Open Access Article
Open Access ArticleRetracted Article: Icariin enhances intestinal barrier function by inhibiting NF-κB signaling pathways and modulating gut microbiota in a piglet model†
Wen Xiong a,
Haoyue Mab,
Zhu Zhanga,
Meilan Jina,
Jian Wanga,
Yuwei Xua and
Zili Wang*a
a,
Haoyue Mab,
Zhu Zhanga,
Meilan Jina,
Jian Wanga,
Yuwei Xua and
Zili Wang*a
aCollege of Animal Science and Technology, Southwest University, Chongqing, China. E-mail: pansquito@hotmail.com; zhuzi625@sina.com; meilan0622@swu.edu.cn; jane0931@126.com; xuyuwei@swu.edu.cn; wzl9698@126.com
bCollege of Parmaceutical Sciences, Southwest University, Chongqing, China. E-mail: mhy950708@vip.qq.com
First published on 20th November 2019
Abstract
This study investigated the effects of icariin on intestinal barrier function and its underlying mechanisms. The icariin diet improved the growth rate and reduced the diarrhea rate in piglets. The icariin diet also reduced the levels of plasma and colonic IL-1β, -6, -8, TNF-α, and MDA but increased the plasma and colonic activity of SOD, GPx, and CAT. Besides, the levels of plasma and colonic endotoxin, DAO, D-lactate, and zonulin were markedly reduced in icariin groups. Meanwhile, dietary intake icariin significantly increased the gene and protein expression of ZO-1, Occludin, and Claudin-1 in the colon. Furthermore, the gene and protein expressions of TLR4, MyD88, and NF-κB were significantly inhibited in the colon of icariin fed piglets. The intestinal microbiota composition and function was changed by the icariin diet. Collectively, these findings increase our understanding of the mechanisms by which ICA enhances the intestinal barrier function and promotes the development of nutritional intervention strategies.
Introduction
The intestine is not only the main digestive and absorbing organ of nutrients but is also an effective barrier against various pathogenic infections.1,2 The main function of the intestinal epithelium is to isolate the digesta, microorganisms, and its metabolites from the intestinal epithelial cells, preventing harmful substances in the intestinal lumen from damaging the intestinal epithelial cells and then entering the lymph or blood circulation.3 Under normal physiological conditions, the intestinal tract can selectively penetrate nutrients. However, under pathological conditions, harmful metabolic substances in the intestinal lumen may escape into the blood circulation due to the destruction of intestinal integrity, causing local or systemic diseases.4 Thus, to prevent harmful metabolites from damaging the intestinal or distant organs, the structural and functional integrity of the mucosal epithelium needs to be maintained.Impaired intestinal integrity increases intestinal permeability, inflammatory response, and oxidative stress, leading to bacterial translocation and subsequent intestinal disease.5 Excessive pro-inflammatory cytokines in the intestinal mucosa can cause inflammatory bowel disease and tissue damage.6 In addition, intestinal oxidative stress disrupts the intestinal barrier by reducing redox sensitivity, resulting in increased intestinal permeability.7 Microorganisms that inhabit the gut and their beneficial metabolites can regulate intestinal health.8 Increasing evidences suggest that gut microbiota disorder is closely related to inflammation and oxidative stress. A previous study has shown that gut microbiota disorder redisposed to inflammatory bowel diseases such as ulcerative colitis, Crohn disease, and indeterminate colitis.9 Besides, it has been reported that change in the gut microbiota may lead to apoptosis and further affect the intestinal barrier by affecting oxidative stress.10 Therefore, the inhibition of pathogenic proliferation by means of nutritional intervention is necessary to alleviate inflammation and oxidative stress. Microbial metabolites such as short-chain fatty acids (SCFAs) could serve as an energy source for intestinal epithelial cells, and could also act as signaling molecules to regulate metabolic and immune functions.11,12 NF-κB is a nuclear transcription factor that has been extensively studied, and many reports have demonstrated that NF-κB mediates inflammatory responses and oxidative stress.13,14 Therefore, maintaining the homeostasis of the gut microbiota and limiting the expression of NF-κB is critical for protecting intestinal health.
Evidence in rodents and humans has revealed that natural product medicines have good health effects, such as alleviation of type 2 diabetes, chronic liver damage, and irritable bowel syndrome.15–17 As a common natural product, the academic community has conducted a comprehensive study on the chemical constituents of epimedium. Among them, icariin (ICA) is a representative active ingredient of epimedium.18 Emerging data suggest that ICA exerts its beneficial effects by regulating some biological processes, such as inhibiting inflammatory responses and oxidative stress.19,20 However, it is unclear whether ICA contributes to maintaining or enhancing the intestinal barrier function.
Pigs are omnivores with similar organ functions and metabolic pathways as humans. Importantly, the gut microbiota of these two species shares a comparatively broad range of gut microbiota.21,22 Therefore, pigs are generally considered to be excellent model species for studying human digestion and nutrition.23,24 In the current study, we employed piglets as a model to investigate the effects of ICA on intestinal barrier function. In addition, the potential mechanisms of ICA for intestinal barrier function were elucidated by studying gut microbiota and NF-κB signaling pathways.
Results
Effect of diet supplemented with ICA on growth performance in weaned piglets
To demonstrate the effect of diet supplemented with ICA on growth performance, we examined BW, ADG, and diarrhea rate (Table 1). On days 1 and 14 of the experimental period, there was no significant change in the BW between the two groups (P > 0.05). However, on day 28, the BW of piglets in the ICA group was significantly higher than that in the CON group (P < 0.05). In addition, there was no difference in the ADG and diarrhea rates between the two groups during the first two weeks of the experimental period (P > 0.05). During the last two weeks of the experimental period, the ADG of the ICA group was significantly higher than that of the CON group (P < 0.05) and the diarrhea rate was significantly lower than that of the CON group (P < 0.05).| Items | Groups | P-value | |
|---|---|---|---|
| CON | ICA | ||
| a Data were shown as mean ± SEM (n = 12). | |||
| BW, kg | |||
| 1 d | 7.0 ± 0.1 | 7.0 ± 0.1 | 0.98 |
| 14 d | 13.1 ± 0.2 | 13.6 ± 0.1 | 0.28 |
| 28 d | 17.8 ± 0.2 | 19.5 ± 0.2 | 0.02 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 1–14 d | |||
| ADG, g | 320 ± 18 | 333 ± 22 | 0.21 |
| Diarrhea rate, % | 5.0 ± 0.1 | 4.5 ± 0.1 | 0.33 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 14–28 d | |||
| ADG, g | 401 ± 24 | 453 ± 28 | 0.03 |
| Diarrhea rate, % | 6.8 ± 0.1 | 4.4 ± 0.1 | 0.01 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| 1–28 d | |||
| ADG, g | 360 ± 14 | 393 ± 18 | 0.04 |
| Diarrhea rate, % | 5.8 ± 0.1 | 4.3 ± 0.1 | 0.04 |
Effect of diet supplemented with ICA on plasma and colonic cytokines and oxidative status in weaned piglets
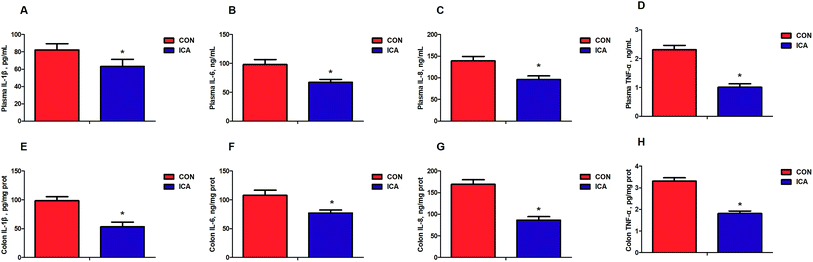
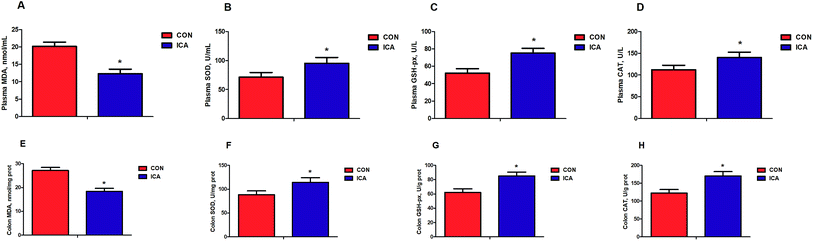
We detected biomarkers related to the inflammation and oxidative stress in the piglets. These biomarkers included IL-1β, IL-6, IL-8, and TNF-α as markers for inflammation and MDA, SOD, GSH-px, and CAT as markers for oxidative stress. The levels of plasma and colonic IL-1β, IL-6, IL-8, and TNF-α were markedly reduced (P < 0.05) in the piglets from the ICA groups (Fig. 1). The levels of plasma and colonic MDA were dramatically decreased and the enzymatic activity of SOD, GSH-px, and CAT were significantly increased (P < 0.05) in the piglets from the ICA groups (Fig. 2).Effect of diet supplemented with ICA on intestinal permeability in weaned piglets
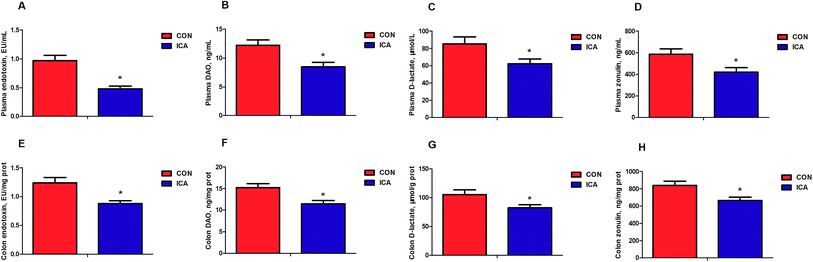
We further detected biomarkers related to the intestinal permeability of piglets. These biomarkers included endotoxin, DAO, D-lactate, and zonulin. The levels of plasma and colonic endotoxin, DAO, D-lactate, and zonulin were markedly reduced (P < 0.05) in the piglets from the ICA groups (Fig. 3).Effect of diet supplemented with ICA on intestinal tight junction protein in weaned piglets
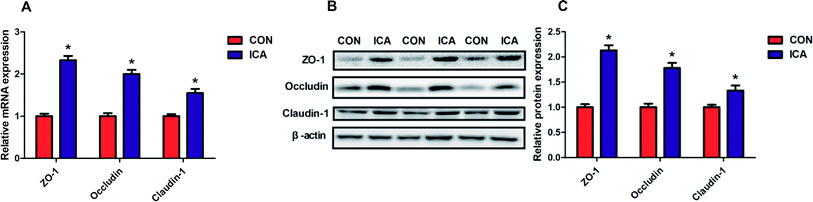
To investigate the effect of diet supplemented with ICA on intestinal tight junction protein, we determined the gene and protein expression of ZO-1, Occludin, and Claudin-1 in the colonic mucosa. The gene and protein abundance of ZO-1, Occludin, and Claudin-1 was dramatically increased (P < 0.05) in the piglets from ICA groups (Fig. 4).Effect of diet supplemented with ICA on NF-κB signal pathway in weaned piglets
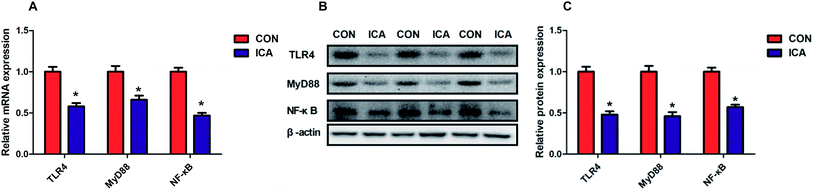
To investigate the potential mechanism by which diet supplemented with ICA increases the intestinal barrier function, we examined the NF-κB signaling pathway in the colonic mucosa. The gene and protein abundance of TLR4, MyD88, and NF-κB was dramatically increased (P < 0.05) in the piglets from the ICA groups (Fig. 5).Effect of diet supplemented with ICA on microbial metabolites in weaned piglets
To analyze the effect of diet supplemented with ICA on intestinal microbiota metabolism, we focused on the colonic and plasma SCFAs of piglets from either CON or ICA groups (ESI Fig. 1†). The results showed that the levels of colonic (ESI Fig. 1A†) and plasma (ESI Fig. 1B†) acetate, propionate, butyrate, and total SCFAs in the piglets from ICA group were higher (P < 0.05) than those from the CON group.Effect of diet supplemented with ICA on intestinal microbial community in weaned piglets
The microbiota composition and diversity of the colonic digesta samples in the piglets were assessed by deep sequencing of the V3–V4 region of the 16S rRNA genes. A total of 533![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 524 high-quality 16S rRNA gene sequences were generated from twelve colonic digesta samples. The average numbers of high-quality sequences generated per sample were 44
524 high-quality 16S rRNA gene sequences were generated from twelve colonic digesta samples. The average numbers of high-quality sequences generated per sample were 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 460 from the microbial populations. The colonic digesta samples of piglets in the CON and ICA groups showed 819 and 897 OTUs, respectively. Among them, 781 shared common OTUs and total 154 individual OTUs were isolated in the two groups (ESI Fig. 2E†). The differences in the microbial α-diversity in the colonic digesta samples between two groups are shown in ESI Fig. 2A–D.† The Sobs, Shannon, Ace and Chao index increased significantly (P < 0.05) in the ICA group. Besides, PCoA based on the Bray–Curtis distances showed a shift in the microbial beta diversity of the colonic digesta samples between two groups (ESI Fig. 2F†).
460 from the microbial populations. The colonic digesta samples of piglets in the CON and ICA groups showed 819 and 897 OTUs, respectively. Among them, 781 shared common OTUs and total 154 individual OTUs were isolated in the two groups (ESI Fig. 2E†). The differences in the microbial α-diversity in the colonic digesta samples between two groups are shown in ESI Fig. 2A–D.† The Sobs, Shannon, Ace and Chao index increased significantly (P < 0.05) in the ICA group. Besides, PCoA based on the Bray–Curtis distances showed a shift in the microbial beta diversity of the colonic digesta samples between two groups (ESI Fig. 2F†).
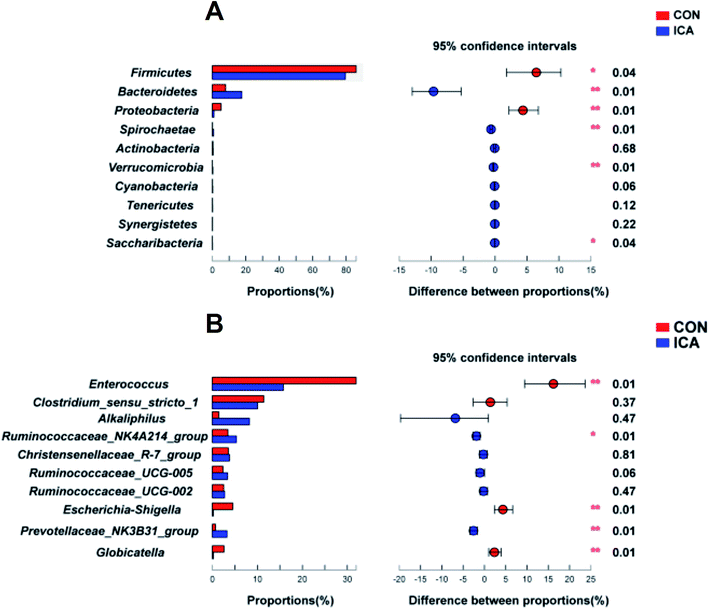
The top 15 phyla and the top 30 genera in relative abundance of the colonic digesta microbiota that are present in piglets are displayed in ESI Fig. 3.† Firmicutes and Bacteroidetes were the most dominant phyla in both the CON and ICA piglets, followed by Proteobacteria, Spirochaetae, and Actinobacteria. Other phyla were present at very low relative abundances (ESI Fig. 3A†). At the genus level, we also revealed the predominant genera that are presented in ESI Fig. 3B.† Significant differences in the relative abundance of the phylum and genus between the colonic digesta samples in the CON and ICA groups were further investigated (Fig. 6). We performed differential analysis of the top 10 bacteria between the two groups at the phylum and genus level. The relative abundances of the phyla Bacteroidetes, Spirochaetae, Verrucomicrobia, and Saccharibacteria were significantly elevated and the phyla Firmicutes and Proteobacteria were significantly reduced in piglets from the ICA group compared with those from the CON group (Fig. 6A). At the genus level, Ruminococcaceae_NK4A214_group and Prevotellaceae_NK3B31_group exhibited increased relative abundances in piglets from the ICA group. In contrast, there was a remarkable decrease in the relative abundances of the genera Enterococcus, Escherichia-Shigella, and Globicatella in pigs from the ICA group (Fig. 6B).
We further performed LEfSe analysis to identify the bacteria that were significantly different between the CON and ICA groups in this study (ESI Fig. 4†). All differential bacteria were demonstrated from the phylum to genus level in the cladogram of LEfSe between the CON and ICA groups (ESI Fig. 4A†). The circle from inside to outside represents distinct bacteria from the phylum to the genus level, respectively. The yellow dots inserted in the circle suggest no significant difference in bacteria among the two groups. Our LEfSe analysis revealed that the phyla Firmicutes and Proteobacteria were significantly enriched in colonic digesta samples of piglets from the CON group and phylum Bacteroidetes was significantly enriched in colonic digesta samples of piglets from the ICA group. At the genus level, genera, Enterococcus, Escherichia_Shigella, and Globicatella were significantly enriched in colonic digesta samples of piglets from the CON group and genus Prevotellaceae_NK3B31_group was significantly enriched in the colonic digesta samples of piglets from the ICA group (ESI Fig. 4B†).
Functional profiling of the microbial communities
To investigate the functional profiles of the colonic digesta bacterial community, we used PICRUSt to predict the gene family abundances of bacterial communities. The results demonstrated that the relative abundances of the genes involved in replication and repair, amino acid metabolism, energy metabolism, metabolism of cofactors and vitamins, genetic information processing, enzyme families, cell growth and death, endocrine system, environmental adaptation, nervous system, immune system, digestive system, and excretory system were significantly elevated and involved in infectious diseases, and immune system diseases were significantly reduced in the ICA group compared with those from the CON group (P < 0.05, Fig. 7).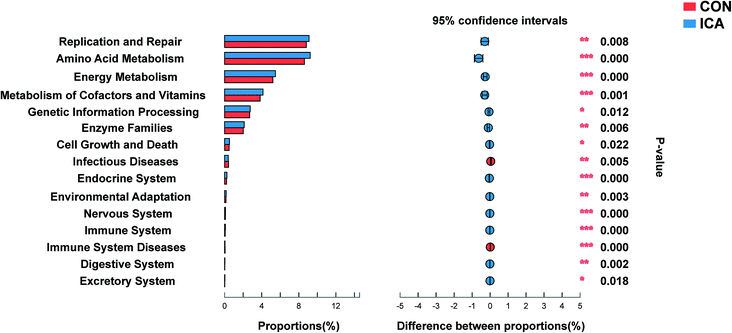 | ||
| Fig. 7 Abundances of KEGG pathways in the functional prediction by PICRUSt. *, P < 0.05; **, P < 0.01; ***, P < 0.001. | ||
Discussion
The integrity of the intestinal barrier is important to ensure the absorption of nutrients and the prevention of harmful substances.25 Due to their high safety and low cost, natural products are increasingly used to improve bodily functions. ICA has been shown to have excellent preventive effects against many disease processes by exerting their potent anti-inflammatory and antioxidant properties.26,27 In this study, we used weaned piglets as a model to explore the effect of ICA on intestinal barrier function. Besides, we systematically evaluated the local inflammatory response, oxidative stress, NF-κB signaling pathway, and the composition and function of the intestinal microbiota to explore the underlying mechanisms of ICA affecting intestinal barrier function.Intestinal integrity is critical for the health and survival of newborns and animals. Damage to intestinal integrity increases intestinal permeability, leading to bacterial translocation and subsequent intestinal infection.28 The chemical markers (endotoxin, DAO, D-lactate, and zonulin) that are usually low in the circulation system in healthy individuals will show a significant increase in the circulation system during the destruction of the intestinal barrier.29 Our data showed that the levels of endotoxin, DAO, D-lactate, and zonulin in plasma and colonic mucosa were significantly decreased in the ICA group, which implied decreased intestinal permeability of the ICA piglets. Tight junctions (TJs) are the primary component of the intestinal epithelium that maintain selective permeability.30 The TJs mainly include the Claudin, Occludin, and ZO families, which together constitute the main physical barrier against intestinal pathogenic microbial invasion and spreading.1 In the current study, our data showed that dietary intake of ICA significantly elevated the gene and protein expression of ZO-1, Occludin, and Claudin-1 in colonic mucosa of piglets. Impaired intestinal barrier function is the direct cause of diarrhea in piglets. We observed that the diarrhea rate of piglets in the ICA group was significantly lower than that of the control group. These results indicated that the dietary intake of ICA effectively enhances colonic barrier function and reduces diarrhea in piglets.
To reveal the underlying mechanisms by which ICA enhances intestinal barrier function, we investigated the effects of ICA on the inflammatory responses and oxidative stress. Previous studies have shown that intestinal invasion by pathogens is accompanied by high expression of pro-inflammatory cytokines such as IL-1β, IL-6, IL-8, and TNF-α.5 TNF-α has a significant destructive effect on the expression and distribution of TJ protein. IL-1β, IL-6, and IL-8 not only regulate the reorganization of cytoskeletal proteins but also directly lead to the rearrangement of TJs proteins, thereby reducing barrier function.31,32 In the current study, our data showed that the dietary intake of ICA significantly decreased the concentrations of IL-1β, IL-6, IL-8, and TNF-α in the plasma and colonic mucosa of piglets. Oxidative stress is one of the most important potential mechanisms by which toxic substances affect mammalian cells.33 When animals and humans are under oxidative stress, the antioxidant system will be destroyed, leading to oxidative damage to DNA and abnormal protein expression, and finally physical damage.34 A previous study has shown that oxidative stress is a common mechanism that destroys intestinal tight junction.35 Our data clearly showed that dietary intake ICA significantly reduced the products of oxidative stress and increased the antioxidant capacity of piglets. As a common nuclear transcription factor, NF-κB has been shown to have the potential to regulate inflammatory responses and oxidative stress.36,37 A previous study demonstrated that ICA can effectively regulate the activation of NF-κB.38 In this study, we found that the TLR4-MyD88-NF-κB signaling pathway was significantly inhibited in the colonic mucosa of piglets in the ICA group. These results strongly suggested that ICA suppresses inflammatory responses and oxidative stress by inhibiting the NF-κB signaling pathway, thereby enhancing intestinal barrier function.
Intestinal microbial diversity is closely related to the health status of humans and animals.39 Our results suggested that the addition of ICA to diet significantly increased the diversity of pigs' gut microbiota. As an important component of the intestinal epithelial barrier, intestinal microbes and their metabolites exert beneficial effects on the host by various means such as regulation of mucosal immunity, inhibition of intestinal pathogen colonization, and production of antimicrobial peptides.40–42 Similar to the previous report,43 Firmicutes and Bacteriodetes represented approximately 90% of total gene sequences in the colonic digesta samples of piglets. A study of gut microbial transplantation showed that the transfer of fecal microbes from lean pigs (Landrace pigs) to antibiotic-treated mice caused significant changes in the intestinal bacterial community structure, mainly leading to a decrease in the ratio of Firmicutes to Bacteriodetes.44 Moreover, it has been recently reported that Lean pigs have a higher abundance of Verrucomicrobia than obese pigs.45 Our data showed that the ratio of Firmicutes to Bacteriodetes in the colonic digesta of pigs in the ICA group was significantly reduced, while Verrucomicrobia was significantly elevated in the ICA group, suggesting that the addition of ICA to the diet may increase the lean rate of piglets. Proteobacteria has been shown to be a mucosa-associated pathogen that increases the risk of colon-related diseases in humans.46 In our study, the ICA group showed a lower abundance of Proteobacteria compared to the CON group. The relative abundance of the phylum Saccharibacteria was significantly elevated in pigs from the ICA group, similar to previous findings in the intestinal microbiota of pigs after feeding with prebiotics.47 In addition, our data suggested that the functional alteration of the intestinal bacterial community in the ICA piglets was characterized by the significant increase in the metabolism, immunity, digestive function, and marked decrease in the infectious and immune system diseases.
Our study revealed that two genera of intestinal microbiota in piglets fed ICA were significantly elevated (Ruminococcaceae_NK4A214_group and Prevotellaceae_NK3B31_group), while the three genera were markedly reduced (Enterococcus, Escherichia-Shigella, and Globicatella). Ruminococcaceae_NK4A214_group belongs to the Firmicutes phylum, which was considered as a fibrolytic bacteria that ferments complex components of plant cell wall into the SCFAs.48 A study has shown that the long-term consumption of foods with high fiber content leads to increased microbial diversity and a predominance of Prevotella over other bacteria.49 Thus, these results led us to analyze intestinal microbiota metabolism in piglets from the ICA group and we focused on the colonic digesta and plasma SCFAs of piglets. Consistently, we found that acetate, propionate, butyrate, and total SCFAs were increased in the colonic digesta and plasma of the piglets. Enterococcus is a large genus of lactic acid bacteria of the phylum Firmicutes, which is a pathogen that can cause diseases such as urinary tract infections, bacteremia, and diverticulitis.50 Globicatella is a non-spore-forming and non-motile genus of bacteria from the family of Aerococcaceae, which was found in the digestive tract of pigs and was associated with the expression of cytokines.51 Escherichia-Shigella is a potential pathogen that was significantly elevated in the feces of diarrhea piglets.52 Regarding the results of functional profiling of microbial communities, the genes involved in immune system diseases were significantly reduced in the ICA group compared with the control group, suggesting that the animals in the ICA group were more resistant to inflammation. The process of infectious diseases is often accompanied by the presence of oxidative stress. Our data showed that the genes involved in the infectious diseases of the ICA group were also significantly lower than the control group, suggesting that the animals in the ICA group may have a stronger ability to resist oxidative stress. In our study, the results of functional profiling of the microbial communities showed that the genes involved in replication and repair, amino acid metabolism, and energy metabolism were relatively abundant but their biological significance still requires further research.
In conclusion, our data highlighted that ICA was an effective natural product for promoting growth performance and reducing diarrhea rates in piglet. Because of its excellent ability to regulate NF-κB signaling pathways and intestinal microbiota, ICA effectively promoted intestinal barrier function. Therefore, our findings help to better understand the mechanisms of ICA that enhance the gut health and provide new ideas for the rational formulation of nutritional intervention strategies.
Materials and methods
Ethical approval
All animal experiments conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication, Eighth edition, 2011) and was approved by the Laboratory Animal Welfare and Ethics Committee of Southwest University. The experimental regulations and methods were approved and then performed according to relevant criteria.Animals and dietary treatments
Twenty four Duroc × (Landrace × Yorkshire) crossbred piglets were allotted into 2 dietary groups randomly by considering the sex and weight of pigs. The piglets were 28 days old (weaned at 21 d and fed creep feed after weaning for one week). The temperature of the pig house was maintained at 25–28 °C and the humidity at 65–75%. This experiment lasted 28 d, feed and water were provided ad libitum, all piglets were healthy, and none received antibiotic treatment during the experimental period. The piglets in the two groups were fed either a control diet based on corn-soybean meal (CON group), or a control diet supplemented with icariin (1 g kg−1 BW; ICA group). Before feeding the animals, we mixed ICA into the diet in proportion. In addition, we ensured that the sufficient ICA dose (1 g kg−1 BW) of each pig was adhered to by recording the feed intake of each pig and feeding them to the appropriate weight of the diet. The diets were formulated according to the nutritional requirements of the National Research Council (NRC, 1998, the United States) for piglets.Sample collection
On day 28, blood and colon samples were collected. Blood samples were collected from pigs by vena jugularis, with a minimum amount of stress, into heparinized tubes (5 mL). Plasma samples were then obtained by centrifuging the blood samples at 3000×g and 4 °C for 10 min, and were stored at −80 °C until analysis. The colonic segments were collected from approximately the middle positions in the intestinal tracts. The intestinal epithelium was separated from the muscular layers by blunt dissection and then stored at −80 °C prior to further analysis. Digesta from the proximal colon was aseptically collected and kept on ice until storage at −80 °C until analysis.Determination of growth performance
The BW of piglets and feed intake per pen were measured to measure the weight gain (ADG) of piglets on day 0, 14, and 28 of the trial. The diarrhea frequency of the piglets was also estimated according to a previous report.53Determination of short-chain fatty acids (SCFAs)
The short-chain fatty acid (acetate, propionate, butyrate, and total SCFAs) concentrations in colonic digesta and plasma were analyzed by using the method described in a previous report.47 Briefly, approximately 1.5 g of colonic digesta was first homogenized in 1.5 mL of deionized water. The samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 10 min. The supernatants of colonic digesta and plasma samples were acidified with 25% meta-phosphoric acid at a 1
000×g for 10 min. The supernatants of colonic digesta and plasma samples were acidified with 25% meta-phosphoric acid at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio. The samples were then centrifuged at 12
5 ratio. The samples were then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g 10 min. The supernatant (1 μL) was separated and analyzed using a gas chromatograph (GC 2010; Shimadzu, Japan) equipped with a CP-Wax 52 CB column of 30.0 m by 0.53 mm internal diameter. SCFAs were quantified using external standard curves from 0.5 to 100 μmol mL−1 of the respective authentic organic acids (Fluka, Switzerland).
000×g 10 min. The supernatant (1 μL) was separated and analyzed using a gas chromatograph (GC 2010; Shimadzu, Japan) equipped with a CP-Wax 52 CB column of 30.0 m by 0.53 mm internal diameter. SCFAs were quantified using external standard curves from 0.5 to 100 μmol mL−1 of the respective authentic organic acids (Fluka, Switzerland).
Determination of cytokines and oxidative stress indicators
One hundred mg frozen colonic mucosa specimen were minced and homogenized in 1 mL of ice-cold cytoplasm RIPA containing the protease inhibitor cocktail complete EDTA-free (Roche, Penzberg, Germany). The homogenates were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 15 min at 4 °C and then the supernatant was collected. Protein concentration was determined using a BCA Protein Assay kit (Pierce, Rockford, IL, USA) and then diluted to the same concentration for subsequent analysis. The plasma and colonic interleukin (IL)-1β, -6, -8, tumor necrosis factor (TNF)-α, superoxide dismutase (SOD), glutathione peroxidase (GSH-Px), malondialdehyde (MDA), and catalase (CAT) were determined by using porcine enzyme-linked immunosorbent assay kits (Shanghai Enzyme-linked Biotechnology Co. Ltd, Shanghai, China), according to the manufacturer's instructions.
000×g for 15 min at 4 °C and then the supernatant was collected. Protein concentration was determined using a BCA Protein Assay kit (Pierce, Rockford, IL, USA) and then diluted to the same concentration for subsequent analysis. The plasma and colonic interleukin (IL)-1β, -6, -8, tumor necrosis factor (TNF)-α, superoxide dismutase (SOD), glutathione peroxidase (GSH-Px), malondialdehyde (MDA), and catalase (CAT) were determined by using porcine enzyme-linked immunosorbent assay kits (Shanghai Enzyme-linked Biotechnology Co. Ltd, Shanghai, China), according to the manufacturer's instructions.
Determination of intestinal permeability indicators
One hundred mg frozen colonic mucosa specimen were minced and homogenized in 1 mL of ice-cold cytoplasm RIPA containing the completely EDTA-free protease inhibitor cocktail (Roche, Penzberg, Germany). The homogenates were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 15 min at 4 °C and then the supernatant was collected. Protein concentration was determined using a BCA Protein Assay kit (Pierce, Rockford, IL, USA) and then diluted to the same concentration for subsequent analysis. The plasma and colonic endotoxin, diamine peroxidase (DAO), D-lactate, and zonulin were determined by using porcine enzyme-linked immunosorbent assay kits (Shanghai Enzyme-linked Biotechnology Co. Ltd, Shanghai, China), according to the manufacturer's instructions.
000×g for 15 min at 4 °C and then the supernatant was collected. Protein concentration was determined using a BCA Protein Assay kit (Pierce, Rockford, IL, USA) and then diluted to the same concentration for subsequent analysis. The plasma and colonic endotoxin, diamine peroxidase (DAO), D-lactate, and zonulin were determined by using porcine enzyme-linked immunosorbent assay kits (Shanghai Enzyme-linked Biotechnology Co. Ltd, Shanghai, China), according to the manufacturer's instructions.
DNA extraction, 16S rRNA gene amplification, and sequencing
Total DNA was extracted from 200 mg of each colonic digesta specimen by using the QIAamp R Fast DNA Stool Mini Kit (Qiagen Ltd., Germany) in accordance with the manufacturer's instructions. The V3–V4 region of the 16S rRNA gene was amplified with universal primers 341F (5′-ACTCCTACGGGA GGCAGCAG-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′), as described in a previous study.54 The amplified products were detected using agarose gel electrophoresis (2% agarose), recovered by AxyPrep DNA Gel Recovery Kit (Axygen Biosciences, Union City, CA, United States), and then quantified by Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) to pool into equimolar amounts. Amplicon libraries were sequenced on the Illumina MiSeq 2500 platform (Illumina, San Diego, CA, United States) for paired-end reads of 250 bp.RNA isolation, cDNA synthesis, and real-time quantitative PCR
Total RNA was extracted from 100 mg of each colonic mucosa specimen with Trizol reagent. The concentration and quality of the RNA were measured by NanoDrop ND-1000 Spectrophotometer (Thermo, USA). Then, two micrograms of total RNA were treated with RNase-Free DNase and reverse transcribed according to the manufacturer's instructions. Two microliters of diluted cDNA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, vol/vol) were used for real-time PCR, which was performed in Mx3000P (Stratagene, USA). GAPDH, which is not affected by the experimental factors, was chosen as the reference gene. All the primers that were used in this study are listed in Table S1† and were synthesized by Generay Company (Shanghai, China). The method of 2−ΔΔCt was used to analyze the real-time PCR results and gene mRNA levels were expressed as the fold change relative to the mean value of the control group.
20, vol/vol) were used for real-time PCR, which was performed in Mx3000P (Stratagene, USA). GAPDH, which is not affected by the experimental factors, was chosen as the reference gene. All the primers that were used in this study are listed in Table S1† and were synthesized by Generay Company (Shanghai, China). The method of 2−ΔΔCt was used to analyze the real-time PCR results and gene mRNA levels were expressed as the fold change relative to the mean value of the control group.
Protein expression analysis by western blotting
Colonic tissue was minced and homogenized in 1 mL of ice-cold homogenization buffer RIPA containing the completely EDTA-free protease inhibitor cocktail (Roche, Penz-berg, Germany). The homogenates were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min at 4 °C, and the supernatant fraction was collected. Protein concentration was determined using a BCA Protein Assay kit (Pierce, Rockford, IL, USA). Protein extract from each sample amounting to 80 μg was then loaded onto 7.5% and 15% SDS-PAGE gels, and the separated proteins were transferred onto nitrocellulose membranes (Bio Trace, Pall Co, USA). Post-transfer, the membranes were blocked for 2 h at room temperature in a blocking buffer and then incubated with the following primary antibodies: Claudin-1, Occludin, ZO-1, and GAPDH in a dilution buffer overnight at 4 °C. After several washes in Tris-Buffered-Saline with Tween (TBST), the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies in a dilution buffer for 2 h at room temperature. Finally, the blot was washed and detected by enhanced chemiluminescence (ECL) using the LumiGlo substrate (Super Signal West Pico Trial Kit, Pierce, USA), the signals were recorded with the help of an imaging system (Bio-Rad, USA), and analyzed using Quantity One software (Bio-Rad, USA).
000 rpm for 20 min at 4 °C, and the supernatant fraction was collected. Protein concentration was determined using a BCA Protein Assay kit (Pierce, Rockford, IL, USA). Protein extract from each sample amounting to 80 μg was then loaded onto 7.5% and 15% SDS-PAGE gels, and the separated proteins were transferred onto nitrocellulose membranes (Bio Trace, Pall Co, USA). Post-transfer, the membranes were blocked for 2 h at room temperature in a blocking buffer and then incubated with the following primary antibodies: Claudin-1, Occludin, ZO-1, and GAPDH in a dilution buffer overnight at 4 °C. After several washes in Tris-Buffered-Saline with Tween (TBST), the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies in a dilution buffer for 2 h at room temperature. Finally, the blot was washed and detected by enhanced chemiluminescence (ECL) using the LumiGlo substrate (Super Signal West Pico Trial Kit, Pierce, USA), the signals were recorded with the help of an imaging system (Bio-Rad, USA), and analyzed using Quantity One software (Bio-Rad, USA).
Analysis of sequencing data
The raw paired-end reads were assembled into longer sequences and quality filtered by PANDAseq (version 2.9) to remove the low-quality reads with a length of <220 nucleotides (nt) or >500 nt, an average quality score of <20, and sequences containing >3 nitrogenous bases.55 The high-quality sequences were clustered into OTUs with a 97% similarity using UPARSE (version 7.0)56 in QIIME (version 1.8)57 and the chimeric sequences were removed using UCHIME.58 Taxonomy was assigned to the OTUs using the RDP classifier59 against the SILVA 16S-rRNA gene database,60 with a confidence threshold of 70%. The data were analyzed on the free online platform of Majorbio I-Sanger Cloud Platform (www.i-sanger.com).Statistical analysis
Data are presented as mean ± SEM. Statistical significance was assessed by the independent sample t-test using SPSS (SPSS v. 20.0, SPSS Inc., Chicago, IL, USA) software packages. The data were considered statistically significant when P < 0.05. The number of replicates used for statistics are noted in the Figures.Authors' contributions
W. X. designed the research. W. X. and H. M. conducted the research. Z. Z., M. J., J. W., and Y. X. analyzed the data. The manuscript was mainly written by W. X. and edited by Z. W. All the authors have read and approved the final manuscript.Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Abbreviations
| ADG | Weight gain |
| BW | Body weight |
| CAT | Catalase |
| DAO | Diamine peroxidase |
| GSH-Px | Glutathione peroxidase |
| ICA | Icariin |
| IL | Interleukin |
| MDA | Malondialdehyde |
| OTUs | Operational taxonomic units |
| SCFAs | Short-chain fatty acids |
| SOD | Superoxide dismutase |
| TNF | Tumor necrosis factor |
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (31902313).References
- J. R. Turner, Nat. Rev. Immunol., 2009, 9, 799–809 CrossRef CAS PubMed.
- F. Sanchez de Medina, I. Romero-Calvo, C. Mascaraque and O. Martinez-Augustin, Inflammatory Bowel Dis., 2014, 20, 2394–2404 CrossRef PubMed.
- L. W. Peterson and D. Artis, Nat. Rev. Immunol., 2014, 14, 141–153 CrossRef CAS PubMed.
- F. E. O. Holmberg, J. Pedersen, P. Jorgensen, C. Soendergaard, K. B. Jensen and O. H. Nielsen, J. Tissue Eng. Regener. Med., 2018, 12, 923–935 CrossRef CAS PubMed.
- H. Zhu, H. Wang, S. Wang, Z. Tu, L. Zhang, X. Wang, Y. Hou, C. Wang, J. Chen and Y. Liu, Mol. Nutr. Food Res., 2018, 62, e1700814 CrossRef PubMed.
- G. Bevivino and G. Monteleone, Expert Rev. Gastroenterol. Hepatol., 2018, 12, 907–915 CrossRef CAS PubMed.
- P. Sheth, S. Basuroy, C. Li, A. P. Naren and R. K. Rao, J. Biol. Chem., 2003, 278, 49239–49245 CrossRef CAS PubMed.
- J. R. Marchesi, D. H. Adams, F. Fava, G. D. Hermes, G. M. Hirschfield, G. Hold, M. N. Quraishi, J. Kinross, H. Smidt, K. M. Tuohy, L. V. Thomas, E. G. Zoetendal and A. Hart, Gut, 2016, 65, 330–339 CrossRef PubMed.
- G. Tomasello, M. Mazzola, A. Leone, E. Sinagra, G. Zummo, F. Farina, P. Damiani, F. Cappello, A. Gerges Geagea, A. Jurjus, T. Bou Assi, M. Messina and F. Carini, Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia, 2016, vol. 160, pp. 461–466 Search PubMed.
- L. Sun, H. Jia, J. Li, M. Yu, Y. Yang, D. Tian, H. Zhang and Z. Zou, Front. Microbiol., 2019, 10, 1745 CrossRef PubMed.
- E. Puertollano, S. Kolida and P. Yaqoob, Curr. Opin. Clin. Nutr. Metab. Care, 2014, 17, 139–144 CrossRef CAS PubMed.
- M. Sun, W. Wu, Z. Liu and Y. Cong, J. Gastroenterol., 2017, 52, 1–8 CrossRef CAS PubMed.
- T. Liu, W. H. Liu, J. S. Zhao, F. Z. Meng and H. Wang, Cell Biol. Toxicol., 2017, 33, 57–67 CrossRef CAS PubMed.
- X. Ren, X. Li, L. Jia, D. Chen, H. Hou, L. Rui, Y. Zhao and Z. Chen, FASEB J., 2017, 31, 711–718 CrossRef CAS PubMed.
- P. Qiu, Y. Dong, T. Zhu, Y. Y. Luo, X. J. Kang, M. X. Pang, H. Z. Li, H. Xu, C. Gu, S. H. Pan, W. F. Du and W. H. Ge, Phytomedicine, 2019, 52, 40–50 CrossRef CAS PubMed.
- Q. Nie, H. Chen, J. Hu, S. Fan and S. Nie, Crit. Rev. Food Sci. Nutr., 2018, 1–16, DOI:10.1080/10408398.2018.1536646.
- H. T. Xiao, L. Zhong, S. W. Tsang, Z. S. Lin and Z. X. Bian, Am. J. Chin. Med., 2015, 43, 1–23 CrossRef PubMed.
- W. Xiong, X. Ma, Y. Wu, Y. Chen, L. Zeng, J. Liu, W. Sun, D. Wang and Y. Hu, BMC Vet. Res., 2015, 11, 205 CrossRef PubMed.
- X. Jing, T. Du, K. Chen, J. Guo, W. Xiang, X. Yao, K. Sun, Y. Ye and F. Guo, J. Cell. Physiol., 2019, 234(7), 10123–10137 CrossRef CAS PubMed.
- L. Kong, X. Liang, A. Liu, X. Yang, Q. Luo, Y. Lv and J. Dong, Clin. Exp. Dermatol., 2019, 44(2), 144–152 CrossRef CAS PubMed.
- F. Meurens, A. Summerfield, H. Nauwynck, L. Saif and V. Gerdts, Trends Microbiol., 2012, 20, 50–57 CrossRef CAS PubMed.
- M. M. Swindle, A. Makin, A. J. Herron, F. J. Clubb Jr and K. S. Frazier, Vet. Pathol., 2012, 49, 344–356 CrossRef CAS PubMed.
- P. Guilloteau, R. Zabielski, H. M. Hammon and C. C. Metges, Nutr. Res. Rev., 2010, 23, 4–22 CrossRef PubMed.
- S. N. Heinritz, R. Mosenthin and E. Weiss, Nutr. Res. Rev., 2013, 26, 191–209 CrossRef PubMed.
- S. Tao, Y. Duanmu, H. Dong, Y. Ni, J. Chen, X. Shen and R. Zhao, PLoS One, 2014, 9, e111596 CrossRef PubMed.
- W. Hua, Y. Zhang, X. Wu, L. Kang, J. Tu, K. Zhao, S. Li, K. Wang, Y. Song, R. Luo, Z. Shao, S. Yang and C. Yang, Curr. Pharm. Des., 2018, 23, 6071–6078 CrossRef PubMed.
- Y. H. Song, H. Cai, Z. M. Zhao, W. J. Chang, N. Gu, S. P. Cao and M. L. Wu, Biomed. Pharmacother., 2016, 83, 1089–1094 CrossRef CAS PubMed.
- S. Tao, Y. Duanmu, H. Dong, J. Tian, Y. Ni and R. Zhao, BMC Vet. Res., 2014, 10, 235 CrossRef PubMed.
- S. Tao, Y. Bai, T. Li, N. Li and J. Wang, FASEB J., 2019, 33(9), 9897–9912 CrossRef CAS PubMed.
- E. E. Schneeberger and R. D. Lynch, Am. J. Physiol.: Cell Physiol., 2004, 286, C1213–C1228 CrossRef CAS PubMed.
- K. Y. Kim, T. W. Oh, H. J. Do, J. H. Yang, I. J. Yang, Y. H. Jeon, Y. H. Go, S. C. Ahn, J. Y. Ma and K. I. Park, J. Immunol. Res., 2018, 2018, 5718396 Search PubMed.
- H. Y. Zhou, H. Zhu, X. M. Yao, J. P. Qian, J. Yang, X. D. Pan and X. D. Chen, Eur. Rev. Med. Pharmacol. Sci., 2017, 21, 5239–5246 Search PubMed.
- Y. Minamiyama, H. Ichikawa, S. Takemura, H. Kusunoki, Y. Naito and T. Yoshikawa, Free Radical Res., 2010, 44, 1398–1406 CrossRef CAS PubMed.
- Q. Zeng, H. Yi, L. Huang, Q. An and H. Wang, Toxicology, 2019, 411, 122–132 CrossRef CAS.
- R. Gangwar, A. S. Meena, P. K. Shukla, A. S. Nagaraja, P. L. Dorniak, S. Pallikuth, C. M. Waters, A. Sood and R. Rao, Biochem. J., 2017, 474, 731–749 CrossRef CAS PubMed.
- A. Kauppinen, T. Suuronen, J. Ojala, K. Kaarniranta and A. Salminen, Cell. Signalling, 2013, 25, 1939–1948 CrossRef CAS PubMed.
- K. Lingappan, Current opinion in toxicology, 2018, 7, 81–86 CrossRef PubMed.
- B. Mi, J. Wang, Y. Liu, J. Liu, L. Hu, A. C. Panayi, G. Liu and W. Zhou, Front. Pharmacol., 2018, 9, 605 CrossRef PubMed.
- A. Karkman, J. Lehtimaki and L. Ruokolainen, Ann. N. Y. Acad. Sci., 2017, 1399, 78–92 CrossRef PubMed.
- J. M. Natividad and E. F. Verdu, Pharmacol. Res., 2013, 69, 42–51 CrossRef CAS PubMed.
- A. J. Baumler and V. Sperandio, Nature, 2016, 535, 85–93 CrossRef CAS PubMed.
- G. den Besten, R. Havinga, A. Bleeker, S. Rao, A. Gerding, K. van Eunen, A. K. Groen, D. J. Reijngoud and B. M. Bakker, PLoS One, 2014, 9, e107392 CrossRef PubMed.
- B. He, Y. Bai, L. Jiang, W. Wang, T. Li, P. Liu, S. Tao, J. Zhao, D. Han and J. Wang, Int. J. Mol. Sci., 2018, 19(8), 2407 CrossRef PubMed.
- H. Yang, Y. Xiang, K. Robinson, J. Wang, G. Zhang, J. Zhao and Y. Xiao, Front. Microbiol., 2018, 9, 3045 CrossRef PubMed.
- M. R. Panasevich, U. D. Wankhade, S. V. Chintapalli, K. Shankar and R. S. Rector, Physiol. Genomics, 2018, 50, 355–368 CrossRef CAS PubMed.
- Y. Litvak, M. X. Byndloss, R. M. Tsolis and A. J. Baumler, Curr. Opin. Microbiol., 2017, 39, 1–6 CrossRef CAS PubMed.
- C. Cheng, H. Wei, C. Xu, X. Xie, S. Jiang and J. Peng, Appl. Environ. Microbiol., 2018, 84(17), e01047-18 CrossRef PubMed.
- J. Zhao, P. Liu, Y. Wu, P. Guo, L. Liu, N. Ma, C. Levesque, Y. Chen, J. Zhao, J. Zhang and X. Ma, J. Agric. Food Chem., 2018, 66, 7995–8004 CrossRef CAS PubMed.
- B. R. Hamaker and Y. E. Tuncil, J. Mol. Biol., 2014, 426, 3838–3850 CrossRef CAS PubMed.
- K. Fisher and C. Phillips, Microbiology, 2009, 155, 1749–1757 CrossRef CAS PubMed.
- J. Xu, X. Chen, S. Yu, Y. Su and W. Zhu, PLoS One, 2016, 11, e0162461 CrossRef PubMed.
- P. Bin, Z. Tang, S. Liu, S. Chen, Y. Xia, J. Liu, H. Wu and G. Zhu, BMC Vet. Res., 2018, 14, 385 CrossRef CAS PubMed.
- C. Yu, S. Zhang, Q. Yang, Q. Peng, J. Zhu, X. Zeng and S. Qiao, Arch. Anim. Nutr., 2016, 70, 263–277 CrossRef PubMed.
- X. Hong, J. Chen, L. Liu, H. Wu, H. Tan, G. Xie, Q. Xu, H. Zou, W. Yu, L. Wang and N. Qin, Sci. Rep., 2016, 6, 26621 CrossRef CAS PubMed.
- A. P. Masella, A. K. Bartram, J. M. Truszkowski, D. G. Brown and J. D. Neufeld, BMC Bioinf., 2012, 13, 31 CrossRef CAS PubMed.
- R. C. Edgar, Nat. Methods, 2013, 10, 996–998 CrossRef CAS PubMed.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Pena, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld and R. Knight, Nat. Methods, 2010, 7, 335–336 CrossRef CAS PubMed.
- R. C. Edgar, B. J. Haas, J. C. Clemente, C. Quince and R. Knight, Bioinformatics, 2011, 27, 2194–2200 CrossRef CAS PubMed.
- G. Bacci, A. Bani, M. Bazzicalupo, M. T. Ceccherini, M. Galardini, P. Nannipieri, G. Pietramellara and A. Mengoni, J Genomics, 2015, 3, 36–39 CrossRef PubMed.
- E. Pruesse, C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies and F. O. Glockner, Nucleic Acids Res., 2007, 35, 7188–7196 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07176h |
| This journal is © The Royal Society of Chemistry 2019 |