Open Access Article
Open Access ArticleCharacterization of an SSB–dT25 complex: structural insights into the S-shaped ssDNA binding conformation
Yen-Hua Huanga,
I-Chen Chena and
Cheng-Yang Huang *ab
*ab
aSchool of Biomedical Sciences, Chung Shan Medical University, No. 110, Sec. 1, Chien-Kuo N. Rd., Taichung City, Taiwan. E-mail: cyhuang@csmu.edu.tw; Tel: +886-4-24730022 ext. 11472
bDepartment of Medical Research, Chung Shan Medical University Hospital, No. 110, Sec. 1, Chien-Kuo N. Rd., Taichung City, Taiwan
First published on 5th December 2019
Abstract
Single-stranded DNA (ssDNA)-binding proteins (SSBs) play an important role in all DNA-dependent cellular processes, such as DNA replication, recombination, repair, and replication restart. The N-terminal domain of SSBs forms an oligonucleotide/oligosaccharide-binding (OB) fold for ssDNA binding. The SSB–dC35 complex structure has revealed how an Escherichia coli SSB (EcSSB) tetramer binds to 65-nucleotide (nt)-long ssDNA, namely, the (SSB)65 binding mode. Knowledge on whether the ssDNA-binding mode for EcSSB is typical for all SSBs or is bacterial strain and length dependent is limited. Here, we studied the ssDNA-binding properties of a Pseudomonas aeruginosa SSB (PaSSB) and investigated its interaction mode through crystallographic analysis. The complex crystal structure containing a PaSSB tetramer with two ssDNA chains was solved at a resolution of 1.91 Å (PDB entry 6IRQ). Results revealed that each bound ssDNA dT25 adopts an S-shaped conformation. This binding mode, as shown by the complex structure of PaSSB, differs significantly from (SSB)65. ssDNA-binding contributions from aromatic residues in PaSSB, except the contribution of Trp54, were not significant. Using electrophoretic mobility shift analysis, we characterized the stoichiometry of PaSSB complexed with a series of ssDNA homopolymers. The minimal length of ssDNA required for PaSSB tetramer binding and the size of the ssDNA-binding site were 25 and 29 nt, respectively. These observations through structure–function analysis suggested that only two OB folds rather than four OB folds in PaSSB are enough for the formation of a stable complex with ssDNA. The PaSSB noninteracting OB folds proposed here may allow sliding via reptation in a dynamic ssDNA binding process.
Introduction
Single-stranded DNA (ssDNA)-binding proteins (SSBs) play essential roles in DNA replication, recombination, repair, and replication restart.1–3 SSBs typically recognize ssDNA via a highly conserved oligonucleotide/oligosaccharide-binding (OB) fold formed from a five-stranded β-barrel capped by an α-helix.4,5 SSBs exhibit high affinity for ssDNA with no significant sequence specificity.3 Bacterial SSB consists of an N-terminal ssDNA-binding domain and flexible C-terminal protein–protein interaction domain.6 The C-terminal acidic peptide by which SSB binds to many nucleoproteins constitutes the SSB interactome.6,7 The intrinsically disordered linker in the C-terminal domain of SSB is involved in the mediation of interactions between SSB and the partner proteins.8–10 The entire C-terminal domain of SSB is disordered even in the presence of ssDNA.11The structures, DNA-binding properties, and functions of SSB have been studied extensively in Escherichia coli (EcSSB).2,3,6 EcSSB forms a stable homotetramer that binds and wraps ssDNA around its subunit.12 EcSSB consists of 177 amino acid residues, of which residues 1–112 in each subunit co-feature together a structured DNA-binding domain possessing four OB folds. The ssDNA binding modes of EcSSB are dependent on salt concentration in a solution.13 In the (SSB)35 mode, which is favored in low salt concentrations, only two subunits in an EcSSB tetramer bind 35 nucleotides (nts) to form the SSB35 complex. In the (SSB)65 mode, which is favored in moderately high salt concentrations, all four subunits of EcSSB participate in ssDNA binding to form the SSB65 complex. Crystallographic studies on SSB bound with two molecules of ssDNA dC35 homopolymer solved at a resolution of 2.8 Å suggest a binding model for the SSB65 complex in a binding topology resembling seams on a baseball.12 In the (SSB)65 mode, four essential aromatic residues, Trp40, Trp54, Phe60, and Trp88, participate in ssDNA binding and allow nucleic acids to wrap around the entire SSB via stacking interactions.
The intermediate ssDNA binding states of EcSSB are found by the structural changes between SSB35 and SSB65 complexes.14 Given that multiple barriers are observed between discrete wrapping conformations, EcSSB–ssDNA interactions are not smooth.14 ssDNA unwrapping analysis shows that EcSSB can diffuse along ssDNA in different binding modes. The structural dynamics of EcSSB reveals four possible modes for EcSSB wrapping, namely, (SSB)65, (SSB)56, (SSB)35, and (SSB)17. Additional complexed crystal structures of SSB with different ssDNA lengths15 or with inhibitor16 are required to elucidate how SSB stably binds ssDNA in different and intermediate wrapping states.
Apart from the EcSSB–dC35 complex, the EcPriB–dT15 complex is another complex structure of SSB-like protein possessing an OB-fold domain from E. coli.17 PriB is a primosomal protein required for replication restart1,18 and shares structural similarity with the DNA-binding domain of EcSSB.19,20 Although structural resemblance suggests that EcPriB may bind ssDNA in the same manner as EcSSB, they have different ssDNA-binding paths.17 In addition, PriB binds to ssDNA mainly through electrostatic interactions in the L45 loop rather than by stacking interactions. The L23 loop of PriB has three aromatic residues, but only Trp47 is involved in ssDNA binding.17
In this study, we solved the crystal structure of PaSSB bound with the two molecules of ssDNA dT25 at a resolution of 1.91 Å (PDB entry 6IRQ). Through structure–function analysis, we proposed that only two OB folds, rather than four OB folds in PaSSB, are enough for the formation of a stable complex with ssDNA.
Experimental
Protein expression and purification
E. coli BL21(DE3) cells transformed with the expression vector pET21b–PaSSB were grown at 37 °C in LB medium supplemented with 100 μg mL−1 ampicillin.21 PaSSB expression was induced by incubating with 1 mM isopropyl thiogalactopyranoside for 3 h at 37 °C. The recombinant PaSSB protein was purified using the protocol described previously.22 Briefly, the cells overexpressing the protein were chilled on ice, harvested by centrifugation, resuspended in the binding buffer (20 mM Tris–HCl, 5 mM imidazole, 0.5 M NaCl; pH 7.9) and disrupted by sonication with ice cooling between pulses. Specifically, the sonicator power was set to 3.5; each pulse lasted 2 s, with 2 s between pulses. The complete pulse sequence lasted 5 min and was repeated three times. PaSSB was purified from the soluble supernatant by Ni2+-affinity chromatography (HiTrap HP; GE Healthcare Bio-Sciences), eluted with Buffer A (20 mM Tris–HCl, 250 mM imidazole, and 0.5 M NaCl, pH 7.9), and dialyzed against a dialysis buffer (Buffer B; 20 mM HEPES and 100 mM NaCl, pH 7.0). Protein purity remained at >97% as determined by SDS-PAGE.Crystallography
Before crystallization, PaSSB was concentrated to 20 mg mL−1 in Buffer B. PaSSB was incubated with dT25 at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (PaSSB tetramer/dT25) ratio. Despite such ratio, only two ssDNA chains were found per asymmetric unit after structural determination. Crystals were grown at room temperature by hanging drop vapor diffusion in 30% PEG 400, 100 mM MES, 100 mM magnesium chloride, pH 6.5. Diffraction data were collected using an ADSC Quantum-315r CCD area detector at SPXF beamline BL13C1 at NSRRC (Taiwan). All data integration and scaling were carried out using HKL-2000.23 The crystal structure of PaSSB complexed with ssDNA dT25 was determined at 1.91 Å resolution with the molecular replacement software Phaser-MR24 using PaSSB as model (PDB entry 5YUO).9 Four monomers of PaSSB (monomers A, B, C and D) and two ssDNAs (chains E and F) were found per asymmetric unit. A model was built and refined with PHENIX25 and Coot.26 The final structure was refined to an R-factor of 0.213 and an Rfree of 0.248 (Table 1). Atomic coordinates and related structure factors have been deposited in the PDB with accession code 6IRQ.
4 (PaSSB tetramer/dT25) ratio. Despite such ratio, only two ssDNA chains were found per asymmetric unit after structural determination. Crystals were grown at room temperature by hanging drop vapor diffusion in 30% PEG 400, 100 mM MES, 100 mM magnesium chloride, pH 6.5. Diffraction data were collected using an ADSC Quantum-315r CCD area detector at SPXF beamline BL13C1 at NSRRC (Taiwan). All data integration and scaling were carried out using HKL-2000.23 The crystal structure of PaSSB complexed with ssDNA dT25 was determined at 1.91 Å resolution with the molecular replacement software Phaser-MR24 using PaSSB as model (PDB entry 5YUO).9 Four monomers of PaSSB (monomers A, B, C and D) and two ssDNAs (chains E and F) were found per asymmetric unit. A model was built and refined with PHENIX25 and Coot.26 The final structure was refined to an R-factor of 0.213 and an Rfree of 0.248 (Table 1). Atomic coordinates and related structure factors have been deposited in the PDB with accession code 6IRQ.
| a Values in parentheses are for the highest resolution shell.b Rsym = Σ|I − ‘I’|/ΣI, where I is the observed intensity, ‘I’ is the statistically weighted average intensity of multiple observations of symmetry-related reflections. | |
|---|---|
| Data collection | |
| Crystal | PaSSB–dT25 |
| Wavelength (Å) | 0.975 |
| Resolution (Å) | 30–1.91 |
| Space group | P31 |
| Cell parameters | |
| a, b, c (Å) | 60.49, 60.49, 131.32 |
| α, β, γ (°) | 90, 90, 120 |
| Completeness (%)a | 97.4 (97.1) |
| <I/σI> | 24.3 (2.67) |
| Rsym or Rmerge (%)b | 0.047 (0.401) |
| Redundancy | 3.6 (3.6) |
| Refinement | |
| Resolution (Å) | 29.47–1.91 |
| No. reflections | 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 647 647 |
| Rwork/Rfree | 0.213/0.248 |
| No. atoms | |
| Protein | 410 |
| DNA | 26 |
| Water | 115 |
Ratio (polypeptide chain![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ssDNA) ssDNA) |
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| Rms deviation | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.866 |
| Ramachandran plot | |
| In preferred regions | 385 (97.96%) |
| In allowed regions | 8 (2.04%) |
| Outliers | 0 (0%) |
| PDB entry | 6IRQ |
Electrophoretic mobility shift analysis (EMSA)
EMSA for PaSSB was conducted in accordance with a previously described protocol for SSB.27 In brief, various lengths of ssDNA oligonucleotides were radiolabeled with [γ32P] ATP (6000 Ci mmol−1; PerkinElmer Life Sciences, Waltham, MA) and T4 polynucleotide kinase (Promega, Madison, WI, USA). Protein (0, 19, 37, 77, 155, 310, 630, 1250, 2500, and 5000 nM) was incubated at 25 °C for 30 min with 1.7 nM DNA substrates (dT23–60) in a total volume of 10 mL in 20 mM Tris–HCl (pH 8.0) and 100 mM NaCl. Aliquots (5 mL) were removed from each of the reaction solutions and added to 2 mL of gel loading solution (0.25% bromophenol blue and 40% sucrose). The resulting samples were resolved on 8% native polyacrylamide gel at 4 °C in TBE buffer (89 mM Tris borate and 1 mM EDTA) for 1 h at 100 V and visualized through phosphorimaging. A phosphor storage plate was scanned, and data regarding complex and free DNA bands were digitized for quantitative analysis.28,29Results and discussion
X-ray crystal structure of the PaSSB-ssDNA complex
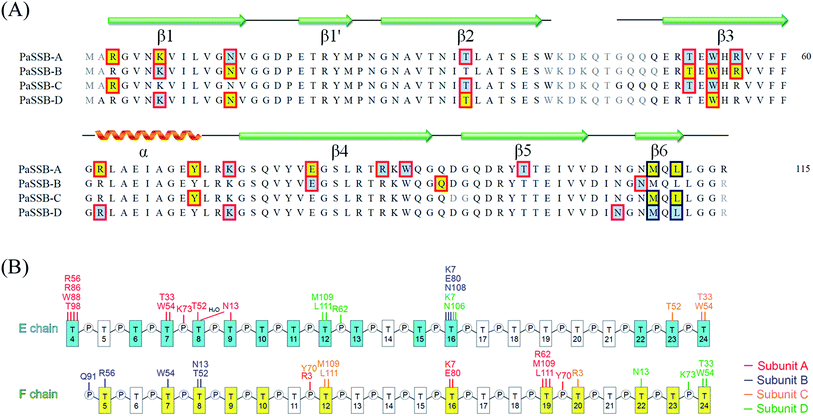
Although the binding of SSB to ssDNA is a dynamic process, stable transition complexes can still be observed.14 In this study, we aimed to identify a new ssDNA-binding mode of SSB via crystallographic analysis. The structural dynamics of EcSSB reveals at least four modes for EcSSB wrapping.14 To understand these intermediate wrapping states of SSB in detail, we generated PaSSB crystal in a complex with homopolymeric ssDNA dT25 through hanging drop vapor diffusion. The structure was determined at a resolution of 1.91 Å by molecular replacement (Fig. 1A). The crystallographic asymmetric unit contained four PaSSB monomers (monomers A, B, C, and D) and two ssDNAs (chains E and F). The PaSSB monomer folded as an OB-fold containing one α-helix and six β-strands (β1–β1′–β2–β3–β4–β5–β6). The core of the OB-fold domain in PaSSB monomer possessed a β-barrel capped with an α-helix. Given that the entire C-terminal domain of SSB is disordered7 even when bound to ssDNA,11 the amino acid residues 114–165 in this complex structure of PaSSB were not observed.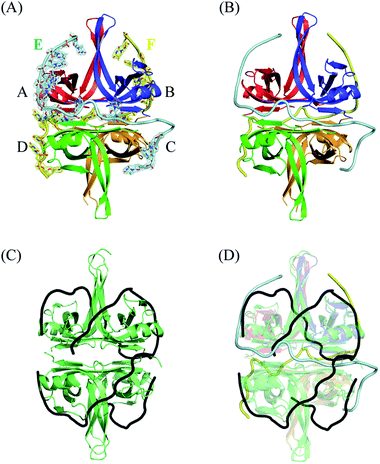 | ||
| Fig. 1 (A) Crystal structure of PaSSB complexed with ssDNA dT25. Four monomers of PaSSB (monomers A, B, C, and D) and two ssDNAs (chains E and F) were found in the asymmetric unit. The composite omit map (at 1.0 σ) indicated the presence of ssDNA. The entire C-terminal domain of PaSSB (aa 114–165) became disordered and disappeared. (B) An S-shaped conformation of bound ssDNA. Evident electron density was observed only for nucleotides T4, T6–13, T15–16, and T22–24 in chain E (14 mers) and T5, T7–10, T12, T16, T19–20, and T22–24 in chain F (12 mers). Undefined nucleotides in chains E and F were modeled on the basis of fragmentary electron density for defining binding paths. As shown in the binding paths, ssDNA chain E or F adopted an S-shaped conformation in the PaSSB complex. (C) The EcSSB–dC35 complex. The structure of EcSSB bound with two molecules of ssDNA dC35 homopolymer (PDB entry 1EYG) suggests a binding model for the SSB65 complex in a binding topology resembling seams on a baseball. (D) The superimposed structures of the EcSSB–dC35 and the PaSSB–dT25 complex. Their ssDNA binding modes differ significantly. | ||
Electron density showed that ssDNA was bound to each PaSSB monomer (Fig. 1A). Given that no sequence specificity occurred to mediate binding, PaSSB needed to bind to ssDNA via specific contact regions to accommodate weak and delicate interactions. A portion of the ssDNA electron density was discontinuous in this complex structure, likely reflecting its disorder. Although dT25 was used for crystallization, evident electron density was observed only for nucleotides T4, T6–13, T15–16, and T22–24 in chain E (14 mers) and T5, T7–10, T12, T16, T19–20, and T22–24 in chain F (12 mers). The resolved ssDNA segments likely represent major binding regions to SSB.30 For clarity, undefined nts in chains E and F were modeled on the basis of fragmentary electron density for defining binding paths (Fig. 1B). As shown in the binding paths, ssDNA chain E or F adopted an S-shaped conformation in the PaSSB complex. The two bound ssDNA chains adopted an H-shaped conformation in the PaSSB complex, which was significantly different from that in the EcSSB complex with two dC35.12 In (SSB)65 mode, ssDNA wrapped around all four SSB subunits with a topology that resembled the seams on a baseball (Fig. 1C). The bound ssDNA in the EcSSB–ssDNA complex grabbed subunit A and preferentially bound to the adjacent subunit B with a binding path of A–B–D–C. In the PaSSB complex, however, the ssDNA chain E comes into contact with an ssDNA binding cavity in subunit A, goes along with the dimer–dimer interface, and then binds subunit C with a binding path of A–C. Similarly, another ssDNA (chain F) binds the SSB dimer via a binding path of B–D. If all four PaSSB subunits are involved in a single ssDNA chain wrapping, a binding path of A–C–B–D is expected. However, whether all four PaSSB subunits are necessary for ssDNA binding remains unclear (see below).
ssDNA-interaction sites
Electron density for two separate segments of ssDNA was clearly observed in the PaSSB–ssDNA complex structure (Fig. 1A), in which each bound ssDNA chain adopted an S-shaped conformation (Fig. 1B). Unlike dC35 bound by EcSSB (Fig. 1C), ssDNA chains E and F in the PaSSB–dT25 complex did not occupy the entire binding sites and were not fully wrapped around each PaSSB subunit (Fig. 1D). Although PaSSB is a homotetramer,21 the ssDNA interaction sites in each monomer were different (Fig. 2A). Only Trp54 in all four subunits always involved ssDNA binding (Fig. 2A). Except in Met109 and Leu111, PaSSB–ssDNA interactions occurred through the side chains of the residues in PaSSB tetramer (Fig. 2A). A schematic of the protein–ssDNA interactions in the PaSSB–dT20 complex is shown in Fig. 2B. Although SSB binds to ssDNA in a sequence-independent manner, many specific interactions with ssDNA bases are observed in the PaSSB–dT25 complex. A detailed view of the PaSSB–ssDNA interactions for T4–10 (Fig. 3A), T11–13 (Fig. 3B), T15–16 (Fig. 3C), and T22–24 (Fig. 3D) in chain E and T5–10 (Fig. 3E), T12 (Fig. 3F), T16 (Fig. 3F), T19–20 (Fig. 3G), and T22–24 (Fig. 3H) in chain F is shown. Structurally, the following residues are involved in ssDNA binding: Arg3 (in subunits A and C), Lys7 (A, B, D), Asn13 (A, B, D), Thr33 (A, C, D), Thr52 (A, B, C), Trp54 (A, B, C, D), Arg56 (A, B), Arg62 (A, D), Tyr70 (A, C), Lys73 (A, D), Arg86 (A), Trp88 (A), Gln91 (B), Thr98 (A), Asn104 (D), Asn106 (B), Met109 (A, C, D), and Leu111 (A, C, D).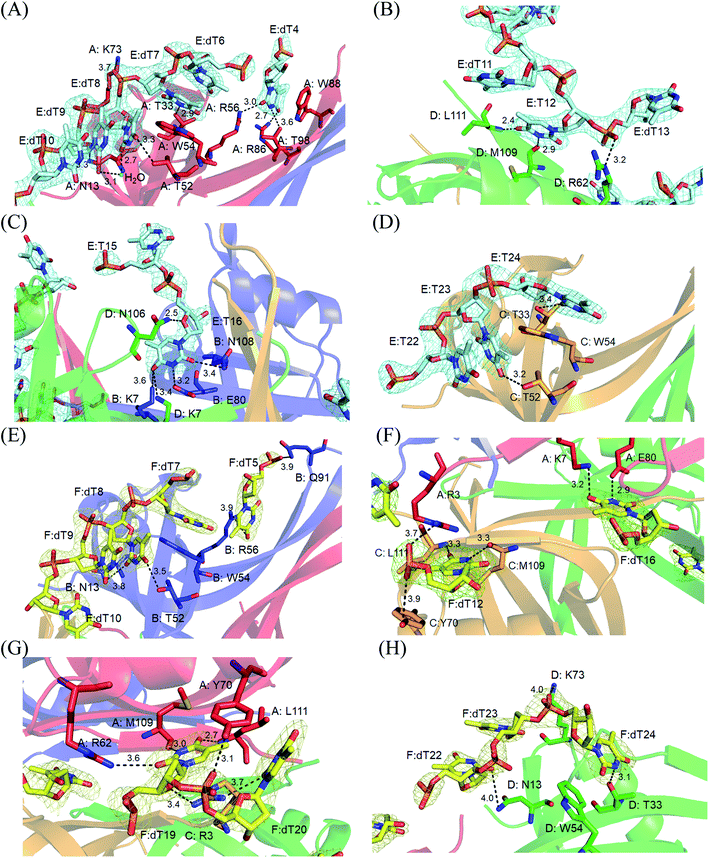 | ||
| Fig. 3 A detailed view of the PaSSB–ssDNA interactions for (A) T4–10, (B) T11–13, (C) T15–16, (D) T22–24 in chain E and (E) T5–10, (F) T12 and T16, (G) T19–20, (H) T22–24 in chain F is shown. | ||
The bound ssDNA in the PaSSB–ssDNA complex adopted an S-shaped conformation. ssDNA chain E comes into contact with an ssDNA binding cavity between β1β2 and β4β5, traverses along β2β3 of subunit A, goes along with the AB–CD dimer–dimer interface, and then crosses through the dimer interface to bind subunit C. The main chains of residues Met109 and Leu111 at the dimer–dimer interface were found to interact with ssDNA and are crucial for directing the S-shaped binding route. Met109 and Leu111 were located at the β6 strand (Fig. 2A). SsbA,31 SsbB,32 and some other SSBs17,19 did not contain this additional β6 strand in contrast to PaSSB. Additional β6 strands in SSB are known to function as a clamp to catch and join two neighboring subunits together in a tetrameric SSB.33 As revealed by our complex structure, we further found that β6 strands in PaSSB guide and define S-shaped binding route via nonspecific interactions by Met109 and Leu111 at the β6 strand.
Contributions of ssDNA binding by aromatic residues
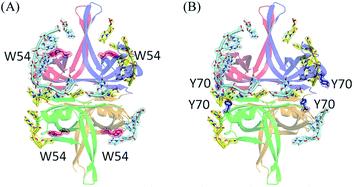
In the EcSSB–ssDNA complex, four essential aromatic residues, Trp40, Trp54, Phe60, and Trp88, participate in ssDNA binding.12 These residues are conserved in most SSB families, and the corresponding residues in PaSSB are Trp40, Trp54, Phe60, and Trp88 (Fig. 2A). Trp40, Trp54, and Phe60 located at the L23 loop of EcSSB are strongly involved in ssDNA binding and guide the ssDNA to wrap around the tetramer through base stacking interactions.12 Although these residues in PaSSB are also located at the L23 loop, only Trp54 is crucial for ssDNA binding (Fig. 2A). Unlike that in the EcSSB–ssDNA complex, Trp40 and Phe60 in the PaSSB–dT25 complex was extremely far and thus could not interact with ssDNA (Table 2). In addition, only one Trp88 in subunit A was involved in ssDNA binding. The distances between Trp88 and the nearest ssDNA were 3.6 Å (subunit A and dT4 in chain E), 8.3 Å (subunit B and dT5 in chain F), 13.3 Å (subunit C and dT24 in chain E), and 13.6 Å (subunit D and dT24 in chain F). These distances between Trp88 and ssDNA suggest that only one Trp88 can serve as an interaction site for ssDNA. Among the four essential aromatic residues in EcSSB, only Trp54 fully participates in ssDNA binding in a PaSSB tetramer (Fig. 4A and Table 2).| Residue | Distance (Å) | ssDNA | Importance (distance < 4 Å) |
|---|---|---|---|
| a The side chains of Trp40 in subunits B and D were unobserved in the PaSSB–dT25 complex due to poor electron density. | |||
| Trp40 (A) | 5.8 | dT13 (E) | |
| Trp40 (C) | 19.3 | dT15 (E) | |
| Trp54 (A) | 3.5 | dT7 (E) | ● |
| Trp54 (B) | 3.6 | dT7 (F) | ● |
| Trp54 (C) | 3.4 | dT24 (E) | ● |
| Trp54 (D) | 3.5 | dT24 (F) | ● |
| Phe60 (A) | 13.5 | dT16 (F) | |
| Phe60 (B) | 12.9 | dT16 (E) | |
| Phe60 (C) | 14.2 | dT16 (F) | |
| Phe60 (D) | 14.1 | dT15 (E) | |
| Tyr70 (A) | 3.1 | dT20 (F) | ● |
| Tyr70 (B) | 13.9 | dT22 (E) | |
| Tyr70 (C) | 3.9 | dT12 (F) | ● |
| Tyr70 (D) | 4.1 | dT11 (E) | |
| Trp88 (A) | 3.6 | dT4 (E) | ● |
| Trp88 (B) | 8.3 | dT5 (F) | |
| Trp88 (C) | 13.3 | dT24 (E) | |
| Trp88 (D) | 13.6 | dT24 (F) | |
Through structural analysis, we noted another aromatic residue, namely, Tyr70, which may interact with ssDNA (Fig. 4B and Table 2). Tyr70 is located at the helix between β3 and β4 strand (Fig. 2A). The distance between Tyr70 and the nearest ssDNA was 3.1 Å (subunit A and dT20 in chain F) or 3.9 Å (subunit C and dT12 in chain F). These distances suggest the role of Tyr70 in PaSSB for ssDNA interaction. In summary, the contribution from aromatic residues in PaSSB tetramer for ssDNA binding is not equal for each subunit. Contributions of ssDNA binding by aromatic residues were obviously found in the subunit A of PaSSB tetramer (Trp54, Tyr70, and Trp88).
Comparison of ssDNA-binding modes of other SSBs
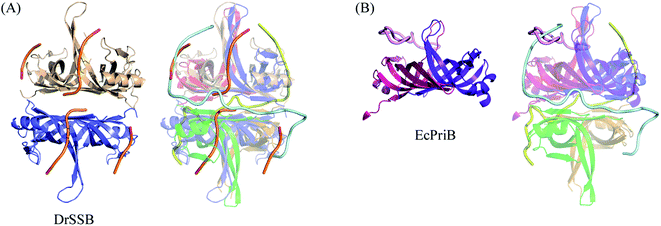
The N-terminal domain of EcSSB and PaSSB is similar in terms of sequence and structure.9,21 Therefore, they may be concluded to be similar in complex structure and function. However, their ssDNA-binding modes were very different (Fig. 1D). The ssDNA-binding mode of PaSSB also significantly differed from that of Deinococcus radiodurans SSB (DrSSB),34 a homodimeric SSB in which each monomer contains two OB folds (Fig. 5A). We also compared the ssDNA-binding mode of PaSSB with that of PriB, an SSB-like protein (Fig. 5B). PriB is a basic accessory protein in PriA-directed DNA replication restart primosome.18,35 The PriB monomer has an OB-fold structure19,20 and shares structural similarity with the DNA-binding domain of EcSSB. Although PriB and PaSSB have a classical OB-fold ssDNA-binding surface,17 they differentially bind DNA (Fig. 5B). Unlike EcSSB and PaSSB, PriB dimers bind a single ssDNA chain dT15 with the highly electrostatic positive L45 loop surface rather the aromatic residues on the L23 loop and β3 strand. The L23 loop of PriB also has three aromatic residues, but only Trp47 is involved in ssDNA binding.17 In summary, the ssDNA-binding mode of PaSSB revealed by the PaSSB–dT25 complex also differs from those of the other SSB proteins, such as PriB and DrSSB, not only EcSSB.25-mer ssDNA (dT25) is sufficient for the stable formation of the PaSSB–ssDNA complex
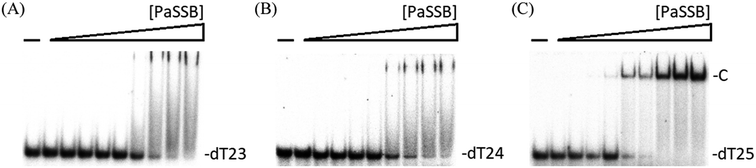
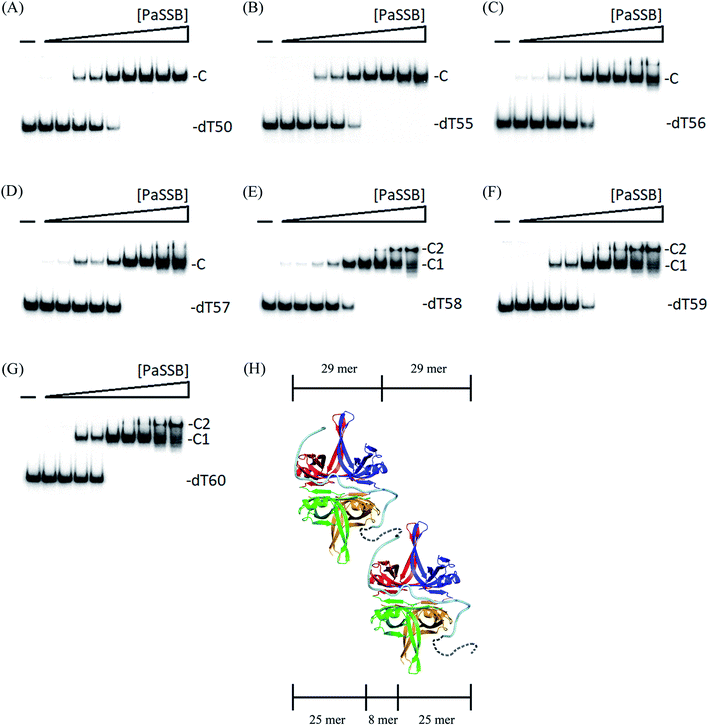
We previously demonstrated that the size of the ssDNA binding site of PaSSB is 29 ± 2 nt.22 In this study, the complex structure further reveals that a PaSSB tetramer may be bound by two ssDNA dT25. To experimentally investigate whether both or just one of these two dT25 is sufficient for the stable formation of the PaSSB–ssDNA complex, we studied the binding of PaSSB to ssDNA of different lengths and with different protein concentrations by using EMSA (Fig. 6). EMSA is a well-established approach in studies of molecular biology,27 allowing the detection of the distinct protein–DNA complex(es).36 When we incubated PaSSB with dT23 (Fig. 6A) or dT24 (Fig. 6B), no significant band shift was observed, indicating that PaSSB could not form a stable complex with these homopolymers. Given that some smears were observed, PaSSB could still interact with dT23 or dT24. However, the binding activity was not strong enough to form a stable protein–DNA complex in solution. We then used dT25 to test whether the complex can be stably formed. Although dT25 was only 1 nt longer than dT24, its pattern of the PaSSB–ssDNA complex observed using EMSA was very different. In contrast to dT23 or dT24, dT25 complexed with PaSSB produced a very significant band shift (Fig. 6C). Thus, the EMSA results suggested that the minimal length of ssDNA required for a PaSSB tetramer binding was 25 nt. This result thereby raises an interesting possibility whether one of the two ssDNA chains in complex structure of PaSSB (Fig. 1A) is an artifact, that is, just bound via a crystal packing (see below).Two different complexes formed when PaSSB binds to ssDNA dT58
To further examine the minimal nucleotide length necessary for the binding of a second PaSSB tetramer to ssDNA already prebound to a PaSSB, we studied the binding of PaSSB to long dT homopolymers of 50–60 nt. Incubation of PaSSB with dT50 (Fig. 7A), dT55 (Fig. 7B), dT56 (Fig. 7C), or dT57 (Fig. 7D) formed a single complex. The binding behavior of >58-long dT homopolymers to PaSSB was different. At low protein concentrations, PaSSB formed a single complex with dT58 (Fig. 7E), similar to that observed with dT55 (Fig. 7B). However, when the PaSSB concentration was increased, another slow-migrating complex appeared. This case was similar to that in dT59 (Fig. 7F) or dT60 (Fig. 7G). The appearance of the second complex resulted from the increasing PaSSB concentration, suggesting that at least two PaSSB tetramers were contained per oligonucleotide. The presence of an additional nt in dT58, as compared with dT57, provided enough interaction space for the binding of a second PaSSB tetramer, which occupied approximately 29 nt (58 ÷ 2 = 29). Given that the minimal length of ssDNA required for PaSSB binding is 25 nt (Fig. 6C), the ssDNA linker for steric consideration between two PaSSB tetramer should be 8 nt (58 − 25 − 25 = 8). On the basis of these structural and functional analyses, we proposed and sketched a possible binding model for PaSSB to ssDNA (Fig. 7H). In this model, we assumed that the second ssDNA chain packed in the complex structure of PaSSB, probably the F chain (Fig. 1A), is not required for forming a stable complex. However, this second (or “backup”) site for ssDNA binding in PaSSB is still needed due to the dynamic binding process of SSB during different conformational change states.14Two OB folds in PaSSB are adequate for the formation of a stable complex with ssDNA
Although two ssDNA chains (dT25) were found in the complex structure of PaSSB (Fig. 1), EMSA analysis further indicated that the minimal length of ssDNA required for PaSSB binding is only 25 nt, that is, only two OB folds in PaSSB may participate in ssDNA binding. The appearance of the second complex resulted from increasing PaSSB concentrations, suggesting at least two PaSSB tetramers occur per dT58 (Fig. 7H). In addition, the binding of PaSSB to dT58 (Fig. 7E), dT59 (Fig. 7F), or dT60 (Fig. 7G) appeared to be nearly noncooperative because essentially, these long dT homopolymers shift into the first complex (C1) before the appearance of the second complex (C2). Thus, the S-shaped conformation of ssDNA revealed by our complex structure of PaSSB (Fig. 1) may indicate a noncooperative binding mode. This mode in structure was not similar to that of EcSSB because ssDNA did not fully wrap around PaSSB (Fig. 1D) and only the Trp54 among all aromatic residues in PaSSB is involved in ssDNA binding (Table 2). These significant differences may explain why distinct ssDNA binding mechanisms can be used for SSBs.Given the structural and experimental evidence in this study, we speculated that only two OB folds in PaSSB are essential for ssDNA binding. In many cases, OB folds can be broad ligand binders to both ssDNA and protein.4 For the tumor suppressor BRCA2,37 two OB folds bind ssDNA, and a third OB fold is involved in protein–protein interactions. For RPA, a eukaryotic homolog of SSB, two distinct binding modes involve two OB folds and four OB folds.38 For PriB, an SSB-like protein, only one of two OB folds binds ssDNA.17 Whether another OB fold in PriB is open for other primosomal protein binding remains unclear.17 Thus, all four OB folds in PaSSB are not necessary to simultaneously participate in the binding to ssDNA. Empty OB folds in PaSSB may allow sliding, as described in a single-molecule experiment.39
We previously found that P. aeruginosa does not contain any of the recognizable homologs of priB, priC, dnaT, and dnaC in its genome.18 These gene products are primosomal proteins serving as different kinds of DnaB helicase loader for DNA replication restart.1 Only the replication restart initiator PriA and SSB are found in P. aeruginosa. Thus, P. aeruginosa following the replication restart strategy of E. coli is impossible. In conclusion, our structure–function analyses indicate different ssDNA-binding modes between PaSSB and EcSSB (Fig. 1D). The ssDNA-binding contributions by their aromatic residues are significantly different (Fig. 4 and Table 2). According to EMSA results, two OB folds in PaSSB are enough to form a stable complex with ssDNA (Fig. 6 and 7). One of the two ssDNA chains in the complex of PaSSB may be bound through crystal packing (Fig. 1B). Further research can directly focus on determining how ssDNA can be differently interacted and recognized by homotetrameric SSB. Given the absence of homologs to priB, priC, dnaT, and dnaC, whether and how PaSSB participates in PriA-directed primosome for DNA replication restart remains unknown.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank the experimental facility and the technical services provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology, Taiwan, ROC. This research was supported by a grant from the Ministry of Science and Technology, Taiwan (MOST 108-2320-B-040-010 to C.·Y. Huang)References
- T. A. Windgassen, S. R. Wessel, B. Bhattacharyya and J. L. Keck, Nucleic Acids Res., 2018, 46, 504–519 CrossRef CAS.
- E. Antony and T. M. Lohman, Semin. Cell Dev. Biol., 2018, 86, 102–111 CrossRef.
- R. R. Meyer and P. S. Laine, Microbiol. Rev., 1990, 54, 342–380 CAS.
- T. H. Dickey, S. E. Altschuler and D. S. Wuttke, Structure, 2013, 21, 1074–1084 CrossRef CAS PubMed.
- A. G. Murzin, EMBO J., 1993, 12, 861–867 CrossRef CAS.
- R. D. Shereda, A. G. Kozlov, T. M. Lohman, M. M. Cox and J. L. Keck, Crit. Rev. Biochem. Mol. Biol., 2008, 43, 289–318 CrossRef CAS PubMed.
- P. R. Bianco, Prog. Biophys. Mol. Biol., 2017, 127, 111–118 CrossRef CAS PubMed.
- R. Nigam, M. Mohan, G. Shivange, P. K. Dewangan and R. Anindya, Mol. Biol. Rep., 2018, 45, 865–870 CrossRef CAS PubMed.
- Y. H. Huang and C. Y. Huang, RSC Adv., 2018, 8, 35280–35288 RSC.
- P. R. Bianco, S. Pottinger, H. Y. Tan, T. Nguyenduc, K. Rex and U. Varshney, Protein Sci., 2017, 26, 227–241 CrossRef CAS PubMed.
- S. N. Savvides, S. Raghunathan, K. Futterer, A. G. Kozlov, T. M. Lohman and G. Waksman, Protein Sci., 2004, 13, 1942–1947 CrossRef CAS PubMed.
- S. Raghunathan, A. G. Kozlov, T. M. Lohman and G. Waksman, Nat. Struct. Biol., 2000, 7, 648–652 CrossRef CAS PubMed.
- T. M. Lohman and M. E. Ferrari, Annu. Rev. Biochem., 1994, 63, 527–570 CrossRef CAS PubMed.
- S. Suksombat, R. Khafizov, A. G. Kozlov, T. M. Lohman and Y. R. Chemla, eLife, 2015, 4, e08193 CrossRef PubMed.
- Y. H. Huang, E. S. Lin and C. Y. Huang, Biochem. Biophys. Res. Commun., 2019, 520, 353–358 CrossRef CAS PubMed.
- C. Y. Huang, Biochem. Biophys. Res. Commun., 2018, 504, 704–708 CrossRef CAS PubMed.
- C. Y. Huang, C. H. Hsu, Y. J. Sun, H. N. Wu and C. D. Hsiao, Nucleic Acids Res., 2006, 34, 3878–3886 CrossRef CAS PubMed.
- Y. H. Huang and C. Y. Huang, BioMed Res. Int., 2014, 2014, 195162 Search PubMed.
- Y. H. Huang, Y. H. Lo, W. Huang and C. Y. Huang, Genes Cells, 2012, 17, 837–849 CrossRef CAS PubMed.
- J. H. Liu, T. W. Chang, C. Y. Huang, S. U. Chen, H. N. Wu, M. C. Chang and C. D. Hsiao, J. Biol. Chem., 2004, 279, 50465–50471 CrossRef CAS PubMed.
- H. C. Jan, Y. L. Lee and C. Y. Huang, Protein J., 2011, 30, 20–26 CrossRef CAS PubMed.
- Y. H. Huang and C. Y. Huang, BioMed Res. Int., 2014, 2014, 573936 Search PubMed.
- Z. Otwinowski and W. Minor, Methods Enzymol., 1997, 276, 307–326 CAS.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed.
- J. J. Headd, N. Echols, P. V. Afonine, R. W. Grosse-Kunstleve, V. B. Chen, N. W. Moriarty, D. C. Richardson, J. S. Richardson and P. D. Adams, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 381–390 CrossRef CAS PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- C. Y. Huang, Determination of the binding site-size of the protein–DNA complex by use of the electrophoretic mobility shift assay, in Stoichiometry and Research: The Importance of Quantity in Biomedicine, ed. Innocenti A., InTech Press, Rijeka, Croatia, 2012, pp. 235–242 Search PubMed.
- Y. H. Huang, M. J. Lin and C. Y. Huang, Genes Cells, 2013, 18, 1007–1019 CrossRef CAS PubMed.
- Y. H. Huang, Y. L. Lee and C. Y. Huang, Protein J., 2011, 30, 102–108 CrossRef CAS PubMed.
- K. Dubiel, A. R. Myers, A. G. Kozlov, O. Yang, J. Zhang, T. Ha, T. M. Lohman and J. L. Keck, J. Mol. Biol., 2019, 431, 178–195 CrossRef CAS PubMed.
- Y. H. Huang, H. H. Guan, C. J. Chen and C. Y. Huang, PLoS One, 2017, 12, e0182060 CrossRef PubMed.
- K. L. Chen, J. H. Cheng, C. Y. Lin, Y. H. Huang and C. Y. Huang, RSC Adv., 2018, 8, 28367–28375 RSC.
- T. Paradzik, N. Ivic, Z. Filic, B. A. Manjasetty, P. Herron, M. Luic and D. Vujaklija, Nucleic Acids Res., 2013, 41, 3659–3672 CrossRef CAS PubMed.
- N. P. George, K. V. Ngo, S. Chitteni-Pattu, C. A. Norais, J. R. Battista, M. M. Cox and J. L. Keck, J. Biol. Chem., 2012, 287, 22123–22132 CrossRef CAS.
- M. M. Cox, M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler and K. J. Marians, Nature, 2000, 404, 37–41 CrossRef CAS PubMed.
- B. Dey, S. Thukral, S. Krishnan, M. Chakrobarty, S. Gupta, C. Manghani and V. Rani, Mol. Cell. Biochem., 2012, 365, 279–299 CrossRef CAS PubMed.
- H. Yang, P. D. Jeffrey, J. Miller, E. Kinnucan, Y. Sun, N. H. Thoma, N. Zheng, P. L. Chen, W. H. Lee and N. P. Pavletich, Science, 2002, 297, 1837–1848 CrossRef CAS.
- C. A. Brosey, C. Yan, S. E. Tsutakawa, W. T. Heller, R. P. Rambo, J. A. Tainer, I. Ivanov and W. J. Chazin, Nucleic Acids Res., 2013, 41, 2313–2327 CrossRef CAS PubMed.
- R. Zhou, A. G. Kozlov, R. Roy, J. Zhang, S. Korolev, T. M. Lohman and T. Ha, Cell, 2011, 146, 222–232 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |