase |
| PBMC | Peripheral blood mononuclear cells |
| ROS | Reactive oxygen species; |
| RTL | Rhodamine-thiolactone |
| QA | Quinacrine |
| SB | Sudan Black |
Acknowledgements
The authors wish to acknowledge the following contributors: Dr L. Bianchi and Prof. G. Petrillo for the generous gift of a reference of compound 10; Dr P. Leal for the gift of xanthoxyline; Prof. S. Renshaw and Prof. G. Lutfalla for providing the zebrafish transgenic Tg(mpx:GAL4.VP16)i22 and Tg(mfap4:mCherryF) lines, respectively; Dr F. Niedergang for the gift of Cy5-zymosan; T. M. R. Gianeti and Dr A. Maciuk for their practical help regarding NIR fluorescence and HRMS analysis, respectively; A. Blin for taking pictures of UVA-irradiated cuvettes in Fig. 4; Dr N. Bizat for preliminary labelling experiments in C. elegans; V. Briolat, Y. Rolin and N. Aimar for fish husbandry, as well as L. Boucontet for helping with zebrafish embryo preparation; Dr M. Anderson for the preparation of isolated human neutrophils; Dr A. Cras for fruitful discussions regarding the biology of human blood cells. R. Duval gratefully acknowledges Prof. R. J. Nunes, Prof. B. Rossi-Bergmann and Prof. N. Leulliot for permitting certain experiments to be conducted in their laboratories. Grants from the Conselho Nacional de Desenvolvimento Cientifico e Tecnologico, CNPq (401897/2010-9 to R. D.), the Institut Pasteur (GPF Microbes and Brain Myconeuro, S-PI14032-02G to E. C.-G.) and the Fondation pour la Recherche Médicale (Equipe FRM DEQ20120323714 to P. H.) are acknowledged, as well as Dr J. Moreira for translating the CNPq scientific proposal into Portuguese.References
- S. Masud, V. Torraca and A. H. Meijer, Curr. Top. Dev. Biol., 2017, 124, 277–329 CrossRef PubMed.
- G. J. Lieschke, A. C. Oates, M. O. Crowhurst, A. C. Ward and J. E. Layton, Blood, 2001, 98, 3087–3096 CrossRef CAS PubMed.
- D. Traver, P. Herbomel, E. E. Patton, R. D. Murphey, J. A. Yoder, G. W. Litman, A. Catic, C. T. Amemiya, L. I. Zon and N. S. Trede, Adv. Immunol., 2003, 81, 253–330 Search PubMed.
- P. Herbomel, B. Thisse and C. Thisse, Development, 1999, 126, 3735–3745 CAS.
- D. Le Guyader, M. J. Redd, E. Colucci-Guyon, E. Murayama, K. Kissa, V. Briolat, E. Mordelet, A. Zapata, H. Shinomiya and P. Herbomel, Blood, 2008, 111, 132–141 CrossRef CAS PubMed.
- K. M. Henry, C. A. Loynes, M. K. B. Whyte and S. A. Renshaw, J. Leukocyte Biol., 2013, 94, 633–642 CrossRef CAS PubMed.
- E. A. Harvie and A. Huttenlocher, J. Leukocyte Biol., 2015, 98, 523–537 CrossRef CAS PubMed.
- E. Suzaki, H. Kobayashi, Y. Kodama, T. Masujima and S. Terakawa, Cell Motil. Cytoskeleton, 1997, 38, 215–228 CrossRef CAS PubMed.
- C. F. Bassoe, N. Y. Li, K. Ragheb, G. Lawler, J. Sturgis and J. P. Robinson, Cytometry B Clin Cytom., 2003, 51, 21–29 CrossRef PubMed.
- R. Duval and C. Duplais, Nat. Prod. Rep., 2017, 34, 161–193 RSC.
- N. DiCesare and J. R. Lakowicz, Tetrahedron Lett., 2002, 43, 2615–2618 CrossRef CAS.
- J. Prabhu, K. Velmurugan and R. Nandhakumar, Spectrochim. Acta, Part A, 2015, 144, 23–28 CrossRef CAS PubMed.
- C. G. Niu, A. L. Guan, G. M. Zeng, Y. G. Liu and Z. W. Li, Anal. Chim. Acta, 2006, 577, 264–270 CrossRef CAS PubMed.
- S. C. Lee, N. Y. Kang, S. J. Park, S. W. Yun, Y. Chandran and Y. T. Chang, Chem. Commun., 2012, 48, 6681–6683 RSC.
- M. Tomasch, J. S. Schwed, L. Weizel and H. Stark, Front. Syst. Neurosci., 2012, 6, 14 CAS.
- B. Zhou, P. Jiang, J. Lu and C. Xing, Arch. Pharm., 2016, 349, 539–552 CrossRef CAS PubMed.
- T. Fuchigami, Y. Yamashita, M. Haratake, M. Ono, S. Yoshida and M. Nakayama, Bioorg. Med. Chem., 2014, 22, 2622–2628 CrossRef CAS PubMed.
- H. G. O. Alvim, E. L. Fagg, A. L. de Oliveira, H. C. B. de Oliveira, S. M. Freitas, M. A. E. Xavier, T. A. Soares, A. F. Gomes, F. C. Gozzo, W. A. Silva and B. A. D. Neto, Org. Biomol. Chem., 2013, 11, 4764–4777 RSC.
- A. Bessette, T. Auvray, D. Desilets and G. S. Hanan, Dalton Trans., 2016, 45, 7589–7604 RSC.
- P. Alexander and H. Moronson, Nature, 1962, 194, 2 Search PubMed.
- D. Kulms and T. Schwarz, Skin Pharmacol. Appl. Skin Physiol., 2002, 15, 342–347 CrossRef CAS PubMed.
- S. J. Lord, H. L. Lee, R. Samuel, R. Weber, N. Liu, N. R. Conley, M. A. Thompson, R. J. Twieg and W. E. Moerner, J. Phys. Chem. B, 2010, 114, 14157–14167 CrossRef CAS PubMed.
- S. D. Barker, K. Wilson and R. K. Norris, Aust. J. Chem., 1995, 48, 1969–1979 CrossRef CAS.
- P. G. E. Alcorn and P. R. Wells, Aust. J. Chem., 1965, 18, 1377–1389 CrossRef CAS.
- V. Balasubramaniyan, Chem. Rev., 1966, 66, 567–641 CrossRef CAS.
- D. Skalamera, L. P. Cao, L. Isaacs, R. Glaser and K. Mlinaric-Majerski, Tetrahedron, 2016, 72, 1541–1546 CrossRef CAS.
- A. Niemann, J. Baltes and H. P. Elsasser, J. Histochem. Cytochem., 2001, 49, 177–185 CrossRef CAS PubMed.
- X. J. Yang, C. M. Shi, R. Tong, W. P. Qian, H. E. Zhau, R. X. Wang, G. D. Zhu, J. J. Cheng, V. W. Yang, T. M. Cheng, M. Henary, L. Strekowski and L. W. K. Chung, Clin. Cancer Res., 2010, 16, 2833–2844 CrossRef CAS PubMed.
- R. A. Duval, R. L. Allmon and J. R. Lever, J. Med. Chem., 2007, 50, 2144–2156 CrossRef CAS PubMed.
- P. Boeck, C. A. B. Falcao, P. C. Leal, R. A. Yunes, V. Cechinel, E. C. Torres-Santos and B. Rossi-Bergmann, Bioorg. Med. Chem., 2006, 14, 1538–1545 CrossRef CAS PubMed.
- X. L. Zi and A. R. Simoneau, Cancer Res., 2005, 65, 3479–3486 CrossRef CAS PubMed.
- M. J. Waring, Bioorg. Med. Chem. Lett., 2009, 19, 2844–2851 CrossRef CAS PubMed.
- I. Ziegler, T. McDonald, C. Hesslinger, I. Pelletier and P. Boyle, J. Biol. Chem., 2000, 275, 18926–18932 CrossRef CAS PubMed.
- C. W. Higdon, R. D. Mitra and S. L. Johnson, PLoS One, 2013, 8, e67801 CrossRef CAS PubMed.
- Q. T. Phan, T. Sipka, C. Gonzalez, J. P. Levraud, G. Lutfalla and N. C. Mai, PLoS Pathog., 2018, 14, e1007157 CrossRef PubMed.
- F. Ellett, L. Pase, J. W. Hayman, A. Andrianopoulos and G. J. Lieschke, Blood, 2011, 117, E49–E56 CrossRef CAS PubMed.
- S. A. Renshaw, C. A. Loynes, D. M. I. Trushell, S. Elworthy, P. W. Ingham and M. K. B. Whyte, Blood, 2006, 108, 3976–3978 CrossRef CAS PubMed.
- M. O. Crowhurst, J. E. Layton and G. J. Lieschke, Int. J. Dev. Biol., 2002, 46, 483–492 CAS.
- C. T. Pham, Nat. Rev. Immunol., 2006, 6, 541–550 CrossRef CAS PubMed.
- K. Wang, X. Fang, N. Ma, Q. Lin, Z. Huang, W. Liu, M. Xu, X. Chen, W. Zhang and Y. Zhang, Fish Shellfish Immunol., 2015, 44, 109–116 CrossRef CAS PubMed.
- B. J. Bain, Am. J. Hematol., 2010, 85, 707 CrossRef PubMed.
- W. van den Ancker, T. M. Westers, D. C. de Leeuw, Y. F. van der Veeken, A. Loonen, E. van Beckhoven, G. J. Ossenkoppele and A. A. van de Loosdrecht, Cytometry, Part B, 2013, 84, 114–118 CrossRef PubMed.
- E. Colucci-Guyon, J. Y. Tinevez, S. A. Renshaw and P. Herbomel, J. Cell Sci., 2011, 124, 3053–3059 CrossRef CAS PubMed.
- E. K. Macrae and K. B. Pryzwansky, Carlsberg Res. Commun., 1984, 49, 315–322 CrossRef.
- P. M. Elks, M. van der Vaart, V. van Hensbergen, E. Schutz, M. J. Redd, E. Murayama, H. P. Spaink and A. H. Meijer, PLoS One, 2014, 9, e100928 CrossRef PubMed.
- Y. H. Chen, W. H. Wang, Y. H. Wang, Z. Y. Lin, C. C. Wen and C. Y. Chern, Molecules, 2013, 18, 2052–2060 CrossRef CAS PubMed.
- B. P. Bandgar, S. A. Patil, R. N. Gacche, B. L. Korbad, B. S. Hote, S. N. Kinkar and S. S. Jalde, Bioorg. Med. Chem. Lett., 2010, 20, 730–733 CrossRef CAS PubMed.
- J. Z. Wu, J. L. Li, Y. P. Cai, Y. Pan, F. Q. Ye, Y. L. Zhang, Y. J. Zhao, S. L. Yang, X. K. Li and G. Liang, J. Med. Chem., 2011, 54, 8110–8123 CrossRef CAS PubMed.
- D. J. Kwon, S. M. Ju, G. S. Youn, S. Y. Choi and J. Park, Food Chem. Toxicol., 2013, 58, 479–486 CrossRef CAS PubMed.
- N. Wu, J. Liu, X. Z. Zhao, Z. Y. Yan, B. Jiang, L. J. Wang, S. S. Cao, D. Y. Shi and X. K. Lin, Tumor Biol., 2015, 36, 9667–9676 CrossRef CAS PubMed.
- S. Hatziieremia, A. I. Gray, V. A. Ferro, A. Paul and R. Plevin, Br. J. Pharmacol., 2006, 149, 188–198 CrossRef CAS PubMed.
- M. Hachet-Haas, K. Balabanian, F. Rohmer, F. Pons, C. Franchet, S. Lecat, K. Y. C. Chow, R. Dagher, P. Gizzi, B. Didier, B. Lagane, E. Kellenberger, D. Bonnet, F. Baleux, J. Haiech, M. Parmentier, N. Frossard, F. Arenzana-Seisdedos, M. Hibert and J. L. Galzi, J. Biol. Chem., 2008, 283, 23189–23199 CrossRef CAS PubMed.
- F. de Campos-Buzzi, J. P. de Campos, P. P. Tonini, R. Correa, R. A. Yunes, P. Boeck and V. Cechinel-Filho, Arch. Pharm., 2006, 339, 361–365 CrossRef CAS PubMed.
- M. S. Cooper, L. A. D'Amico and C. A. Henry, Methods Cell Biol., 1999, 59, 179–204 CrossRef CAS PubMed.
- E. Murayama, M. Sarris, M. Redd, D. Le Guyader, C. Vivier, W. Horsley, N. Trede and P. Herbomel, Nat. Commun., 2015, 6, 1–14 Search PubMed.
- D. Dalle Nogare, K. Somers, S. Rao, M. Matsuda, M. Reichman-Fried, E. Raz and A. B. Chitnis, Development, 2014, 141, 3188–3196 CrossRef CAS PubMed.
- N. Yakuwa, T. Inoue, T. Watanabe, K. Takahashi and F. Sendo, Microbiol. Immunol., 1989, 33, 843–852 CrossRef CAS PubMed.
- K. Ley, H. M. Hoffman, P. Kubes, M. A. Cassatella, A. Zychlinsky, C. C. Hedrick and S. D. Catz, Sci Immunol, 2018, 3, eaat4579 CrossRef PubMed.
- J. L. Eyles, A. W. Roberts, D. Metcalf and I. P. Wicks, Nat. Clin. Pract. Rheumatol., 2006, 2, 500–510 CrossRef CAS PubMed.
- M. C. Anderson, T. Chaze, Y. M. Coic, L. Injarabian, F. Jonsson, N. Lombion, D. Selimoglu-Buet, J. Souphron, C. Ridley, P. Vonaesch, B. Baron, E. T. Arena, J. Y. Tinevez, G. Nigro, K. Nothelfer, E. Solary, V. Lapierre, T. Lazure, M. Matondo, D. Thornton, P. J. Sansonetti, F. Baleux and B. S. Marteyn, Cell Chem. Biol., 2018, 25, 483–493 CrossRef CAS PubMed.
- X. Chen, K. A. Lee, E. M. Ha, K. M. Lee, Y. Y. Seo, H. K. Choi, H. N. Kim, M. J. Kim, C. S. Cho, S. Y. Lee, W. J. Lee and J. Yoon, Chem. Commun., 2011, 47, 4373–4375 RSC.
- D. D. Li, N. Ropert, A. Koulakoff, C. Giaume and M. Oheim, J. Neurosci., 2008, 28, 7648–7658 CrossRef CAS PubMed.
- B. Lemieux, M. D. Percival and J. P. Falgueyret, Anal. Biochem., 2004, 327, 247–251 CrossRef CAS PubMed.
- T. Sasaki, S. S. Lian, J. Qi, P. E. Bayliss, C. E. Carr, J. L. Johnson, S. Guha, P. Kobler, S. D. Catz, M. Gill, K. L. Jia, D. J. Klionsky and S. Kishi, PLoS Genet., 2014, 10, e1004409 CrossRef PubMed.
- A. Pierzynska-Mach, P. A. Janowski and J. W. Dobrucki, Cytometry, Part A, 2014, 85, 729–737 CrossRef PubMed.
- M. Wu, T. Baumgart, S. Hammond, D. Holowka and B. Baird, J. Cell Sci., 2007, 120, 3147–3154 CrossRef CAS PubMed.
- H. Y. Chen, D. M. Chiang, Z. J. Lin, C. C. Hsieh, G. C. Yin, I. C. Weng, P. Guttermann, S. Werner, K. Henzler, G. Schneider, L. J. Lai and F. T. Liu, Sci. Rep., 2016, 6, 34879 CrossRef CAS PubMed.
- A. W. Segal, Annu. Rev. Immunol., 2005, 23, 197–223 CrossRef CAS PubMed.
- M. A. Kolber and P. A. Henkart, Biochim. Biophys. Acta, 1988, 939, 459–466 CrossRef CAS.
- J. P. Crow, Nitric Oxide, 1997, 1, 145–157 CrossRef CAS PubMed.
- Y. Qin, M. Lu and X. G. Gong, Cell Biol. Int., 2008, 32, 224–228 CrossRef CAS PubMed.
- L. Yuan, L. Wang, B. K. Agrawalla, S. J. Park, H. Zhu, B. Sivaraman, J. Peng, Q. H. Xu and Y. T. Chang, J. Am. Chem. Soc., 2015, 137, 5930–5938 CrossRef CAS PubMed.
- A. Budnjo, S. Narawane, C. Grauffel, A. S. Schillinger, T. Fossen, N. Reuter and B. E. Haug, J. Med. Chem., 2014, 57, 9396–9408 CrossRef CAS PubMed.
- W. C. Groutas, D. Dou and K. R. Alliston, Expert Opin. Ther. Pat., 2011, 21, 339–354 CrossRef CAS PubMed.
- A. Parks, X. Charest-Morin, M. Boivin-Welch, J. Bouthillier and F. Marceau, PeerJ, 2015, 3, e1314 CrossRef PubMed.
- A. T. Sumner, Histochemistry, 1986, 84, 566–574 CAS.
- P. Huang, Y. Zou, X. Z. Zhong, Q. Cao, K. Zhao, M. X. Zhu, R. Murrell-Lagnado and X. P. Dong, J. Biol. Chem., 2014, 289, 17658–17667 CrossRef CAS PubMed.
- T. Sasaki, S. Lian, J. Qi, P. E. Bayliss, C. E. Carr, J. L. Johnson, S. Guha, P. Kobler, S. D. Catz, M. Gill, K. Jia, D. J. Klionsky and S. Kishi, PLoS Genet., 2014, 10, e1004409 CrossRef PubMed.
- E. Dona, J. D. Barry, G. Valentin, C. Quirin, A. Khmelinskii, A. Kunze, S. Durdu, L. R. Newton, A. Fernandez-Minan, W. Huber, M. Knop and D. Gilmour, Nature, 2013, 503, 285–289 CrossRef CAS PubMed.
- I. Vermes and C. Haanen, Adv. Clin. Chem., 1994, 31, 177–246 CAS.
- J. A. Udovich, D. G. Besselsen and A. F. Gmitro, J. Microsc., 2009, 234, 124–129 CrossRef CAS PubMed.
- T. J. van Ham, J. Mapes, D. Kokel and R. T. Peterson, FASEB J., 2010, 24, 4336–4342 CrossRef CAS PubMed.
- B. Tucker and M. Lardelli, Zebrafish, 2007, 4, 113–116 CrossRef PubMed.
- M. Poot, Y. Z. Zhang, J. A. Kramer, K. S. Wells, L. J. Jones, D. K. Hanzel, A. G. Lugade, V. L. Singer and R. P. Haugland, J. Histochem. Cytochem., 1996, 44, 1363–1372 CrossRef CAS PubMed.
- X. Wang, H. Fang, Z. Huang, W. Shang, T. Hou, A. Cheng and H. Cheng, J. Mol. Med., 2013, 91, 917–927 CrossRef CAS PubMed.
- I. D. Trayner, A. P. Rayner, G. E. Freeman and F. Farzaneh, J. Immunol. Methods, 1995, 186, 275–284 CrossRef CAS PubMed.
- J. P. Crow, Nitric Oxide, 1997, 1, 145–157 CrossRef CAS PubMed.
- Y. Koide, Y. Urano, K. Hanaoka, T. Terai and T. Nagano, J. Am. Chem. Soc., 2011, 133, 5680–5682 CrossRef CAS PubMed.
- B. J. Bain, in Dacie and Lewis Practical Haematology, ed. B. J. Bain, I. Bates and M. A. Laffan, Elsevier, 12th edn, 2017 Search PubMed.
- S. Bedouhene, F. Moulti-Mati, M. Hurtado-Nedelec, P. M. Dang and J. El-Benna, Am. J. Blood Res., 2017, 7, 41–48 Search PubMed.
- G. Lauter, I. Soll and G. Hauptmann, Methods Mol. Biol., 2014, 1082, 175–185 CrossRef CAS PubMed.
- S. Kossodo, J. Zhang, K. Groves, G. J. Cuneo, E. Handy, J. Morin, J. Delaney, W. Yared, M. Rajopadhye and J. D. Peterson, Int. J. Mol. Imaging, 2011, 2011, 581406 CrossRef PubMed.
- P. Kasperkiewicz, Y. Altman, M. D'Angelo, G. S. Salvesen and M. Drag, J. Am. Chem. Soc., 2017, 139, 10115–10125 CrossRef CAS PubMed.
- M. Westerfield, The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), University of Oregon Press, 1995 Search PubMed.
- V. Monceaux, C. Chiche-Lapierre, C. Chaput, V. Witko-Sarsat, M. C. Prevost, C. T. Taylor, M. N. Ungeheuer, P. J. Sansonetti and B. S. Marteyn, Blood, 2016, 128, 993–1002 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures regarding compound synthesis, structural and spectral characterization, pKa prediction, bioimaging experiments, staining dynamics of HAB in 72 hpf zebrafish larvae (Videos) and complementary figures. See DOI: 10.1039/c8sc05593a |
| ‡ These co-authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |