Open Access Article
Open Access ArticleMetabolomics study of the anti-inflammatory effects of endogenous omega-3 polyunsaturated fatty acids†
Yu Peng ,
Huixia Ren,
Hongxun Tao,
Chengwei He,
Peng Li,
Jian-Bo Wan
,
Huixia Ren,
Hongxun Tao,
Chengwei He,
Peng Li,
Jian-Bo Wan * and
Huanxing Su*
* and
Huanxing Su*
State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Taipa, Macau, China. E-mail: huanxingsu@umac.mo; jbwan@umac.mo
First published on 17th December 2019
Abstract
Low-grade inflammation is usually defined as the chronic production and a low-grade state of inflammatory factors, it often does not have symptoms, and has been associated with neurodegenerative disease, obesity, and diabetes. Omega-3 polyunsaturated fatty acids (n-3 PUFAs) are the precursors of many anti-inflammatory metabolites, such as resolvins and neuroprotectins. It is of interest to study the metabolic profile of endogenous n-3 PUFAs in low-grade inflammatory conditions. To evaluate the protective effects of endogenous n-3 PUFAs on low-grade inflammation with the metabolomics approach, we fed fat-1 mice with an n-6 PUFAs rich diet for a long time to induce a low-grade inflammatory condition. Multi-analysis techniques, including structural analysis using quadrupole time-of-flight mass spectrometry with MSE mode, were applied in untargeted metabolomics to search for meaningful metabolites with significant variance in fat-1 mice under low-grade inflammation. Following the untargeted metabolomics screening, several meaningful metabolites were selected which were associated with anti-inflammatory effects generated from endogenous n-3 PUFAs for further analysis. The results revealed that the purine metabolism, fatty acid metabolism and oxidative stress response pathways through insulin resistance were involved in anti-inflammatory mechanisms of n-3 PUFA in low-grade inflammatory conditions. For the first time, this study explored the highlighted pathways as contributors to the anti-inflammatory effects of endogenous n-3 PUFAs in low-grade inflammatory conditions.
1. Introduction
Low-grade inflammation has received considerable attention recently. It is usually defined as the chronic production and a low-grade state of inflammatory factors, but often does not have symptoms. In addition, low-grade inflammation contributes to the establishment of obesity, type 2 diabetes, cancer, cardiovascular diseases,1 neurodegeneration disease2 and so on. High-fat meals may induce the inflammation, and this is exaggerated by saturated fatty acids (SFA) and trans-monounsaturated fatty acids (trans-MUFA).3 Polyunsaturated fatty acids (PUFAs) are involved in the regulation of immunological and inflammatory responses. n-6 PUFAs, such as linoleic acid (LA) and arachidonic acid (AA), are associated with pro-inflammatory.4 Conversely, Omega-3 polyunsaturated fatty acids (n-3 PUFAs) have been reported to be able to modulate inflammatory responses to exert beneficial effects in a variety of neurological disorders.5–10 n-3 PUFAs are essential fatty acids for normal metabolism in mammals and play key roles in maintaining structural and functional integrity of cellular membrane. Mammals are not able to endogenously synthesize n-3 PUFAs and fail to convert n-6 PUFA to n-3 PUFAs, on account of lacking n-3 desaturase, the enzyme that catalyzes this reaction.11 In fat-1 transgenic mice fed a diet rich in n-6 and deficient in n-3 PUFAs, the tissue n-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-3 ratio is approximately 1
n-3 ratio is approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, whereas the ratio is 50
1, whereas the ratio is 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in wild type (WT) animals under this diet condition.12,13 In WT mammals, n-6 PUFAs often undergo transformation by the action of the enzyme n-6-desaturase and γ-linolenic acid is elongated to form dihomo-γ-linolenic acid and arachidonic acid, the precursor of pro-inflammatory cytokines.14 Diet rich in n-6 and deficient in n-3 PUFAs has the ability to enhance proinflammatory cytokine production.15 The modern Western diet is poor in n-3 PUFAs with a n-3
1 in wild type (WT) animals under this diet condition.12,13 In WT mammals, n-6 PUFAs often undergo transformation by the action of the enzyme n-6-desaturase and γ-linolenic acid is elongated to form dihomo-γ-linolenic acid and arachidonic acid, the precursor of pro-inflammatory cytokines.14 Diet rich in n-6 and deficient in n-3 PUFAs has the ability to enhance proinflammatory cytokine production.15 The modern Western diet is poor in n-3 PUFAs with a n-3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-6 PUFAs ratio of 1
n-6 PUFAs ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15–20, whereas the diet of our ancestors is estimated to have had a ratio of 1
15–20, whereas the diet of our ancestors is estimated to have had a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.14,16,17
1.14,16,17
It is of interest to provide detailed information about the role of n-3 PUFAs in the low-grade inflammatory conditions. The transgenic fat-1 mouse carries a fat-1 gene that encodes an n-3 fatty acid desaturase which can convert n-6 PUFAs to n-3 PUFAs, leading to an increased abundance of n-3 PUFAs and a decrease in n-6 PUFAs in animal tissues.18–21 The majority of studies adopt low-grade inflammation models with various diet formula, but it is difficult to control the fat composition between experimental groups. Fat-1 mice thus provide a desired model to investigate the biological functions of n-3 PUFAs under the low-grade inflammation condition induced by a diet rich in n-6 PUFAs.
Metabolomics has been widely applied to identify biomarkers and investigate mechanisms of biological effects.22 Currently, ultra performance liquid chromatography quadrupole time of flight mass spectrometry (UPLC-Q-TOFMS) has become an indispensable analytical platform of metabolomics due to its high sensitivity, high throughput and wide coverage of metabolites detected.23 LC-MS-based metabolomics offers untargeted and targeted strategies. Untargeted metabolomics aims to analyze all metabolites without prior knowledge of the components by full-scan mode. The MSE, an effective MS technique, can perform parallel alternating scans for acquisition to obtain precursor ion information at low collision energy and to obtain full-scan accurate mass fragment, precursor ion and neutral loss information at high collision energy.24 The MSE mode allows almost simultaneous acquisition of both MS and MS/MS fragmentation data in a single analytical run.25 Thus MSE provides significant benefits in metabolite identification, which is important to simplify untargeted metabolomics studies.26 Additionally, Metabolite Set Enrichment Analysis (MSEA) based on web-database is frequently used to identify and reveal the biological significance of patterns of changes in metabolite concentration.5
Herein, in the present study, the metabolomics was conducted to investigate the anti-inflammatory effects of endogenous n-3 PUFAs in a low-grade inflammation animal model induced on transgenic fat-1 mice. Additionally, we identified the metabolite changes which are responsible for the anti-inflammatory effect of endogenous n-3 PUFAs. It is worth mentioning that this study explored the metabolism profile of the anti-inflammatory effects of endogenous n-3 PUFAs for the first time.
2. Methods
2.1 Chemicals and materials
HPLC-grade acetonitrile, formic acid and methanol were purchased from Thermo Fisher Scientific (Waltham, MA, USA). EDTA K2-treated glass blood collecting tubes were purchased from Kangjie Company (Jiangsu, China). Mouse IL-6 enzyme-linked immunosorbent assay (ELISA) kit and Mouse TNF-α ELISA kit were purchased from Ready-SET-Go® (eBioscience Inc., Thermo Fisher Scientific, San Diego, USA). Eicosapentaenoic acid (EPA), arachidonic acid (ARA), prostaglandin E2 (PGE2), palmitic acid, resolvin E1, 5-hydroxyeicosatetraenoic acid, 20-hydroxyeicosatetraenoic acid-d6 (20-HETE-d6) were purchased from Cayman Chemical (Ann Arbor, MI, USA).2.2 Animal, modeling and verification
Fat-1 transgenic mice breeders with a C57BL/6 background were obtained from Dr Jing X. Kang (Harvard Medical School, Boston, MA, USA). The male heterozygous fat-1 mice mated with female C57BL/6 mice. The offspring would be heterozygous fat-1(±) mice and their wild type (WT) littermates. To get the fat-1(±) transgenic mice, the progeny was identified by PCR. All experimental animals were housed in controlled conditions (temperature 25 ± 2 °C, humidity 50 ± 10%, 12 h light/dark circle) and fed ad libitum. A total of 12 fat-1 transgenic mice and 12 wild type (WT) mice, designated as a fat-1 group and WT group, were fed a modified diet immediately after weaning until they were 1 year old to induce the low-grade inflammation model. The modified diet contained 10% corn oil (TROPHIC Animal Feed High-tech Co., Ltd, China), with a fatty acid profile high in n-6 PUFAs (mainly linoleic acid) and low in n-3 PUFAs (∼0.1% of the total fat content). An additional group of 7 WT mice, which were designated as the control group, were fed a normal chow diet during the corresponding experimental period. All animal experiments were performed in strict accordance with the NIH guidelines for the care and use of laboratory animals (NIH publication no. 85-23 Rev. 1985) and were approved by the Institutional Animal Care and Use Committee, University of Macau, China.The low-grade inflammation model was verified by determining the plasma levels of IL-6 and TNF-α. The levels of these two inflammatory cytokines were tested by antigen-specific ELISA with mouse IL-6 ELISA kit and mouse TNF-α ELISA kit.
2.3 Blood sample collection and preparation
Blood samples (about 400 μL of each sample) were drawn from the tail vein and collected in EDTA K2-treated glass tubes. Plasma samples were collected after centrifugation of the blood samples for 10 min at 6000 × g using a refrigerated centrifuge. The samples were maintained at 0–4 °C during the processing period. A mixture of equal amounts (20 μL) of each sample was prepared for use as quality control (QC) sample. All the plasma samples were stored at −80 °C until further analysis.To maximize the component profile in samples used for metabolomics analysis, the solvent in the protein precipitation step was optimized. Methanol covered more metabolites and had better performance than other solvents, including acetonitrile, methanol with 0.1% formic acid and acetonitrile with 0.1% formic acid, respectively. Thus, methanol was used for protein precipitation during the extraction of plasma metabolites. First, 150 μL of methanol was added into a 50 μL aliquot of plasma sample. The mixture was mixed by vortexing for 2 min and then centrifuged twice at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min. Subsequently, the supernatant was transferred into an Eppendorf tube and analyzed by untargeted metabolomics.
000 × g for 15 min. Subsequently, the supernatant was transferred into an Eppendorf tube and analyzed by untargeted metabolomics.
The metabolite 20-HETE-d6 was used as the internal standard (I.S.) and was prepared in acetonitrile at 10 μg mL−1, as a stock solution. All the samples prepared for multiple reaction monitoring (MRM) analysis were mixed with 5 μL of I.S. solution and 145 μL of methanol. The mixtures were then centrifuged twice at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min, and supernatants were transferred into Eppendorf tubes.
000 × g for 15 min, and supernatants were transferred into Eppendorf tubes.
2.4 UPLC-Q-TOFMS conditions for untargeted metabolomics
Chromatographic separation was conducted on a Waters ACQUITY UPLC system (Waters Corp., Milford, MA, USA) with an ACQUITY HSS T3 column (2.1 × 100 mm, i.d. 1.8 μm) at 50 °C. The mobile phase was a mixture of 0.1% formic acid water solution as solvent A and methanol as solvent B. An 18 min gradient elution program was used as follows: 0–2 min, 5–55% B (v/v); 2–10 min, 55–90% B (v/v); 10–15 min, 90–100% B (v/v); 15–17 min, 100% B (v/v). The column was equilibrated and eluted under isocratic conditions with a flow rate of 0.4 mL min−1, and the injection volume was 3 μL.The LC eluent was introduced into a Waters SYNAPT G2-Si High Definition Mass Spectrometry (HDMS) equipped with an electrospray ionization (ESI) interface (Waters Corp., Manchester, UK). Ionization was achieved with the following parameters: capillary voltage, 3.0 kV in the positive mode, 2.5 kV in the negative mode; sampling cone voltage, 40 V in the positive mode and 30 V in the negative mode; source temperature, 140 °C and 120 °C, in the positive and the negative mode, respectively; desolvation temperature, 450 °C; cone gas flow, 10 L h−1; desolvation gas, nitrogen; desolvation gas flow, 900 L h−1 in the positive mode and 600 L h−1 in the negative mode; nebuliser gas flow pressure, 5.9 bar. A Leucine-Enkephalin solution (LE, 200 pg mL−1) was continuously infused into MS at 10 μL min−1 for real-time calibration. In addition, the samples were also analyzed in the MSE mode with mass range of 50 to 1000 Da, with a data acquisition rate set to 0.25 s for TOF-MS scan, the collision energy ramp range was set from 0 to 30 V.
2.5 Statistical analysis
The raw data were processed with MarkerLynx XS software (Waters, Milford, MA, USA). Pretreatment procedures of variables extraction, alignment, and normalization were performed to process the raw data. The resulting matrix was composed of sample name, m/z – retention time, and normalized peak intensity. Ions were refined by “80% rule”, in which the ions that were detected in <80% of the all injections were removed from the dataset.56 The data were then processed using EZinfo 3.0 (MKS Umetrics, Umeå, Sweden) with pareto scaling and mean centering before multivariate data analysis to perform the unsupervised principal component analysis (PCA) and supervised orthogonal partial least squares-discriminant analysis (OPLS-DA). In these two models, cumulative goodness of fit (R2) and the cumulative goodness of prediction (Q2) were used to evaluate separates classes of observations. In the OPLS-DA, the variable importance in projection (VIP) was weighted to estimate the importance of metabolites. The metabolites with a VIP value above 1 were responsible for the apparent discrimination in OPLS-DA models. One-way ANOVA and significance analysis of microarray (SAM) were also performed to identify important metabolites. The metabolites selected by VIP with both p value less than 0.05 in one-way ANOVA and FDR equal 0.001 in SAM.The structural characterization information of the metabolites as potential biomarkers was performed by MSE analysis. To illuminate the concentration change, interaction, and core pathways of potential biomarkers, the Metaboanalyst 4.0 software (http://www.metaboanalyst.ca/), the high-quality Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways as the backend knowledgebase were applied. A one-column marker candidates list with human metabolome database (HMDB) ID input type was imported into the online software. During the process, the Mus musculus (mouse) pathway library was chosen for the following enrichment pathway analysis.
2.6 Selected metabolites analysis conditions
20-HETE-d6 was used as the internal standard. For uric acid, PC16, eicosapentaenoic acid, prostaglandin E2, arachidonic acid, palmitic acid, the lower limit of determination (LLOD) was selected by S/N > 10 and showed in Table S3.† In addition, LLOQs of RvE1 and 5-HETE were selected by S/N > 3. Chromatographic conditions were same as the untargeted metabolomics. Ionization was achieved using electrospray in negative mode with capillary voltages at 3000 V, sampling cone voltage at 30 V, source temperature at 120 °C, desolvation temperature at 450 °C, cone gas flow at 50 L h−1, desolvation gas flow at 900 L h−1, and nebuliser had the flow pressure at 5.9 bar. Quantification was performed using the MRM mode at m/z 325.2 → 275.3 for EPA, m/z 303.2 → 259.25 for ARA, m/z 353.2 → 289.2 for PGE2, m/z 255.2 → 239.2 for palmitic acid, m/z 351.2 → 303.2 for RvE1, m/z 319.2 → 303.2 for 5-HETE, m/z 327.4 → 253.2 for I.S. The cone voltages for EPA, ARA, PGE2, palmitic acid, RvE1, 5-HETEand I.S. were set at 30, 30, 30, 15, 20, 20 and 20 V, respectively. The trap CE voltages of each chemical were 20, 20, 20, 15, 0, 0 and 0 V, respectively.The data sets from the MRM determination analysis were then processed with the Metaboanalyst 4.0 software to obtain the relative determination by metabolite set enrichment analysis (MSEA).
3. Results and discussion
3.1 Biological change of the low grad inflammation in normal and fat-1 animal model
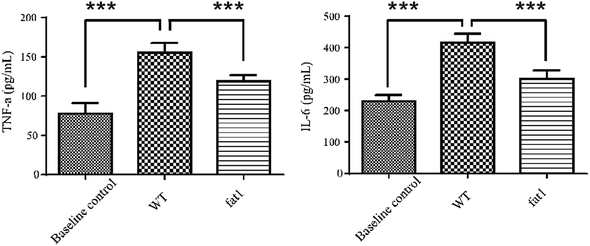
Low-grade inflammation is recognized as a pathological feature of a wide range of chronic conditions in a growing number of chronic diseases.27 Various biomarkers have been shown to correlate with the morbidity biological profile. The most prominent ones are the interleukin (IL) cytokines and tumor necrosis factor (TNF).28 Circulating IL-6 and TNF-α levels are associated with the state of systematic low-grade inflammation.29–31 In this study, IL-6 and TNF-α levels in plasma were measured by high-sensitivity ELISA to validate the low-grade inflammation model. As shown in Fig. 1, both IL-6 and TNF-α level in WT mice fed with a modified diet were much higher than those in the animals which were fed with a normal diet, indicating that long-term intake of n-6 PUFAs could lead to low-grade inflammation. Compared with the WT mice, fat-1 mice showed less plasma levels of IL-6 and TNF-α (Fig. 1), which is consistent with previous studies that endogenous n-3 PUFAs significantly reduces inflammation.32–343.2 Metabolic variations of the low grad inflammation in normal and fat-1 animal
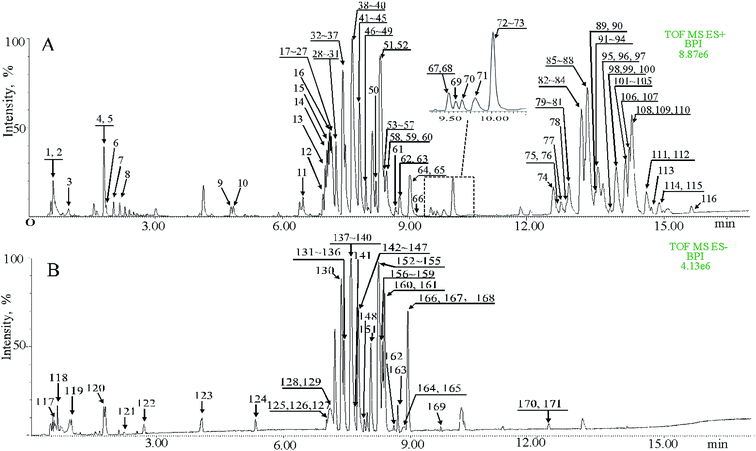
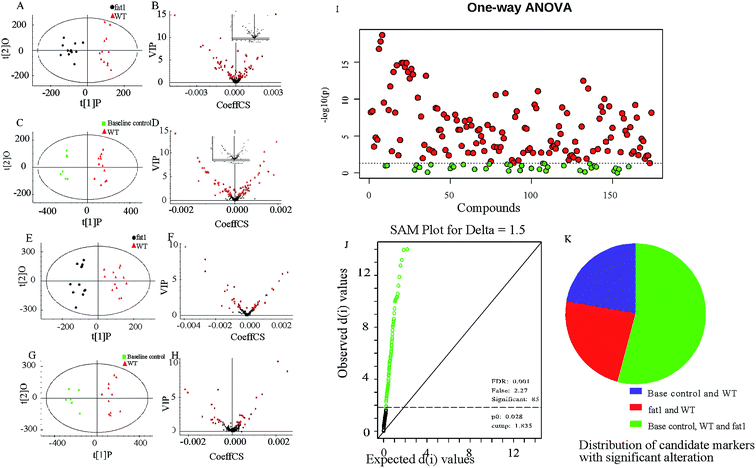
The procedure and result of validation of untargeted metabolomics method are shown in ESI.† The base peak intensity (BPI) chromatograms obtained in the positive and negative modes are depicted in Fig. 2. After data extraction alignment, normalization and miss value elimination, 721 peak indices (RT-m/z pairs) and 501 ions were acquired from plasma sample in positive and negative modes, respectively. The resultant matrix with the exact mass-retention time pair, intensity and sample code was generated for further statistical analyses. After pareto scaling and mean centering, OPLS-DA score plot was constructed using the datasets from fat-1 and WT groups (Fig. 3A and E). The separation between the fat-1 and WT group was observed in OPLS-DA score plots derived from the data in either positive or negative mode. The OPLS-DA on plasma samples from the WT and baseline control group was also performed to monitor the metabolite changes in the low-grade inflammation situation (Fig. 3C and G). The scatter plots of the variable importance in projection (VIP) values in these two OPLS models are shown in Fig. 3B, D, F and H. All VIP values of variables greater than one are marked with red dots and labeled in the BPI chromatograms, and a total of 171 metabolites are marked (Fig. 2). One-way ANOVA results of these highlighted metabolites are shown in Fig. 3I. As a way of conceptualizing the rate of true null hypothesis incorrect rejection, false discovery rate (FDR) procedures are used under conditions of multiple comparisons. Among the various FDR analytical methods, the microarray is a typical example to solve the calculation of matrix with multi-classes. Significance analysis of microarray (SAM) identifies statistically significant variables by carrying out t-tests for 2 groups35 and F-tests for multi-groups.36 It provides a tuning parameter, namely the delta, to obtain significant variables along with acceptable FDR.37 In this study, 171 metabolites derived from the OPLS-DA of plasma samples analyzed in the ESI positive and negative mode were mixed in one matrix for SAM. In the analysis procedure, the delta was adjusted to control the FDR to no more than 0.001 and a p-value of less than 0.05 (Fig. 3J). The more a variable deviated from the expected line, the more likely it was to be significant. A total of 85 significant metabolites were selected from the ones obtained from the OPLS-DA model. As shown in Fig. 3K, 54% of the metabolites showed significant alteration among the baseline control, WT, and fat-1 group. However, only 24% of the metabolites showed significant alteration between the WT and fat-1 group. Those metabolites contributing most to the significant variance between the baseline control and WT group could be considered as potential biomarkers for low-grade inflammation, and those contributing to the variance between the WT and fat-1 group could be considered to play roles in the mechanism of the anti-inflammatory effects of n-3 PUFAs.The highlighted metabolites were tentatively identified by analyses of the MS spectra and then by searching those data in available biochemical libraries, including the HMDB (http://www.hmdb.ca/spectra/ms/search), METLIN (http://metlin.scripps.edu/) and KEGG (http://www.genome.jp/kegg/) using a 50 ppm tolerance. The candidate markers were tentatively assigned (Table 1).
| No. | HMDB | RT | m/z | Adduct | Adduct MW (Da) | Compound MW (Da) | Delta | Chemical formula | Compound name |
|---|---|---|---|---|---|---|---|---|---|
| 1 | HMDB61890 | 0.59 | 187.0797 | M + H | 187.0713 | 186.0641 | 0.0084 | C7H10N2O4 | Pyroglutamylglycine |
| 2 | HMDB40275 | 0.60 | 203.0547 | M + H | 203.0550 | 202.0477 | 0.0003 | C8H10O6 | Ethyl aconitate |
| 3 | HMDB00289 | 0.95 | 169.0369 | M + H | 169.0356 | 168.0283 | 0.0013 | C5H4N4O3 | Uric acid |
| 4 | HMDB29737 | 1.79 | 146.0612 | M + H | 146.0600 | 145.0528 | 0.0012 | C9H7NO | 1H-Indole-3-carboxaldehyde |
| 5 | HMDB00734 | 1.79 | 188.0721 | M + H | 188.0706 | 187.0633 | 0.0015 | C11H9NO2 | Indoleacrylic acid |
| 6 | HMDB32857 | 1.84 | 261.1335 | M + H | 261.1333 | 260.1260 | 0.0002 | C12H20O6 | Glycerol tripropanoate |
| 14 | HMDB02904 | 7.06 | 343.2267 | M + H | 343.2115 | 342.2042 | 0.0152 | C18H30O6 | 2,3-Dinor-TXB2 |
| 18 | HMDB10383 | 7.14 | 494.3279 | M + H | 494.3241 | 493.3168 | 0.0038 | C24H48NO7P | LysoPC (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
| 19 | HMDB10389 | 7.14 | 516.31 | M + H | 516.3085 | 515.3012 | 0.0015 | C26H46NO7P | LysoPC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
| 53 | HMDB10410 | 8.38 | 351.2309 | M + H | 351.2166 | 350.2093 | 0.0143 | C20H30O5 | Resolvin E1 |
| 62 | HMDB01220 | 8.83 | 353.2476 | M + H | 353.2323 | 352.2250 | 0.0154 | C20H32O5 | Prostaglandin E2 |
| 63 | HMDB11651 | 8.83 | 375.2296 | M + H | 375.2166 | 374.2093 | 0.0130 | C22H30O5 | 11beta,20-Dihydroxy-3-oxopregn-4-en-21-oic acid |
| 64 | HMDB10409 | 9.00 | 321.2117 | M + H | 321.2424 | 320.2351 | 0.0307 | C20H32O3 | 11,12-EpETrE |
| 71 | HMDB13213 | 9.83 | 367.3202 | M + H | 367.3445 | 366.3372 | 0.0243 | C23H44NO2 | alpha-Linoleoylcholine |
| 89 | HMDB11134 | 7.01 | 319.23 | M − H | 319.2279 | 320.2351 | 0.0009 | C20H32O3 | 5-HETE |
| 118 | HMDB00822 | 0.64 | 167.0226 | M − H | 167.0350 | 168.0423 | 0.0124 | C8H8O4 | p-Hydroxymandelic acid |
| 119 | HMDB01874 | 0.98 | 191.0211 | M − H | 191.0197 | 192.0270 | 0.0014 | C6H8O7 | D-threo-Isocitric acid |
| 121 | HMDB06357 | 2.24 | 187.0084 | M − H | 187.0248 | 188.0321 | 0.0164 | C7H8O6 | cis-2-Methylaconitate |
| 127 | HMDB01445 | 7.01 | 586.3178 | M − H | 586.2188 | 587.2260 | 0.0990 | C23H43NO12P2 | N-Acetyl-D-glucosaminyldiphosphodolichol |
| 138 | HMDB13206 | 7.59 | 255.2342 | M − H | 255.2204 | 256.2277 | 0.0138 | C15H30NO2 | 9-Decenoylcholine |
| 142 | HMDB01045 | 7.70 | 554.3286 | M − H | 554.2620 | 555.2693 | 0.0666 | C28H37N5O7 | Enkephalin L |
| 148 | HMDB13022 | 7.88 | 616.3632 | M − H | 616.3716 | 617.3788 | 0.0084 | C32H51N5O7 | Neuromedin N |
| 149 | HMDB01999 | 7.97 | 301.2192 | M − H | 301.2173 | 302.2246 | 0.0019 | C20H30O2 | Eicosapentaenoic acid |
| 156 | HMDB59632 | 8.33 | 395.2222 | M − H | 395.2204 | 396.2277 | 0.0018 | C18H37O7P | (9S,10S)-10-Hydroxy-9-(phosphonooxy)octadecanoate |
| 157 | HMDB02183 | 8.34 | 327.2347 | M − H | 327.2330 | 328.2402 | 0.0017 | C22H32O2 | Docosahexaenoic acid |
| 158 | HMDB01043 | 8.36 | 303.2349 | M − H | 303.2330 | 304.2402 | 0.0019 | Arachidonic acid | |
| 159 | HMDB12869 | 8.36 | 371.222 | M − H | 371.2228 | 372.2301 | 0.0008 | C23H32O4 | 9′-Carboxy-gamma-tocotrienol |
| 163 | HMDB00220 | 8.69 | 255.2345 | M − H | 255.2330 | 256.2402 | 0.0012 | C16H32O2 | Palmitic acid |
| 165 | HMDB04708 | 8.76 | 329.2499 | M − H | 329.2333 | 330.2406 | 0.0166 | C18H34O5 | 9,12,13-TriHOME |
| 166 | HMDB41480 | 8.93 | 281.2504 | M − H | 281.2486 | 282.2559 | 0.0018 | Octadecenoic acid | |
| 167 | HMDB13040 | 8.94 | 349.2377 | M − H | 349.2020 | 350.2093 | 0.0357 | C20H30O5 | PGH3 |
| 169 | HMDB00827 | 9.72 | 283.2661 | M − H | 283.2643 | 284.2715 | 0.0088 | C18H36O2 | Stearic acid |
To gain a better understanding of the function of these candidate markers in the mechanism of the anti-inflammatory effect of endogenous n-3 PUFAs, the pathway analysis was performed by the Metaboanalyst 2.1 software (Fig. 4A). The heat map of selected metabolites in the serum in the baseline control, WT and fat-1 group was generated to observe the relative change of the metabolites (Fig. 4B). Red indicates the high intensity of the metabolite relative to its mean intensity (black), and green represents the low intensity. In this study, energy metabolisms, including glycerophospholipid metabolism, starch and sucrose metabolism, and fatty acid metabolism, including biosynthesis of unsaturated fatty acids, linoleic acid metabolism, α-linoleic acid metabolism and fatty acid elongation in mitochondria were found to be involved. As most altered metabolites were lipids and fatty acids, this indicated that lipid and fatty acid metabolisms were severely disrupted in low-grade inflammation model. It was reported that n-3 PUFAs acted as substrates of cyclooxygenase (COX) and lipoxygenase (LOX) enzymes to generate alternative eicosanoids and reduce the generation of eicosanoids from arachidonic acid.38 Accordingly, it was apparent that endogenous n-3 PUFAs could decrease low-grade inflammation.3,39,40 In addition, purine metabolism and tryptophan metabolism were found to be involved in the anti-inflammatory effects of endogenous n-3 PUFAs in this study. As shown in Fig. 3A, the purine metabolism was strongly correlated to low-grade inflammation. It is well-known that purine metabolism is associated with oxidative stress.41,42 However, the relationship between n-3 PUFAs and purine metabolism has rarely been reported. Recently, the study of tryptophan metabolism has become a hotspot for research on low-grade inflammation and inflammatory disorders, such as metabolic syndromes and neuropsychiatric diseases. Altered tryptophan metabolism could be triggered by steroids, cytokines and tryptophan itself under inflammation.43 Previous studies showed that the major route of tryptophan metabolism was the kynurenine pathway,44–46 which was found to be associated with high levels of TNF-α and IL-6 in a systemic inflammatory condition.44 In conclusion, with the global metabolomics analysis, the major pathway changes have been determined in the low-grade inflammation model.
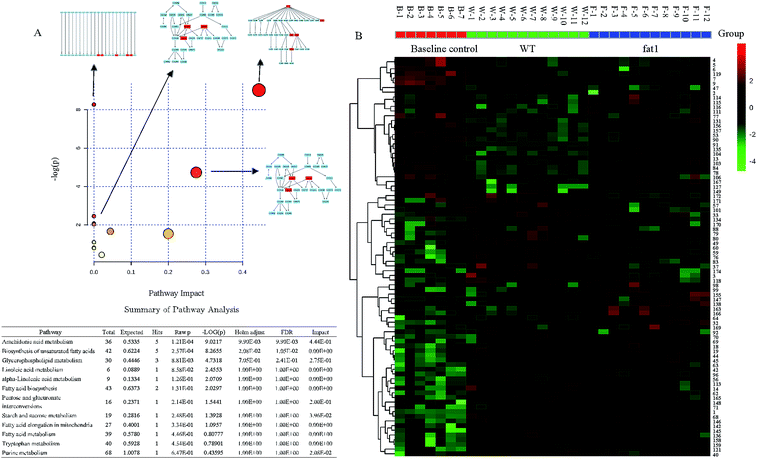 | ||
| Fig. 4 (A) Summary of the pathway analysis results in the low-grade inflammation model (B) heat map of significantly altered metabolites. | ||
3.3 Pathway changes of anti-inflammation effect of endogenous n-3 PUFA
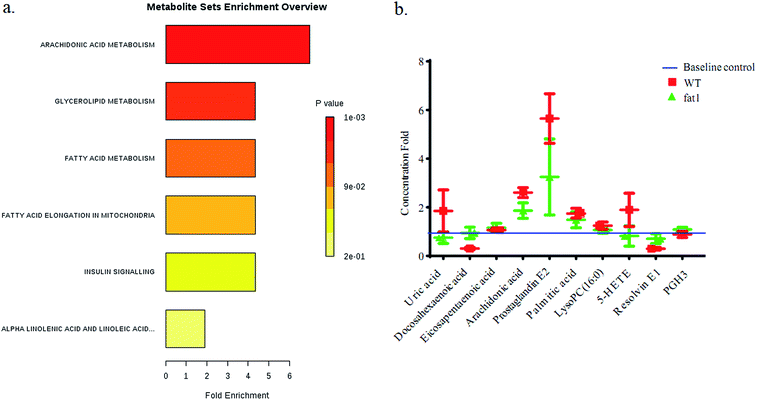
After the selection of metabolomics study, several metabolites, uric acid, PC 16, EPA, ARA, PGE2, palmitic acid, RvE1, 5-HETE, were character makers which were highlighted in an alternated pathway. In this study, these markers were compared between WT group and fat-1 group with quantitative analysis. Stable isotope-labeled 20-HETE-d6 (I.S.) were used to correct error in the determination of the concentration changes of the reference metabolites. All the MRM detected metabolites were compared with the peak area of the I.S. in each sample to obtain the concentration with the reference. Linear responses were obtained for analytes in the range of concentration curves. The regression equations were shown in Table S3.† The concentration data set was analyzed by MSEA with MetaboAnalyst 4.0 (Fig. 5). Alpha-linolenic acid and linoleic acid metabolism, arachidonic acid metabolism, fatty acid elongation in mitochondria, fatty acid metabolism, glycerolipid metabolism and insulin signaling pathways were found to be altered, and the rank of Holm p values are shown as the color depth ranked (Fig. 5).4. Discussion
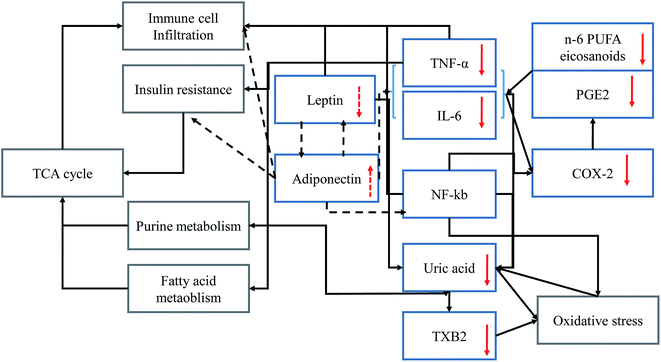
At high n-6 PUFAs concentration increased the serum levels of IL-6 and TNF-α in animals (Fig. 1) consistent with previous reports.47,48 The key intermediate metabolite of n-6 PUFAs is arachidonic acid (ARA)49 as the substrate of COX or prostaglandin H (PGH) synthase, and precursor of prostaglandins such as PGE2.50 PGE2 could also be derived from IL-6 and TNF-α and induced COX enzymes expression.51,52 Previous studies suggested that the gene encoding for IL-6 is regulated by nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB).53,54 However, NF-κB could induce oxidative stress by regulating the level of reactive oxygen species.55 Based on this metabolomics study and a review of the literature, inflammatory mediator interactions occurring during the inflammatory process are summarized (Fig. 6). Solid arrows indicate stimulatory effects and dotted arrows indicate inhibitory effects between inflammatory mediators. Red arrows indicate the effects of n-3 PUFA on the regulation of the level of inflammatory mediators. Uric acid was identified as a biomarker which is closely associated with the oxidative stress response pathways56 and recognized as the final oxidation product of purine metabolism. In addition, the high level of uric acid might act as a strong pro-oxidant which accelerates the copper-induced peroxidation of lipids, even in the presence of endogenous antioxidants.57 Thus, the strengthened oxidative stress would directly activate the transcription factor NF-κB in feedback. Following NF-κB activation, insulin resistance could be the result.58 As a consequence, purine metabolism and fatty acid metabolism would also be affected. Thus, n-3 PUFA-derived eicosanoids, such as EPA and resolvin E1 were subsequently up-regulated, and n-6 PUFA-derived eicosanoids, including PGE2, 5-HETE, cytokines (TNF-α, IL-6), and macrophage tissue infiltration were subsequently downregulated. Presumably, the anti-inflammatory effects of endogenous n-3 PUFAs might be exerted through alteration of the signaling pathway of insulin resistance,59 purine metabolism, fatty acid metabolism and oxidative stress response pathways.605. Conclusion
A metabolomics study was conducted to investigate the anti-inflammatory effects of endogenous n-3 PUFAs response to a low-grade inflammation induced on transgenic fat-1 mice in this work. Multi-analysis techniques, including OPLS-DA, one-way ANOVA and SAM, were used to optimize the identification of 1222 candidate markers. Structural analysis using the quadrupole time-of-flight mass spectrometry specifically targeted the altered pathways associated with the low-grade inflammation. After analyzing the MS/MS spectrum obtained using the MSE mode, candidate markers related to anti-inflammatory effects of endogenous n-3 PUFAs were identified by further MSEA analysis. Ultimately, the altered pathways involved in the anti-inflammatory effects of endogenous n-3 PUFAs were identified, indicating that insulin signalling and fatty acid alteration are responsible for anti-inflammatory effects of endogenous n-3 PUFAs.Author contributions
The experimental set-up was designed by Yu Peng and Huanxing Su. The samples were prepared by Huixia Ren and Yu Peng. The experiments of UPLC-Q-TOF were performed by Yu Peng. Data analyses were performed by Yu Peng, HongXun Tao, Chengwei He, JianBo Wan, and Peng Li. The manuscript was written by Yu Peng, edited by Huanxing Su and Jianbo Wan, and approved by all the authors.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This study was supported by the Macao Science and Technology Development Fund (020/2017/A1 and 039/2017/AFJ) and multi-year research grant, University of Macau (MYRG2016-00184-ICMS-QRCM and MYRG2018-00242-ICMS).References
- R. Rorato, B. C. Borges, E. T. Uchoa, J. Antunes-Rodrigues, C. F. Elias and L. L. K. Elias, Int. J. Mol. Sci., 2017, 18, 1431 CrossRef PubMed.
- K. A. Walker, Aging, 2018, 11, 3–4 Search PubMed.
- P. C. Calder, N. Ahluwalia, F. Brouns, T. Buetler, K. Clement, K. Cunningham, K. Esposito, L. S. Jonsson, H. Kolb, M. Lansink, A. Marcos, A. Margioris, N. Matusheski, H. Nordmann, J. O'Brien, G. Pugliese, S. Rizkalla, C. Schalkwijk, J. Tuomilehto, J. Warnberg, B. Watzl and B. M. Winklhofer-Roob, Br. J. Nutr., 2011, 106(suppl. 3), S5–S78 CrossRef CAS PubMed.
- E. Scaioli, E. Liverani and A. Belluzzi, Int. J. Mol. Sci., 2017, 18, 2619 CrossRef.
- J. Xia and D. S. Wishart, Nucleic Acids Res., 2010, 38, W71–W77 CrossRef CAS.
- M. J. Zhang and M. Spite, Annu. Rev. Nutr., 2012, 32, 203–227 CrossRef CAS.
- P. C. Calder, Br. J. Clin. Pharmacol., 2013, 75, 645–662 CrossRef CAS PubMed.
- C. Luo, H. Ren, J. B. Wan, X. Yao, X. Zhang, C. He, K. F. So, J. X. Kang, Z. Pei and H. Su, J. Lipid Res., 2014, 55, 1288–1297 CrossRef CAS PubMed.
- C. Luo, H. Ren, X. Yao, Z. Shi, F. Liang, J. X. Kang, J. B. Wan, Z. Pei, K. P. Su and H. Su, EBioMedicine, 2018, 32, 50–61 CrossRef.
- Z. Shi, H. Ren, C. Luo, X. Yao, P. Li, C. He, J. X. Kang, J. B. Wan, T. F. Yuan and H. Su, Mol. Neurobiol., 2016, 53, 6482–6488 CrossRef CAS PubMed.
- S. Qin, J. Wen, X. C. Bai, T. Y. Chen, R. C. Zheng, G. B. Zhou, J. Ma, J. Y. Feng, B. L. Zhong and Y. M. Li, Mol. Med. Rep., 2014, 9, 2097–2104 CrossRef CAS PubMed.
- J. X. Kang, J. Wang, L. Wu and Z. B. Kang, Nature, 2004, 427, 504 CrossRef CAS.
- S. Ma, Y. Ge, X. Gai, M. Xue, N. Li, J. Kang, J. Wan and J. Zhang, Neurosci. Lett., 2016, 611, 28–32 CrossRef CAS.
- S. Marventano, P. Kolacz, S. Castellano, F. Galvano, S. Buscemi, A. Mistretta and G. Grosso, Int. J. Food Sci. Nutr., 2015, 66, 611–622 CrossRef PubMed.
- J. K. Kiecolt-Glaser, M. A. Belury, K. Porter, D. Q. Beversdorf, S. Lemeshow and R. Glaser, Psychosom. Med., 2007, 69, 217–224 CrossRef CAS PubMed.
- M. E. Berger, S. Smesny, S. W. Kim, C. G. Davey, S. Rice, Z. Sarnyai, M. Schlogelhofer, M. R. Schafer, M. Berk, P. D. McGorry and G. P. Amminger, Transl. Psychiatry, 2017, 7, e1220 CrossRef CAS PubMed.
- A. P. Simopoulos, World Rev. Nutr. Diet., 2009, 100, 1–21 CAS.
- J. X. Kang, J. Nutrigenet. Nutrigenomics, 2008, 1, 172–177 CrossRef CAS PubMed.
- Y. Tan, H. Ren, Z. Shi, X. Yao, C. He, J. X. Kang, J. B. Wan, P. Li, T. F. Yuan and H. Su, Mol. Neurobiol., 2016, 53, 3146–3153 CrossRef CAS PubMed.
- H. Ren, Z. Yang, C. Luo, H. Zeng, P. Li, J. X. Kang, J. B. Wan, C. He and H. Su, Mol. Neurobiol., 2017, 54, 3317–3326 CrossRef CAS PubMed.
- H. Ren, C. Luo, Y. Feng, X. Yao, Z. Shi, F. Liang, J. X. Kang, J. B. Wan, Z. Pei and H. Su, FASEB J., 2017, 31, 282–293 CrossRef CAS.
- C. H. Johnson, J. Ivanisevic and G. Siuzdak, Nat. Rev. Mol. Cell Biol., 2016, 17, 451–459 CrossRef CAS.
- W. Lu, B. D. Bennett and J. D. Rabinowitz, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2008, 871, 236–242 CrossRef CAS PubMed.
- T. Wei, L. Zhao, J. Jia, H. Xia, Y. Du, Q. Lin, X. Lin, X. Ye, Z. Yan and H. Gao, Sci. Rep., 2015, 5, 11998 CrossRef PubMed.
- Y. Y. Zhao, P. Lei, D. Q. Chen, Y. L. Feng and X. Bai, J. Pharm. Biomed. Anal., 2013, 81–82, 202–209 CAS.
- B. Bonn, C. Leandersson, F. Fontaine and I. Zamora, Rapid Commun. Mass Spectrom., 2010, 24, 3127–3138 CrossRef CAS.
- C. C. Ledbetter-Nelepovitz, B. D. Rao and A. Fronek, Microvasc. Res., 1991, 41, 164–172 CrossRef CAS.
- D. Calcada, D. Vianello, E. Giampieri, C. Sala, G. Castellani, A. de Graaf, B. Kremer, B. van Ommen, E. Feskens, A. Santoro, C. Franceschi and J. Bouwman, Mech. Ageing Dev., 2014, 136–137, 138–147 CrossRef CAS.
- J. Scheller, A. Chalaris, D. Schmidt-Arras and S. Rose-John, Biochim. Biophys. Acta, 2011, 1813, 878–888 CrossRef CAS.
- M. Maachi, L. Pieroni, E. Bruckert, C. Jardel, S. Fellahi, B. Hainque, J. Capeau and J. P. Bastard, Int. J. Obes. Relat. Metab. Disord., 2004, 28, 993–997 CrossRef CAS PubMed.
- M. Yisireyili, K. Takeshita, M. Hayashi, H. Wu, Y. Uchida, K. Yamamoto, R. Kikuchi, C. N. Hao, T. Nakayama, X. W. Cheng, T. Matsushita, S. Nakamura and T. Murohara, Psychoneuroendocrinology, 2016, 73, 186–195 CrossRef CAS.
- C. A. Hudert, K. H. Weylandt, Y. Lu, J. Wang, S. Hong, A. Dignass, C. N. Serhan and J. X. Kang, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11276–11281 CrossRef CAS PubMed.
- K. H. Weylandt, A. Nadolny, L. Kahlke, T. Kohnke, C. Schmocker, J. Wang, G. Y. Lauwers, J. N. Glickman and J. X. Kang, Biochim. Biophys. Acta, 2008, 1782, 634–641 CrossRef CAS PubMed.
- P. J. White, M. Arita, R. Taguchi, J. X. Kang and A. Marette, Diabetes, 2010, 59, 3066–3073 CrossRef CAS PubMed.
- S. Zang, R. Guo, L. Zhang and Y. Lu, J. Biomed. Inform., 2007, 40, 552–560 CrossRef CAS PubMed.
- D. M. Dziuda, Data Mining for Genomics and Proteomics: Analysis of Gene and Protein Expression Data, JohnWiley & Sons, Inc., 2010 Search PubMed.
- V. G. Tusher, R. Tibshirani and G. Chu, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5116–5121 CrossRef CAS PubMed.
- W. Wang, J. Zhu, F. Lyu, D. Panigrahy, K. W. Ferrara, B. Hammock and G. Zhang, Prostaglandins Other Lipid Mediat., 2014, 113–115, 13–20 CrossRef CAS.
- A. S. Lund, A. L. Hasselbalch, M. Gamborg, K. Skogstrand, D. M. Hougaard, B. L. Heitmann, K. O. Kyvik, T. I. Sorensen and T. Jess, Obes. Facts, 2013, 6, 369–379 CrossRef CAS PubMed.
- S.-k. Kim, Marine Medicinal Foods: Implications and Applications: Animals and Microbes, Elsevier, 1st edn, 2012 Search PubMed.
- T. Ali-Sisto, T. Tolmunen, E. Toffol, H. Viinamaki, P. Mantyselka, M. Valkonen-Korhonen, K. Honkalampi, A. Ruusunen, V. Velagapudi and S. M. Lehto, Psychoneuroendocrinology, 2016, 70, 25–32 CrossRef CAS PubMed.
- W. Baldwin, S. McRae, G. Marek, D. Wymer, V. Pannu, C. Baylis, R. J. Johnson and Y. Y. Sautin, Diabetes, 2011, 60, 1258–1269 CrossRef CAS PubMed.
- Q. Wang, D. Liu, P. Song and M.-H. Zou, Front. Biosci., Landmark Ed., 2015, 20, 1116–1143 CrossRef CAS PubMed.
- Q. Wang, D. Liu, P. Song and M. H. Zou, Front. Biosci., Landmark Ed., 2015, 20, 1116–1143 CrossRef CAS PubMed.
- G. Z. Reus, K. Jansen, S. Titus, A. F. Carvalho, V. Gabbay and J. Quevedo, J. Psychiatr. Res., 2015, 68, 316–328 CrossRef PubMed.
- H. Fujigaki, Y. Yamamoto and K. Saito, Neuropharmacology, 2017, 112, 264–274 CrossRef CAS PubMed.
- T. Tashiro, H. Yamamori, K. Takagi, N. Hayashi, K. Furukawa and N. Nakajima, Nutrition, 1998, 14, 551–553 CrossRef CAS PubMed.
- S. J. Woo, K. Lim, S. Y. Park, M. Y. Jung, H. S. Lim, M. G. Jeon, S. I. Lee and B. H. Park, J. Nutr. Biochem., 2015, 26, 713–720 CrossRef CAS PubMed.
- B. Lands, BioMed Res. Int., 2015, 2015, 285135 Search PubMed.
- O. Morteau, Arch. Immunol. Ther. Exp., 2000, 48, 473–480 CAS.
- R. M. Hinson, J. A. Williams and E. Shacter, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 4885–4890 CrossRef CAS PubMed.
- C. C. Chen, Y. T. Sun, J. J. Chen and K. T. Chiu, J. Immunol., 2000, 165, 2719–2728 CrossRef CAS PubMed.
- A. G. Eliopoulos, M. Stack, C. W. Dawson, K. M. Kaye, L. Hodgkin, S. Sihota, M. Rowe and L. S. Young, Oncogene, 1997, 14, 2899–2916 CrossRef CAS PubMed.
- M. A. Collart, P. Baeuerle and P. Vassalli, Mol. Cell. Biol., 1990, 10, 1498–1506 CrossRef CAS PubMed.
- M. J. Morgan and Z. G. Liu, Cell Res., 2011, 21, 103–115 CrossRef CAS PubMed.
- G. K. Glantzounis, E. C. Tsimoyiannis, A. M. Kappas and D. A. Galaris, Curr. Pharm. Des., 2005, 11, 4145–4151 CrossRef CAS PubMed.
- W. G. Lima, M. E. Martins-Santos and V. E. Chaves, Biochimie, 2015, 116, 17–23 CrossRef CAS PubMed.
- A. Gil, C. Maria Aguilera, M. Gil-Campos and R. Canete, Br. J. Nutr., 2007, 98(suppl. 1), S121–S126 CrossRef CAS PubMed.
- K. Srikanthan, A. Feyh, H. Visweshwar, J. I. Shapiro and K. Sodhi, Int. J. Med. Sci., 2016, 13, 25–38 CrossRef CAS PubMed.
- J. F. Ndisang, Curr. Pharm. Des., 2014, 20, 1318–1327 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra08356a |
| This journal is © The Royal Society of Chemistry 2019 |