Open Access Article
Open Access ArticleThree new coordination polymers based on bis(4-(4H-1,2,4-triazol-4-yl)phenyl)methane: syntheses, structures, multiresponsive luminescent sensitive detection for antibiotics and pesticides, and antitumor activities†
Chang-Sheng Wanga,
Qi Huanga,
Xia Wanga,
Yu-Tong Zhanga,
Dong-Sheng Ma a,
Ying-Hui Yu
a,
Ying-Hui Yu *a and
Jin-Sheng Gao*ab
*a and
Jin-Sheng Gao*ab
aSchool of Chemistry and Materials Science, Heilongjiang University, Harbin 150080, China. E-mail: Yuyinghui@hlju.edu.cn; gaojins@hlju.edu.cn; Fax: +86-451-86609151; Tel: +86-451-86609001
bAgricultural College, Heilongjiang University, Harbin 150080, China
First published on 20th December 2019
Abstract
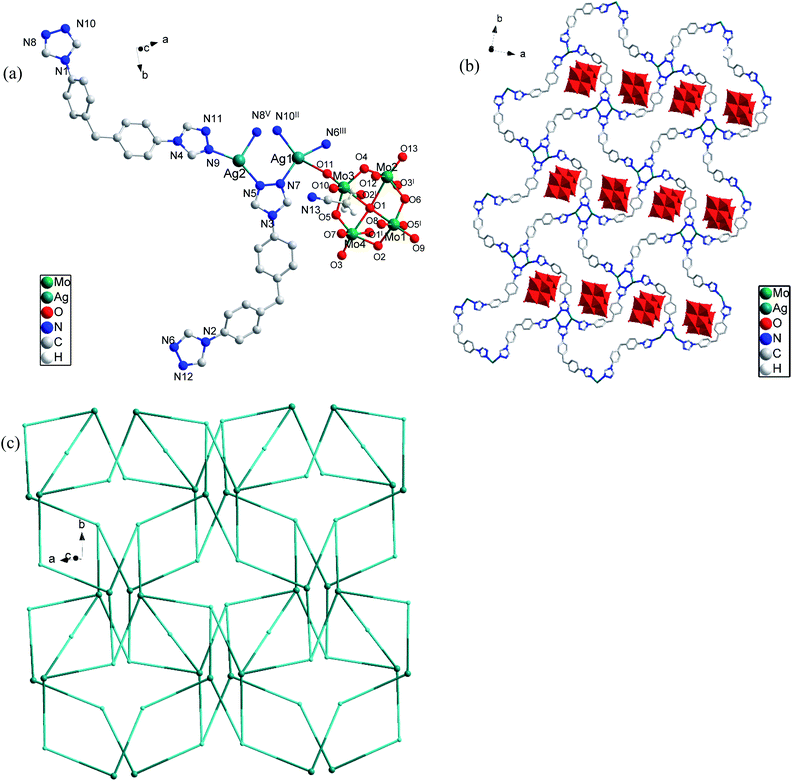
Three novel coordination polymers (CPs), namely, {[Ag2(L)2(Mo4O13)·(CH3CN)]}n (1), {[Zn(L)(1,4-bdc)2·2(1,4-H2bdc)]}n (2), {[Cd(L)(1,4-bdc)0.5]}n (3) have been synthesized under solvothermal conditions by the reaction of bis(4-(4H-1,2,4-triazol-4-yl)phenyl)methane (L) and varied metal salts. Their structures are determined by single X-ray crystal diffraction, and further characterized by elemental analysis, IR, TGA and PXRD. CP 1 with ammonium molybdate as a secondary ligand displays a 2D network with (2,3,3,3,4)-connected net topology and the point symbol of {4·82}6{4·84·10}2{8}, CP 2 and CP 3 with 1,4-H2bdc as a secondary ligand demonstrate 3D structures with different topologies. CP 2 exhibits high sensibility and low detection limit in the recognition of antibiotics (NZF, NFT and FZD) and pesticide (DCN) identification. CP 1 demonstrates good anti-tumor activity toward the tested glioma cells. The possible luminescent sensitivity and anti-tumor mechanisms are also discussed.
1 Introduction
Coordination polymers (CPs) from the assembly of inorganic clusters and tunable organic linkers are extensively investigated as promising materials for applications in various fields,1–4 such as life sciences,5,6 environmental areas,7–10 medicinal science11,12 and the nuclear industry.13 Due to the simultaneous introduction of organic ligands and metal centers, some specific properties have been endowed to the CPs with versatile functions like biological activity,14–16 molecular magnetism,17,18 electronic conductivity,19 second-order nonlinear optics (NLO) activity,20,21 ferromagnetism,22,23 catalysis,24–26 and molecular recognition.27–30 Among these applications, CPs with luminescent recognition properties attracted special attention for their great advantages of structural tunability, high sensitivity and efficiency. A variety of CPs-based luminescence sensors were synthesized for the aim of detecting metal cations, aromatic explosives and organic pollutants.31–33 In the past several decades, antibiotics and pesticides originally used in the medical and agricultural fields have emerged as one of typical stubborn organic pollutants in water, simple and convenient methods for their effective recognition is fully meaningful. The design of luminescence sensing materials represents an attractive way to efficiently detect such organics.N-Heterocyclic compounds such as pyridine, pyrazine and triazole, which are easily obtained and modified, were widely used and reported for the construction of CPs. In 2018, several 3D MOFs based on triazole ligand were found to own excellent stability in water and could be used as recyclable multi-responsive sensors for metal ions, anions and antibiotics.34,35 Interestingly, triazole and its derivatives usually exhibit good biological activities, which could be maintained in their coordination compounds. As Srinivasa Budagumpi research group reported in 2019, several silver(I) and gold(I) CPs with coumarin-functionalized 1,2,4-triazol-5-ylidene ligands, exhibit good performance in cytotoxic studies.36,37
In this article, we choose a bidentate triazole ligand bis(4-(4H-1,2,4-triazol-4-yl)phenyl)methane and ammonium molybdate, 1,4-benzenedicarboxylic acid (1,4-H2bdc) as ligands to build functional CPs (Scheme 1). Three novel CPs, namely, {[Ag2(L)2(Mo4O13)·(CH3CN)]}n (1), {[Zn(L)(1,4-bdc)2·2(1,4-H2bdc)]}n (2), {[Cd(L)(1,4-bdc)0.5]}n (3) were successfully obtained and their structures were characterized by single crystal X-ray diffraction analysis, elemental analysis, IR analysis and thermal gravimetric analysis. The antibiotic and pesticide fluorescence recognition and antitumor activities were studied.
2 Experimental sections
2.1 General methods and materials
All of chemicals and reagents used in the experiment were purchased and without further purification. The IR spectra were obtained on a PerkinElmer Spectrum 100 FT-IR spectrometer equipped with a DGTS detector (32 scans). We used the KBr compression method with the scanning ranging from 4000 to 500 cm−1. The elemental analyses of C, H by the PerkinElmer 2400 elemental analyzer. Thermogravimetric analyses (TGA) were carried out by a PerkinElmer STA 6000 with a heating rate of 10 °C min−1 under atmosphere from 50 to 800 °C. The PXRD data of the samples were collected on a RigakuD/MAX-3B diffractometer using Cu-Kα radiation (λ = 1.5418 Å) and 2θ ranging from 5 to 50°. Luminescence spectra were recorded on a Shimadzu RF-5301 spectrophotometer. UV spectra were recorded on a Shimadzu UV-2700 spectrophotometer. Electrochemical testing was performed on an electrochemical analyzer potentiostat model 600E from CH Instruments.2.2 Synthesis of {[Ag2(L)2(Mo4O13)·(CH3CN)]}n (1)
The mixture of AgNO3 (17 mg, 0.1 mmol), L (30.2 mg, 0.1 mmol), (NH4)6Mo7O24·4H2O (123.6 mg, 0.1 mmol), acetonitrile (8 mL) and H2O (2 mL) were sealed in a 25 mL Teflon-lined stainless steel container and heated at 130 °C for three days. After cooled to room temperature, the colorless and transparent short block crystals of 1 were separated by filtration. Yield: 72% (based on Ag(I)). Elemental analysis (%) calcd for 1 (C36H31N13O13Ag2Mo4): C, 29.73%; H, 2.13%; N, 12.53%; found: C, 30.14%; H, 2.07%; N, 12.32%. IR (solid KBr pellet, cm−1): 3100 m, 3030 m, 2800 m, 1650 m, 1540 w, 1400 s, 870 s, 790 s, 734 m, 680 m, 581 s, 527 w.2.3 Synthesis of {[Zn(L)(1,4-bdc)2·2(1,4-H2bdc)]}n (2)
The mixture of Zn(NO3)2·6H2O (29.8 mg, 0.1 mmol), L (30.2 mg, 0.1 mmol), 1,4-H2bdc (16.6 mg, 0.1 mmol), acetonitrile (8.0 mL) and H2O (2.0 mL) were stirred for 0.5 h in air with adjusting to pH = 8 by dilute sodium hydroxide and then sealed in a Teflon-lined stainless steel container (25 mL), and heated at 130 °C for three days. After cooled to room temperature, the light yellow and transparent block crystals of 2 were separated by filtration. Yield: 52% (based on Zn(II)). Elemental analysis (%) calcd for 2 (C33H20N6O8Zn): C, 57.07%; H, 2.88%; N, 12.11%; found: C, 56.43%; H, 2.91%; N, 12.24%. IR (solid KBr pellet, cm−1): 3123 s, 2757 m, 2611 m, 2483 w, 1715 s, 1605 s, 1560 s, 1385 s, 1248 s, 1047 m, 882 w, 745 s, 571 w, 525 w.2.4 Synthesis of {[Cd(L)(1,4-bdc)0.5]}n (3)
The mixture of Cd(NO3)2·4H2O (64.1 mg, 0.2 mmol), L (30.2 mg, 0.1 mmol), 1,4-H2bdc (16.6 mg, 0.1 mmol), acetonitrile (8.0 mL) and H2O (2.0 mL) were sealed in a 25 mL Teflon lined stainless steel container and heated at 140 °C for three days. After cooled to room temperature, the light blue and transparent strip crystals of 3 were separated by filtration. Yield: 47% (based on Cd(II)). Elemental analysis (%) calcd for 3 (C21H16N6O2Cd): C, 50.72%; H, 3.22%; N, 16.91%; found: C, 49.93%; H, 3.57%; N, 16.74%. IR (solid KBr pellet, cm−1): 3453 s, 3105 w, 1952 w, 1568 s, 1385 s, 1248 m, 1101 s, 983 w, 920 w, 745 s, 644 s, 534 s.2.5 X-ray crystallography
Single crystal X-ray diffraction data of CPs 1–3 were collected on a Bruker APEX CCD area detector imaging plate diffractometer equipped with graphite-monochromated Mo-Kα (λ = 0.71073 Å), and the test temperature of CPs 1–3 is 302 K, 298 K and 98 K respectively. The structures were solved by direct methods and then refined by full-matrix least-squares methods on F2 using SHELXL-2017/1 crystallographic software package. All isolated O atoms have been considered as water atoms. The crystal parameters, refinement results and data collection for 1–3 are listed in Table 1.| 1 | 2 | 3 | |
|---|---|---|---|
| a R1 = Σ ||Fo| − |Fc||/Σ|Fo|.b wR2 = {Σ[w(Fo2 − Fc2)2/Σw(Fo2)2]}1/2. | |||
| Empirical formula | C36H31Ag2Mo4N13O13 | C33H20N6O8Zn | C21H16CdN6O2 |
| Fw | 1453.24 | 693.92 | 496.80 |
| Crystal system | Monoclinic | Triclinic | Monoclinic |
| Space group | C2/c | ![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) 1 1 |
P21/c |
| Temperature (K) | 302 | 298 | 98 |
| a (Å) | 24.1943(9) | 9.6385(2) | 16.6171(5) |
| b (Å) | 23.8328(9) | 12.8687(3) | 8.1566(2) |
| c (Å) | 15.2210(6) | 13.4092(3) | 20.3047(6) |
| α (deg) | 90 | 71.8180(10) | 90 |
| β (deg) | 102.567(2) | 75.2240(10) | 110.4400(10) |
| γ (deg) | 90 | 74.5340(10) | 90 |
| V (Å3) | 8566.4(6) | 1495.94(6) | 2578.80(13) |
| Z | 8 | 2 | 4 |
| Dcalc (g cm−3) | 2.254 | 1.541 | 1.280 |
| μ (mm−1) | 2.113 | 0.886 | 0.871 |
| F(000) | 5632 | 708 | 992 |
| Collected/unique | 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 529/10 529/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656 656 |
33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100/7393 100/7393 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 943/5921 943/5921 |
| R (int) | 0.0611 | 0.0289 | 0.0232 |
| GOF on F2 | 1.090 | 1.017 | 1.060 |
| R1a [I > 2σ(I)] | 0.0855 | 0.0333 | 0.0268 |
| wR2b [I > σ(I)] | 0.1978 | 0.0887 | 0.0776 |
| R1a (all) | 0.1479 | 0.0407 | 0.0311 |
| wR2b (all) | 0.2603 | 0.0933 | 0.0803 |
3 Results and discussion
3.1 Structural description
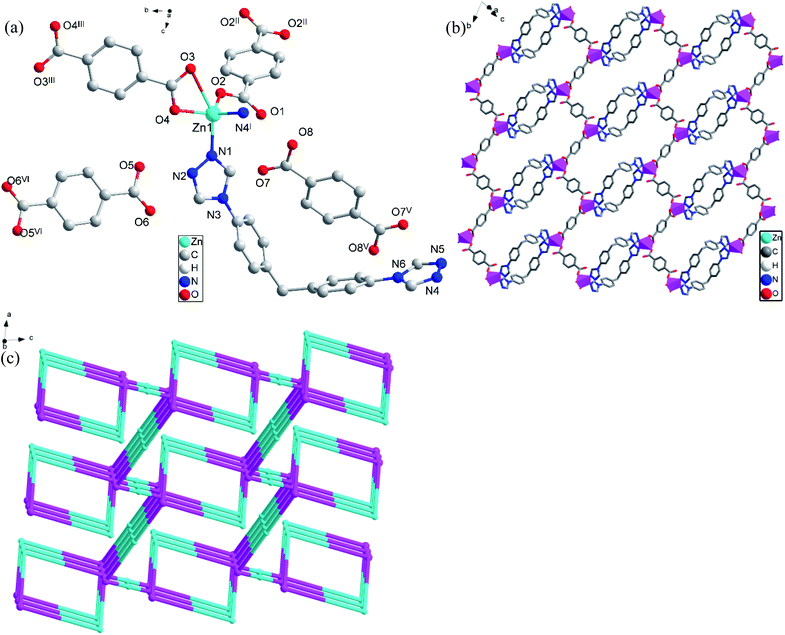
![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) 1 space group. An asymmetric unit of CP 2 consists of one independent Zn(II) atom, one L ligand, two [1,4-bdc] ions with different coordination modes and two free [1,4-H2bdc] molecules. Fig. 2(a) shows the coordination environment of Zn(II) atom in CP 2. The Zn(II) atom is coordinated by two N (N1, N4I) atoms from two different L ligands and three O (O2, O3, O4) atoms from two different [1,4-bdc] ions. The Zn–O bond lengths range from 1.9430(13) to 1.9499(13) Å, the Zn–N bond lengths range from 2.0020(15) to 2.0484(15) Å, the N–Zn–N bond angle is 99.82(6), the N–Zn–O bond angles range from 96.46(6) to 119.60(7), the O–Zn–O bond angle range is 111.17(6), which are all in the normal ranges. The coordination environment of Zn(II) atom can be described as a trigonalbipyramidal geometry.
1 space group. An asymmetric unit of CP 2 consists of one independent Zn(II) atom, one L ligand, two [1,4-bdc] ions with different coordination modes and two free [1,4-H2bdc] molecules. Fig. 2(a) shows the coordination environment of Zn(II) atom in CP 2. The Zn(II) atom is coordinated by two N (N1, N4I) atoms from two different L ligands and three O (O2, O3, O4) atoms from two different [1,4-bdc] ions. The Zn–O bond lengths range from 1.9430(13) to 1.9499(13) Å, the Zn–N bond lengths range from 2.0020(15) to 2.0484(15) Å, the N–Zn–N bond angle is 99.82(6), the N–Zn–O bond angles range from 96.46(6) to 119.60(7), the O–Zn–O bond angle range is 111.17(6), which are all in the normal ranges. The coordination environment of Zn(II) atom can be described as a trigonalbipyramidal geometry.
In CP 2, two L ligands connect two Zn(II) atoms to form a ring structure of [ZnLZnL], the distance of Zn(II)⋯Zn(II) is 14.55 Å. At the same time, each Zn(II) atom is also connected to two [1,4-bdc] ions with different coordination modes. The infinite [ZnLZnL] rings are further connected through [1,4-bdc] ions with different coordination modes to form an infinitely extended 3D structure (Fig. 2(b)). In order to further analyze and understand the 3D structure of CP 2, topological analysis is carried out. If considering the Zn(II) atom as a 4-connected node, L and two [1,4-bdc] ions with different coordination modes as 2-,2-, 2-connected nodes respectively, the 3D structure of CP 2 can be seemed as a (2,2,4)-connected net topology with the point symbol of {12}{4·125}{4} (Fig. 2(c)).
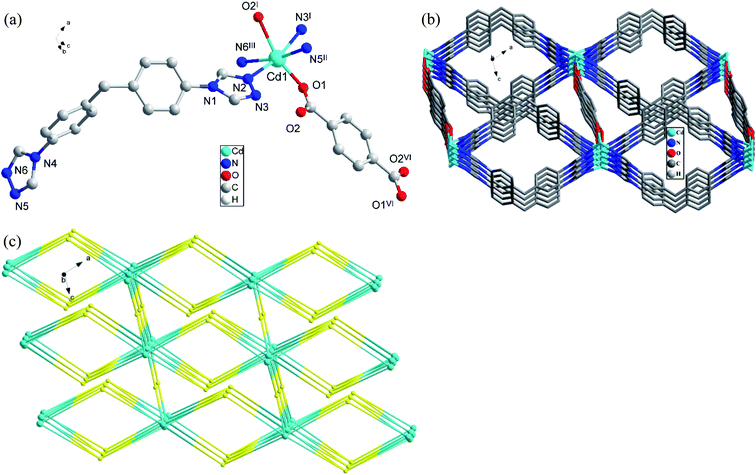
In CP 3, each L ligand connect four different Cd(II) atoms, and each Cd(II) atom is linked by four different ligands, thus forming an alternating 2D [CdLCdL] ring structure, the distance of Cd(II)⋯Cd(II) is 16.25 Å or 16.31 Å. Furthermore, each Cd(II) atom of [CdLCdL] ring is linked by two different [1,4-bdc] ions to generate a 3D structure eventually (Fig. 3(b)). In order to further analyze and understand the 3D structure of CP 3, topological analysis is carried out. If considering the [1,4-bdc] ion as a 4-connected node, L and Cd(II) atom as 4- and 6-connected nodes respectively, the 3D structure of CP 3 can be seemed as a (4,4,6)-connected net topology with the point symbol of {42·83·10}{44·62}2{48·66·8}2 (Fig. 3(c)).
3.2 PXRD patterns and thermal stability analysis
To further determine the purity of CPs 1–3, powder X-ray diffraction (Fig. S2†) was carried out. It is found that the diffraction peaks of the as-synthesized CPs are highly consistent with the simulated PXRD patterns, indicating their excellent phase purity. To study the stability of CPs 1–3, thermogravimetric analysis (TGA) (Fig. S3†) was conducted from 50 to 800 °C with a heating rate of 10 °C min−1 under air atmosphere. For CP 1, the first weight loss of 2.88% from 50 °C to 175 °C can be ascribed to the loss of one free acetonitrile molecule (calcd 2.82%). The second weight loss of 23.85% from 175 °C to 600 °C can be ascribed to the loss of the two diphenylmethane molecules (calcd 23.12%). When the temperature reaches 750 °C, the triazole ring begins to break. It is supposed that the skeleton remains stable under high temperature, which indicates its potential to act as high temperature resistant material.38 For CP 2, almost no weight loss is observed when the temperature is below 300 °C. The first weight loss of 22.76% from 300 °C to 390 °C can be ascribed to the loss of one free [1,4-H2bdc] molecule (calcd 23.92%). The second weight loss of 63.36% from 390 °C to 703 °C is consistent with the loss of coordinated [1,4-bdc] ions and L (calcd 64.34%). The remaining weight of 14.88% corresponds to the formation of ZnO (calcd 11.74%). For CP 3, the first weight loss of 3.78% from 50 °C to 240 °C can be ascribed to the loss of one coordinated water molecule (calcd 3.63%). The second weight loss of 12.59% occurs from 240 °C to 397 °C due to the loss half of coordinated [1,4-bdc] ion (calcd 12.88%). Further decomposition of 57.12% from 397 °C to 740 °C is consistent with the loss of L (calcd 57.73%). The remaining weight of 26.51% was ascribed to the formation of CdO (calcd 25.76%).3.3 Detection of antibiotics
Since the advent of penicillin in the 1950s, antibiotics have been widely used in the field of medicine. In the past several decades, antibiotics have been recognized as a type of serious organic pollutant to environment especially water. Currently, the detection of antibiotics mainly depends on precision apparatus such as mass spectrometry (MS), ion mobility spectrometry (IMS), liquid chromatography and mass spectrometry (LC-MS), etc. All these methods are complex, inconvenient, and expensive.39,40 Therefore, developing simple and effective ways to detect antibiotics is of great importance. As one of ongoing research in our laboratory, we try to explore the application of CPs to identify trace organics such as antibiotics in water. The experimental results indicate that CP 2 owns better fluorescence recognition activity compared with CP 1 and CP 3. Therefore, CP 2 is selected to investigate its sensibility toward analytes (antibiotics) in water medium. First, CP 2 was grounded into powder using a mortar, then stirred in water for 2 hours to prepare a CP 2 suspension. 10 different antibiotics of 5 categories, namely nitrofurans (nitrofurazone, NZF; nitrofurantoin, NFT; furazolidone FZD), nitroimidazoles (metronidazole, MDZ; dimetridazole, DTZ), sulfonamides (sulfadiazine, SDZ; sulfamethazine, SMZ), amide alcohols (chloramphenicol, CAP; thiamphenicol, THI) and β-lactams (cephalexin, LEX) were selected for luminescent recognition. Various antibiotic in DMF solutions with a concentration of 0.30 mM were added to 1 mM well-dispersed suspension of CP 2 in water. As shown in Fig. 4, nitrofuran antibiotics (NZF, NFT and FZD) have very obvious fluorescence quenching effects to the luminescence of CP 2. At the same time, it is also found that the emission peak of CP 2 had a significant red shift when the nitrofuran and nitroimidazole antibiotics were added, indicating the decrease in the energy level of the π orbits of the ligand.40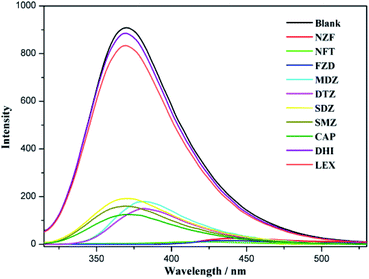 | ||
| Fig. 4 Comparisons of the fluorescence intensities of CP 2 treated by different antibiotics of 0.30 mM. | ||
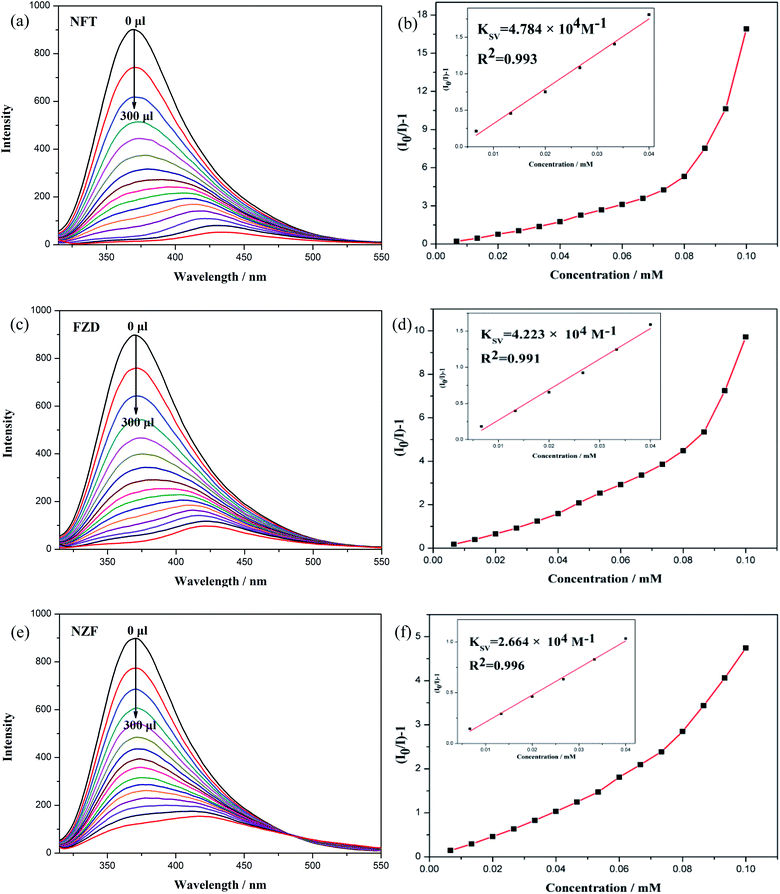
We performed quantitative luminescent titration experiments to assess the sensitivity of CP 2 toward three identifiable nitrofuran antibiotics. According to Fig. 5(a), (c) and (e), the fluorescence emission intensity of the sensor CP 2 drops dramatically as the concentration of antibiotics increases. When the antibiotics concentration is 0.1 mM, the luminescent quenching efficiencies of NFT, FZD and NZF toward CP 2 toward CP 2 reach 98.4%, 96.6% and 86.5%, calculated by (I0 − I)/I0 × 100%. CP 2 demonstrates the best recognition activity to NFT compared with the other antibiotics, suggesting that its potential application as a promising fluorescent probe for nitrofuran antibiotics. The luminescent quenching efficiency of antibiotics towards CP 2 is calculated by the Stern–Volmer (SV) equation: I0/I = 1 + KSV × [C], KSV represents the quenching constant (M−1), [C] represents the molar concentration of the analyte (M), I0 and I represent the fluorescence emission intensity at certain excitation wavelength in the absence and presence of the analyte, respectively. Based on the KSV values, the limitation of detection (LOD) of CP 2 toward NFT, FZD and NZF could be calculated by the formula of 3δ/KSV, where σ is the standard deviation for three repeated tests of the fluorescence emission intensity (δ = 0.012). As showed in Fig. 5(b), (d) and (f), it is found that the SV plots of NFT, FZD and NZF show good linear correlation at lower concentration, deviating from linearity and bending upward at higher concentration. Based on the experimental data, the calculated values of KSV for NFT, FZD and NZF are 4.784 × 104 M−1, 4.223 × 104 M−1 and 2.664 × 104 M−1 respectively. The detection limits for NFT, FZD and NZF are 179 ppb, 192 ppb and 268 ppb, respectively. This work represents an example of a Zn-CP-based luminescence sensor for efficient detection of antibiotics.40
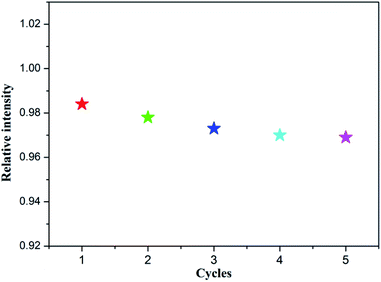
In addition, the recyclability of the CP 2 was also investigated as a fluorescence detector toward nitrofuran antibiotics (NFT taken as an example). After the fluorescence detection experiments are completed, the solid compound of CP 2 is collected from the mixture by physical methods such as washing and centrifugation. It is worth noting that the sample can be reused for five cycles and maintains its original fluorescence sensing activity, which further ensured its potential application as a fluorescent probe in antibiotics detection (Fig. 6).
3.4 Detection of pesticides
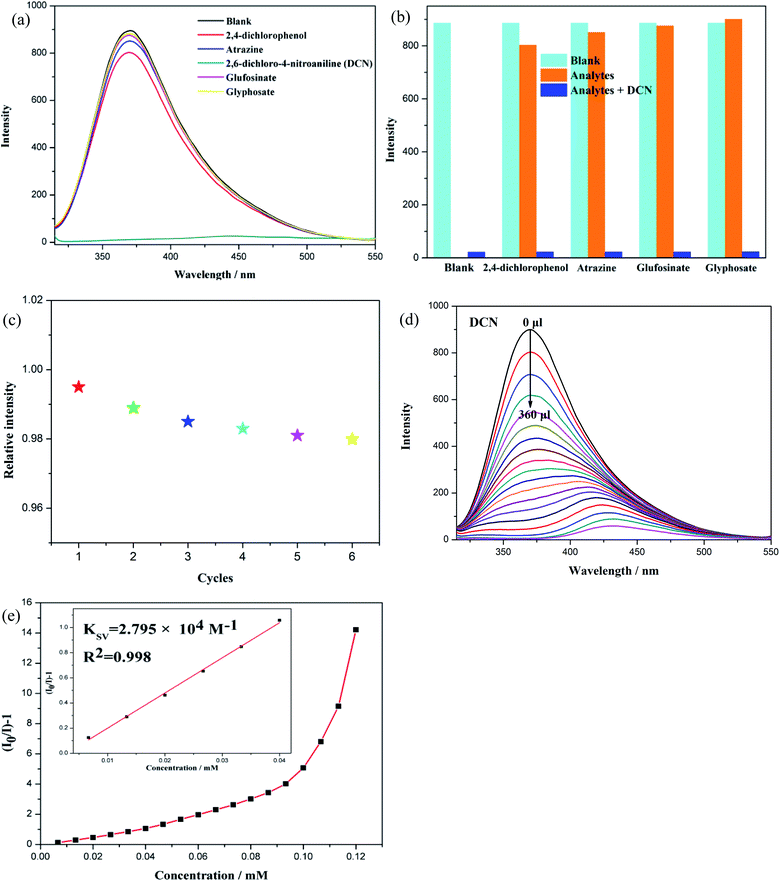
Pesticides play a vital role in agricultural production, however, which not only causes serious environmental pollution, but also becomes a great harm to human health.41–43 Effective detection of pesticides in the environment is needed and CPs-based sensors provided a viable alternative technique. We tried to detect pesticides in water with CP 2 as a fluorescent probe which exhibited good performance in antibiotics recognition. We selected five frequently used pesticides, including 2,4-dichlorophenol, atrazine, 2,6-dichloro-4-nitroaniline (DCN), glufosinate and glyphosate. Fig. 7(a) shows the effect of the presence of pesticides (0.30 mM) on the fluorescence emission intensity of CP 2. Apparently, the fluorescent emission of CP 2 is nearly quenched by DCN, while other pesticides have little influence on the luminescence intensity of CP 2, suggesting that CP 2 has a highly sensitive selectivity for DCN in aqueous solution. In addition, anti-interference experiments showed that the presence of other pesticides did not affect the DCN-induced luminescence quenching response, further confirming its high selectivity for DCN (Fig. 7(b)). After the fluorescence detection experiments were accomplished, the solid compound of CP 2 was collected from the mixture by some physical methods such as washing and centrifugation to investigate its recycle ability. It is found that CP 2 can be reused for six cycles and still maintains the original sensibility toward DCN (Fig. 7(c)). The above results indicate that CP 2 has good anti-interference ability and excellent recycle ability as pesticide sensor. We also performed quantitative luminescent titration experiments to assess the sensitivity of CP 2 toward DCN. As shown in Fig. 7(d), as the concentration of DCN increases, the fluorescence emission intensity of the sensor (CP 2) drops dramatically. When the DCN concentration reached 0.12 mM, DCN produced a maximum quenching efficiency of 99.5%, calculated by (I0 − I)/I0 × 100%. When the concentration of DCN is increased, the fluorescence emission peak of CP 2 has a significant red shift, indicating that the energy level of the π orbital of the ligand is reduced.40 Meanwhile, the fluorescence quenching efficiency is expressed by the Stern–Volmer equation. From Fig. 7(e), calculating the KSV value is 2.795 × 104 M−1, and the limitation of detection (LOD) is 267 ppb that could be calculated by the formula of 3δ/KSV, where δ is the standard deviation for three repeated tests of the fluorescence emission intensity (δ = 0.012). In summary, CP 2 could act as a fluorescent sensing material in the detection of trace pesticide DCN.44,453.5 Mechanism of luminescent sensing
Energy, charge and structure are three important factors that affect fluorescence sensing. We studied the mechanism of CP 2 detection of antibiotics and DCN through subsequent experiments. First, we recovered the CP 2 powder after fluorescence recognition experiments toward nitrofuran antibiotics and DCN. After washing, centrifuging and drying, CP 2 was conducted PXRD analysis and the result was shown in Fig. S4.† No apparent changes were found in the PXRD patterns compared with the original ones, suggesting that the sensing process is not caused by structural collapse of CP 2. According to the previous report, another possible mechanism might be photoinduced electron transfer.46,47 The photoinduced excitation electrons can be transferred from the sensor to the LUMO of the analytes rather than the ground state of the sensor. The lower the LUMO energy of the analytes, the easier the electrons are transferred. Therefore, we tested the electrochemical spectra of 10 antibiotics and 5 pesticide analytes. The map is divided into two stages of oxidation and reduction (Fig. S7†). The HOMO level and LUMO level of the analyte are obtained according to the calculation formula (−|4.78 + x| eV) (Fig. S8†). It is found that NFT, FZD and NZF have the lowest LUMO energy level, which explains the luminescent quenching of CP 2. However, for DCN and other antibiotics with lower quenching ability toward CP 2, the order of their quenching efficiency is not in good accordance with their LUMO energy, indicating that photoinduced electron transfer might not be the only reason for the luminescence recognition in this system. As indicated previously,48 energy allocation and transfer were proposed as another possible mechanism, including Förster resonance energy transfer (FRET) and inner filter effect. Generally, the spectral overlap between the absorbance spectrum and the emission spectrum of the framework structure has an impact on the energy transfer, which could cause the fluorescence quenching phenomenon. As shown in Fig. S9,† the good overlap of the CP 2 emission spectrum with the UV-vis absorption bands of nitrofuran antibiotics and DCN might explain such efficient fluorescence quenching of CP 2. Therefore, the abovementioned co-existence of electron-transfer and energy-transfer processes might be two possible explanations for the sensing activity of CP 2 toward nitrofuran antibiotics and DCN.473.6 Antitumor activity test
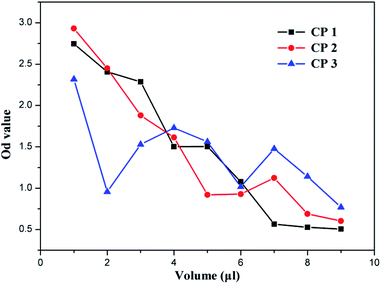
Every year, millions of people are killed by cancer, which has become a major threat to human health. At present, traditional chemotherapy has many limitations, such as severe side effects and multidrug resistance, which would obstruct therapeutic efficacy.49–52 Therefore, the development of antitumor drugs with lower side effects is of urgent need. We used flow cytometer to study the antitumor activities and mechanism of the newly prepared three CPs. In the experiments, antitumor activities were tested by adding CP 1, CP 2 and CP 3 to rat C6 cells (glioma cell line). First, certain amount of CPs was dissolved in water to make a solution of 1 μM. Then, different amounts of the solution were added to the same cell population solution, and then the OD value of the cell solution after 6 hours was measured with an instrument. The final results are shown in Fig. 8, demonstrating that the three CPs do have certain antitumor activities toward the tested glioma cells. The curve of CP 1 is smoother than those of CP 2 and CP 3, indicating that CP 1 is more stable as a bactericidal drug. This further proves the excellent performance of the Ag organic complex exhibited in antitumor activity.53,54 The OD value was significantly reduced when 4 μL of CP 1 was added, and the maximum effect was achieved when about 7 μL was added, at which time only a small amount of tumor cells remained in the solution.3.7 Mechanism of antitumor
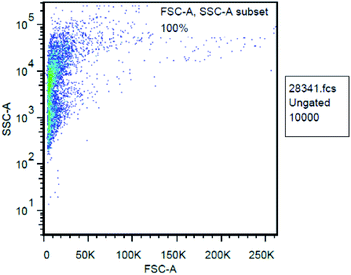
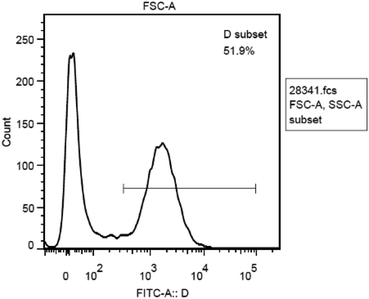
The antitumor mechanism of CP 1 was explored based on the data measured by flow cytometer. Currently, the possible mechanism of anti-malignant drugs can be simply divided into two categories: cytotoxic and indirect cytotoxic.55–57 The action mechanism of cytotoxic drugs is different from each other, some inhibit dihydrofolate reductases such as methotrexate, some destroy intracellular DNA such as carboplatin, some inhibit protein synthesis such as vinblastine, and others produce mitochondria and release toxic reactive oxygen such as arsenic trioxide, and so on. We hypothesized that the antitumor mechanism of these CPs is to produce reactive oxygen species.58 Flow cytometry was used to count the total number of cells, by using forward scattered light and side scattered light to obtain a cell-to-FSC and SSC scattering signal. The FSC-A and SSC-A of all cells are concentrated in the same picture to get the FSC-A, SSC-A scatter plot (Fig. 9). It can be seen that most of the cells are distributed on the left side, indicating that their similar small particle size with little difference. It is proved that the total number of cells is correctly detected, and the initial zero adjustment can be adjusted based on the total number of cells at this time.Since we hypothesized that the antitumor activity of CP 1 derived from mitochondria producing and releasing certain amount of reactive oxygen species that are toxic to the cell itself. Therefore, the fluorescent dye FITC which can respond to the active oxygen component was selected to stain the cells and the instrument was adjusted to the wavelength range in which active oxygen could be detected according to related literature.59 The fluorescence intensity of the cells at this time was examined, as shown in Fig. 10. It shows that the number of cells varies with the activity of reactive oxygen species. There are two peaks in Fig. 10, the distribution of fluorescence intensity is relatively dispersed, demonstrating that active oxygen is widely present in this cell population. Fig. 9 indicates a peak area of 51.9%, indicating that certain amount of active oxygen is produced in 51.9% of the cells. The anticancer mechanism might be the production and release of reactive oxygen species by the cells under the action of the CPs.
4 Conclusions
In conclusion, we have succeeded in preparing three metal–organic compounds CPs 1–3. The presence of different metal ions and second ligands results in CPs with diverse topologies and different properties. CP 2 exhibits excellent luminescence sensing activity toward NFs and DCN which could be applied as an efficient, recyclable, and multiresponsive luminescence sensor. The low detection limits for DCN (267 ppb) and NFs (179 ppb for NFT, 192 ppb for FZD, 268 ppb for NZF) reveal the high sensitivity of CP 2 for trace pesticide and antibiotics. The high luminescence quenching efficiencies of NFs and DCN to CP 2 may be ascribed to a combined effect from photoinduced electron transfer and resonance energy transfer. CP 1 was found to have good antitumor activity, the mechanism of which might be caused by the production of reactive oxygen species from cellular mitochondria, indicating its potential value in antitumor drug research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is financially supported by the National Natural Science Foundation of China (No. 21501050).References
- S. Dissegna, K. Epp, W. R. Heinz, G. Kieslich and R. A. Fischer, Adv. Mater., 2018, 30, 1704501 CrossRef PubMed.
- Q.-L. Zhu and Q. Xu, Chem. Soc. Rev., 2014, 43, 5468–5512 RSC.
- W. P. Lustig, S. Mukherjee, N. D. Rudd, A. V. Desai, J. Li and S. K. Ghosh, Chem. Soc. Rev., 2017, 46, 3242–3285 RSC.
- Y. Liu, X.-Y. Xie, C. Cheng, Z.-S. Shao and H.-S. Wang, J. Mater. Chem. C, 2019, 7, 10743–10763 RSC.
- M. Gimenez-Marques, T. Hidalgo, C. Serre and P. Horcajada, Coord. Chem. Rev., 2016, 307, 342–360 CrossRef CAS.
- J. Zhou, G. Tian, L. Zeng, X. Song and X.-w. Bian, Adv. Healthcare Mater., 2018, 7, 1800022 CrossRef PubMed.
- S.-W. Lv, J.-M. Liu, Z.-H. Wang, H. Ma, C.-Y. Li, N. Zhao and S. Wang, J. Environ. Sci., 2019, 80, 169–185 CrossRef PubMed.
- Y. Liu, Z. Liu, D. Huang, M. Cheng, G. Zeng, C. Lai, C. Zhang, C. Zhou, W. Wang, D. Jiang, H. Wang and B. Shao, Coord. Chem. Rev., 2019, 388, 63–78 CrossRef CAS.
- J. Cao, Z.-h. Yang, W.-p. Xiong, Y.-y. Zhou, Y.-r. Peng, X. Li, C.-y. Zhou, R. Xu and Y.-r. Zhang, Chem. Eng. J., 2018, 353, 126–137 CrossRef CAS.
- S. Mosleh, M. R. Rahimi, M. Ghaedi and K. Dashtian, RSC Adv., 2016, 6, 61516–61527 RSC.
- A. Chaix, E. Cueto-Diaz, A. Delalande, N. Knezevic, P. Midoux, J.-O. Durand, C. Pichon and F. Cunin, RSC Adv., 2019, 9, 31895–31899 RSC.
- W. Chen and C. Wu, Dalton Trans., 2018, 47, 2114–2133 RSC.
- J. Li, X. Wang, G. Zhao, C. Chen, Z. Chai, A. Alsaedi, T. Hayat and X. Wang, Chem. Soc. Rev., 2018, 47, 2322–2356 RSC.
- L. Wang, H. Zhu, Y. Shi, Y. Ge, X. Feng, R. Liu, Y. Li, Y. Ma and L. Wang, Nanoscale, 2018, 10, 11384–11391 RSC.
- M.-X. Wu and Y.-W. Yang, Adv. Mater., 2017, 29, 1606134 CrossRef PubMed.
- M. Esfahanian, M. A. Ghasemzadeh and S. M. H. Razavian, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 2024–2030 CrossRef CAS PubMed.
- W.-N. Liu, W.-Q. Tong, L.-L. Ma, Y. Wang, J.-M. Wang, L. Hou and Y.-Y. Wang, Dalton Trans., 2019, 48, 7786–7793 RSC.
- G. D. Degaga, R. Pandey, C. Gupta and L. Bharadwaj, RSC Adv., 2019, 9, 14260–14267 RSC.
- E. A. Dolgopolova, V. A. Galitskiy, C. R. Martin, H. N. Gregory, B. J. Yarbrough, A. M. Rice, A. A. Berseneva, O. A. Elegbavwo, K. S. Stephenson, P. Kittikhunnatham, S. G. Karakalos, M. D. Smith, A. B. Greytak, S. Garashchuk and N. B. Shustova, J. Am. Chem. Soc., 2019, 141, 5350–5358 CrossRef CAS PubMed.
- X. Liang, F. Zhang, H. Zhao, W. Ye, L. Long and G. Zhu, Chem. Commun., 2014, 50, 6513–6516 RSC.
- Z. Shi, L. Qin and H. Zheng, Dalton Trans., 2017, 46, 4589–4594 RSC.
- R. Dong, Z. Zhang, D. C. Tranca, S. Zhou, M. Wang, P. Adler, Z. Liao, F. Liu, Y. Sun, W. Shi, Z. Zhang, E. Zschech, S. C. B. Mannsfeld, C. Felser and X. Feng, Nat. Commun., 2018, 9, 2637 CrossRef PubMed.
- X. Wang, G. Gou, D. Wang, H. Xiao, Y. Liu, M. Zhang, B. Dkhil, X. Ren and X. Lou, RSC Adv., 2016, 6, 48779–48787 RSC.
- J. Zhao, H. Huang, M. Liu, J.-H. Wang, K. Liu and Z.-Y. Li, RSC Adv., 2019, 9, 26450–26455 RSC.
- C. Li, L. Lu, J. Wang, Q. Yang, D. Ma, A. Alowais, A. Alarifi, A. Kumar and M. Muddassir, RSC Adv., 2019, 9, 29864–29872 RSC.
- W. Mao, L. Xiang, C. A. Lamsfus, L. Maron, X. Leng and Y. Chen, J. Am. Chem. Soc., 2017, 139, 1081–1084 CrossRef CAS PubMed.
- Q. Qiu, H. Chen, Y. Wang and Y. Ying, Coord. Chem. Rev., 2019, 387, 60–78 CrossRef CAS.
- J. Li, C. Ye, Y.-n. Wu, Y. Zhu, J. Xu, Y. Wang, H. Wang, M. Guo and F. Li, Environ. Int., 2019, 125, 135–141 CrossRef CAS PubMed.
- M. Yu, X. Yao, X. Wang, Y. Li and G. Li, Polym, 2019, 11, 21201–21210 CAS.
- Q. Huang, J.-H. Huang, L. Gu, J.-x. Ruan, Y.-H. Yu and J.-S. Gao, RSC Adv., 2018, 8, 557–566 RSC.
- X. Zhang, N.-Y. Xu, Q. Ruan, D.-Q. Lu, Y.-H. Yang and R. Hu, RSC Adv., 2018, 8, 5714–5720 RSC.
- X. Wang, Y. Han, X. X. Han, X. Hou, J.-J. Wang and F. Fu, New J. Chem., 2018, 42, 19844–19852 RSC.
- S. Senthilkumar, R. Goswami, V. J. Smith, H. C. Bajaj and S. Neogi, ACS Sustainable Chem. Eng., 2018, 6, 10295–10306 CrossRef CAS.
- H. He, Q.-Q. Zhu, F. Sun and G. Zhu, Cryst. Growth Des., 2018, 18, 5573–5581 CrossRef CAS.
- H. He, Q.-Q. Zhu, C.-P. Li and M. Du, Cryst. Growth Des., 2019, 19, 694–703 CrossRef CAS.
- D. Aucamp, S. V. Kumar, D. C. Liles, M. A. Fernandes, L. Harmse and D. I. Bezuidenhout, Dalton Trans., 2018, 47, 16072–16081 RSC.
- G. Achar, C. R. Shahini, S. A. Patil, J. G. Malecki and S. Budagumpi, New J. Chem., 2019, 43, 1216–1229 RSC.
- X. Ji, S. Wang, C. Shao and H. Wang, ACS Appl. Mater. Interfaces, 2018, 10, 19712–19720 CrossRef CAS PubMed.
- Z.-W. Zhai, S.-H. Yang, M. Cao, L.-K. Li, C.-X. Du and S.-Q. Zang, Cryst. Growth Des., 2018, 18, 7173–7182 CrossRef CAS.
- X.-Y. Guo, F. Zhao, J.-J. Liu, Z.-L. Liu and Y.-Q. Wang, J. Mater. Chem. A, 2017, 5, 20035–20043 RSC.
- H. Kwon, K. M. Chan and E. T. Kool, Org. Biomol. Chem., 2017, 15, 1801–1809 RSC.
- T. Kumar, M. Venkateswarulu, B. Das, A. Halder and R. R. Koner, Dalton Trans., 2019, 48, 12382–12385 RSC.
- Y. Kim, J. Lee and I.-S. Shin, Sci. Rep., 2019, 9, 10293 CrossRef PubMed.
- C.-L. Tao, B. Chen, X.-G. Liu, L.-J. Zhou, X.-L. Zhu, J. Cao, Z.-G. Gu, Z. Zhao, L. Shen and B. Z. Tang, Chem. Commun., 2017, 53, 9975–9978 RSC.
- Z. Sun, Y. Bao, C. Wang, Z. Lin, A. Shi and H. Li, Inorg. Chim. Acta, 2019, 494, 266–270 CrossRef CAS.
- J.-M. Li, R. Li and X. Li, Crystengcomm, 2018, 20, 4962–4972 RSC.
- N. Xu, Q. Zhang, B. Hou, Q. Cheng and G. Zhang, Inorg. Chem., 2018, 57, 13330–13340 CrossRef CAS PubMed.
- M. Yu, Y. Xie, X. Wang, Y. Li and G. Li, ACS Appl. Mater. Interfaces, 2019, 11, 21201–21210 CrossRef CAS PubMed.
- P. Li, J. Li, X. Feng, J. Li, Y. Hao, J. Zhang, H. Wang, A. Yin, J. Zhou, X. Ma and B. Wang, Nat. Commun., 2019, 10, 2177 CrossRef.
- D. Wang, J. Zhou, R. Chen, R. Shi, G. Xia, S. Zhou, Z. Liu, N. Zhang, H. Wang, Z. Guo and Q. Chen, Biomaterials, 2016, 107, 88–101 CrossRef CAS PubMed.
- I. B. Vasconcelos, T. G. da Silva, G. C. G. Militao, T. A. Soares, N. M. Rodrigues, M. O. Rodrigues, N. B. da Costa Jr, R. O. Freire and S. A. Junior, RSC Adv., 2012, 2, 9437–9442 RSC.
- C. Kim and B. Kim, Nutrients, 2018, 10, 1021 CrossRef PubMed.
- S. Sahin-Bolukbasia and N. Sahin, J. Organomet. Chem., 2019, 891, 78–84 CrossRef.
- M. Monticelli, S. Bellemin-Laponnaz, C. Tubaro and M. Rancan, Eur. J. Inorg. Chem., 2017, 2488–2495 CrossRef CAS.
- I. A. Lazaro, S. Haddad, J. M. Rodrigo-Munoz, C. Orellana-Tavra, V. del Pozo, D. Fairen-Jimenez and R. S. Forgan, ACS Appl. Mater. Interfaces, 2018, 10, 5255–5268 CrossRef PubMed.
- A. R. Simovic, R. Masnikosa, I. Bratsos and E. Alessio, Coord. Chem. Rev., 2019, 398, 113011 CrossRef.
- H. Zhang, W. Jiang, R. Liu, J. Zhang, D. Zhang, Z. Li and Y. Luan, ACS Appl. Mater. Interfaces, 2017, 9, 19687–19697 CrossRef CAS PubMed.
- X. D. Wang, Y. Y. Sun, F. Z. Qu, G. Y. Su and Y. Q. Zhao, Biomed. Pharmacother., 2019, 118, 108589 CrossRef CAS PubMed.
- R. Bellam, D. Jaganyi, A. Mambanda, R. Robinson and M. D. Balakumaran, RSC Adv., 2019, 9, 31877–31894 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1–S5 and Fig. S1–S9. CCDC 1952081–1952083. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra08659e |
| This journal is © The Royal Society of Chemistry 2019 |