Open Access Article
Open Access ArticleRetracted Article: Long noncoding RNA ANRIL knockdown increases sensitivity of non-small cell lung cancer to cisplatin by regulating the miR-656-3p/SOX4 axis
Xianfang Wanga,
Jun Shia,
Ying Chena,
Caihong Wangb,
Huifang Shia and
Xuefang Xie *a
*a
aDepartment of Laboratory, People's Hospital of Rizhao, No. 126 Tai'an Road, Rizhao 276800, Shandong, China. E-mail: lianfan68941@126.com; Tel: +86-0633-3365206
bDepartment of Pharmacy, Rizhao Maternal and Child Health Care Hospital, China
First published on 26th November 2019
Abstract
Long noncoding RNAs (lncRNAs) are implicated in the development of chemoresistance in many cancers. However, the effect and mechanism of lncRNA antisense noncoding RNA in the INK4 locus (ANRIL) on cisplatin (CDDP) resistance in non-small cell lung cancer (NSCLC) remain unclear. The levels of ANRIL, microRNA (miR)-656-3p and sex-determining region Y-related high-mobility group box 4 (SOX4) in NSCLC tissues and cells were detected by quantitative real-time polymerase chain reaction or western blotting. Cell viability, apoptosis, migration and epithelial-to-mesenchymal transition (EMT) were assessed by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT assay), flow cytometry, trans-well assays and western blotting, respectively. The xenograft model was established using CDDP-resistant NSCLC cells. The target association between miR-656-3p and ANRIL or SOX4 was validated by luciferase reporter assay and RNA immunoprecipitation. ANRIL expression was increased in CDDP-resistant NSCLC tissues and cells. Knockdown of ANRIL decreased cell viability, migration and EMT but induced apoptosis in CDDP-resistant NSCLC cells. Moreover, silencing of ANRIL reduced xenograft tumor growth in vivo. miR-656-3p was targeted by ANRIL and its exhaustion attenuated the suppressive role of ANRIL knockdown in CDDP resistance in NSCLC cells. SOX4 acted as a target of miR-656-3p and was positively regulated by ANRIL. Collectively, interference of ANRIL repressed CDDP resistance through promoting apoptosis and inhibiting cell viability, migration and EMT by up-regulating miR-656-3p and down-regulating SOX4, indicating a new target to improve the chemotherapeutic efficacy in NSCLC.
Introduction
Non-small cell lung cancer (NSCLC) is the most common subtype of lung cancer with high mortality.1 With the efforts of the past decades, the diagnosis and treatment of patients with NSCLC have been significantly advanced.2 Cisplatin (CDDP)-based chemotherapy has been the first-line treatment of advanced NSCLC.3 However, the efficacy of chemotherapy has progressively declined because of the development of drug resistance. Hence, challenges remain in exploring novel strategies to improve the chemotherapeutic effect for patients with NSCLC.The drug resistance might be associated with the dysregulation of noncoding RNAs including long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) in cancers.4 LncRNAs with greater than 200 nucleotides play essential roles in the regulation of development and drug resistance in NSCLC.5,6 LncRNA antisense noncoding RNA in the INK4 locus (ANRIL), also known as cyclin-dependent kinase inhibitor 2B antisense RNA 1 (CDKN2B-AS1), has been reported to act as an oncogene to promote the development of multiple cancers.7–9 More importantly, previous studies suggest that ANRIL could encourage the development of drug resistance in gastric cancer, osteosarcoma and ovarian cancer.10–12 Although the carcinogenesis of ANRIL has been exhibited in NSCLC development,13,14 whether this lncRNA could regulate CDDP resistance in NSCLC remains unclear.
The former work also suggests that miRNAs are implicated in CDDP resistance in NSCLC.15 A novel miRNA miR-656-3p has been regarded as a tumor suppressor in glioma and NSCLC.16,17 However, little is known about the role of this miRNA in CDDP resistance in NSCLC. Sex-determining region Y-related high-mobility group box (SOX) family has key roles in tumorigenesis and metastasis of human cancers.18 SOX4, as a member of this family, could regulate epithelial-to-mesenchymal transition (EMT) to be involved in the development and resistance of NSCLC.19–21 Previous study reveals that lncRNAs could exhibit the therapeutic potential in cancers by acting competing endogenous RNAs (ceRNAs) for miRNAs to regulate mRNAs.22 Intriguingly, the database of LncBase predicted and starBase showed the existence of similar complementary sites between miR-656-3p and ANRIL or SOX4, suggesting the potential ceRNA network between them. Here we investigated the function of ANRIL on cell survival rate, apoptosis, migration and EMT in NSCLC after treatment of CDDP and confirmed the ceRNA network of ANRIL/miR-656-3p/SOX4.
Materials and methods
Patient tissues and cell culture
Forty-two NSCLC patients who have received CDDP-based therapy were recruited from People's Hospital of Rizhao. They all signed the informed consents and were divided into CDDP-sensitive (n = 18) or -resistant (n = 24) group according to the patients' responses to CDDP with the standard.23 The tumor tissues and peritumor normal samples were collected and used for RNA isolation. All experiments were performed in accordance with the Guidelines of NIH, and experiments were approved by the ethics committee of People's Hospital of Rizhao. Informed consents were obtained from human participants of this study.NSCLC cell lines (A549 (accession code: CVCL_0023) and H1299 (accession code: CVCL_0060)) and human bronchial epithelioid cell line (16HBE (accession code: CVCL_0112)) were purchased from BeNa Culture Collection (Beijing, China). The cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Solarbio, Beijing, China) containing 10% fetal bovine serum (Sigma, St. Louis, MO, USA) and 1% streptomycin/penicillin (Beyotime, Shanghai, China). The CDDP-resistant cell lines (A549/CDDP and H1299/CDDP) were obtained by exposing to the increasing doses of CDDP (Sigma, St. Louis, MO, USA).24 The resistant cells were maintained in cell medium containing 2 μM CDDP.
Cell transfection
The ANRIL overexpression vector (ANRIL) was generated with pcDNA3.1 empty vector (vector) (Thermo Fisher, Wilmington, DE, USA) as control. The short hairpin RNA (shRNA)-based on lentiviral vector against ANRIL (sh-ANRIL#1, 5′-GCCUAUCAUAUAAGCCCAAAU-3′; sh-ANRIL#2, 5′-GCCCAAUUAUGCUGUGGUAAC-3′; sh-ANRIL#3, 5′-GCACAUAUCCAUCTAUCAUCU-3′), shRNA negative control (sh-NC, 5′-UCUUCCGAACGUGUCACGUTT-3′), miR-656-3p mimics (5′-AAUAUU AUACAGUCAACCUCU-3′) and mimics negative control (NC mimics, 5′-UUUGU ACUACACAAAAGUACUG-3′), miR-656-3p inhibitor (5′-AGAGGUUGACUGUAUAAUAUU-3′), inhibitor negative control (NC inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′) were provided by Fulengen (Guangzhou, China). Following the instructions, A549/CDDP and H1299/CDDP cells were transfected with these conducted oligonucleotides for 24 h.Quantitative real-time polymerase chain reaction (qRT-PCR)
Trizol reagent (Solarbio) was used for total RNA extraction from tissues or cells. The complementary DNA (cDNA) was generated using First Strand cDNA Synthesis Kit (Yeasen, Shanghai, China) or All-in-One™ miRNA qRT-PCR Detection Kit (Fulengen) and then used for qRT-PCR assay with SYBR mix (Vazyme, Nanjing, China) as well as specific primers (Sangon, Shanghai, China) (ANRIL: forward, 5′-GCATGAAGGACACACCAGGG-3′; reverse, 5′-GCCGTTCCATCAGTCCAAGT-3′; SOX4: forward, 5′-CCGAGCTGGTGCAAGACC-3′; reverse, 5′-CCACACCATGAAGGCGTTC-3′; miR-656-3p: forward, 5′-ACACTCCAGCTGGGAATATTATACAGTCA-3′; reverse, Universal Adaptor PCR Primer). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward, 5′-AGACAGCCGCATCTTCTTGT-3′; reverse, 5′-TGATGGCAACAATGTCCACT-3′) and U6 (forward, 5′-CTCGCTTCGGCAGCACATATACT-3′; reverse, Universal Adaptor PCR Primer) were regarded as internal controls. The relative levels of ANRIL, miR-656-3p and SOX4 were assessed by the delta–delta cycle threshold method.253-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay
Cell survival rate was analyzed by MTT assay. Cells (1 × 104 per well) were plated into 96-well plates overnight and then exposed to different concentrations (2, 4, 8, 16, 32 and 64 μM) of CDDP for 24 h. Subsequently, the medium was changed with fresh one with 0.5 mg mL−1 MTT solution (Beyotime) and cells were cultured in normal conditions for 4 h. After addition of dimethyl sulfoxide (Solarbio), the survival rate was analyzed according to optical density at 570 nm through a microplate reader (Potenov, Beijing, China).Flow cytometry
Transfected or non-transfected A549/CDDP and H1299/CDDP cells (1 × 105 per well) were seeded into 24-well plates and challenged by 5 μM CDDP for 24 h. Then cells were collected, washed with phosphate buffer saline (PBS) and resuspended in binding buffer, followed by incubation with Annexin V-fluorescein isothiocyanate (FITC) and Propidium Iodide (PI) (Solarbio) in the dark. The apoptotic cells were determined with a flow cytometry (Countstar, Shanghai, China) and apoptotic rate of A549/CDDP and H1299/CDDP cells was expressed as the percentage of cells (Annexin V-FITC+ and PI+) in the total number of cells.Trans-well assay
The migrated ability was investigated by trans-well assay, A549/CDDP and H1299/CDDP cells (1 × 104) in serum-free RPMI 1640 medium were transferred to upper chambers and 500 μL RPMI 1640 medium containing 10% serum was added to the lower chambers. After treatment of 5 μM CDDP for 12 h, migrated cells were fixed and stained with 0.5% crystal violet (Solarbio). Three random fields were selected for observation of the migrated cells under a microscope (200× magnification, Olympus, Tokyo, Japan).Western blot
Cell lysates were prepared with total protein extraction kit (Solarbio) on ice and the concentration of total protein was measured by using bicinchoninic acid protein assays kit (Beyotime). Proteins were mixed with loading buffer and subjected to the boiled water bath for 5 min, followed by separation through sodium dodecyl sulfate-polyacrylamide gel electrophoresis and membrane transfer using nitrocellulose membranes (Millipore, Billerica, MA, USA). Following the treatment of Western Blocking Buffer (Beyotime), the membranes were interacted with anti-B-cell lymphoma-2 (Bcl-2, ab196495, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution, Abcam, Cambridge, MA, USA), anti-cleaved caspase 3 (c-caspase 3, ab2302, 1
2000 dilution, Abcam, Cambridge, MA, USA), anti-cleaved caspase 3 (c-caspase 3, ab2302, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Abcam), anti-E-cadherin (ab15148, 1
1000 dilution, Abcam), anti-E-cadherin (ab15148, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution, Abcam), anti-matrix metalloprotein-9 (MMP-9, ab38898, 1
500 dilution, Abcam), anti-matrix metalloprotein-9 (MMP-9, ab38898, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Abcam), anti-SOX4 (ab80261, 1
1000 dilution, Abcam), anti-SOX4 (ab80261, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution, Abcam) or anti-GAPDH (ab9485, 1
1000 dilution, Abcam) or anti-GAPDH (ab9485, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 dilution, Abcam) as a loading control and corresponding secondary antibody. Enhanced chemiluminescence reagent (Yeasen Biotech, Shanghai, China) was used for the development of protein signals.
3000 dilution, Abcam) as a loading control and corresponding secondary antibody. Enhanced chemiluminescence reagent (Yeasen Biotech, Shanghai, China) was used for the development of protein signals.
Xenograft model
Animal experiments were approved by the Animal Ethics Committee of People's Hospital of Rizhao. Twenty male BALB/c nude mice (6-week-old) purchased from Charles River (Beijing, China) were randomly divided into four groups (n = 5 per group). A549/CDDP cells (2 × 106 per mouse) stably transfected with sh-ANRIL#1 or sh-NC or non-transfected cells were subcutaneously injected into nude mice. At 5 days, the mice were subjected to intraperitoneal administration of CDDP (5 mg kg−1) or equal volume of PBS (control) every 5 days. The tumor volume was monitored every 5 days with a digital caliper and calculated with a formula: length × width2 × 0.5. At 25 days after the inoculation, mice in each group were killed and tumor tissues were weighed and used for analysis of ANRIL expression.Luciferase reporter assay and RNA immunoprecipitation (RIP)
The targets of ANRIL or miR-656-3p were predicted by LncBase predicted or starBase, respectively. The sequences of lncRNA ANRIL containing wild-type (WT) or mutant (Mut) binding sites of miR-656-3p were synthesized and inserted into psiCHECK-2 vector (Promega, Madison, WI, USA) to generate corresponding luciferase reporter vectors, namely ANRIL-WT and ANRIL-MUT. Similarly, the 3′ untranslated regions (UTR) sequences of SOX4 containing miR-656-3p binding sites and its mutant were inserted into psiCHECK-2 vector to obtain SOX4 3′UTR-WT and SOX4 3′UTR-MUT, respectively. For luciferase reporter assay, A549/CDDP and H1299/CDDP cells were transfected with the constructed luciferase reporter vectors and miR-656-3p mimics or NC mimics and luciferase activity was measured with a luciferase assay system (Promega) at 24 h post-transfection.For RIP assay, A549/CDDP and H1299/CDDP cells were lysed and incubated with Magna RIP Kit (Millipore) following the manufacturer's instructions. The magnetic beads were bound with Ago2 antibody, with IgG as a negative control. The enrichment levels of ANRIL and miR-656-3p in the complex were detected by qRT-PCR.
Statistical analysis
The data were expressed as mean ± standard error of the mean (SEM) and all experiments were performed at least three times. The difference between two or more groups was analyzed by Student's t-test or one-way analysis of variance followed by Tukey's test. The significant difference was determined when P value was less than 0.05.Results
The level of ANRIL is enhanced in CDDP-resistant non-small cell lung cancer
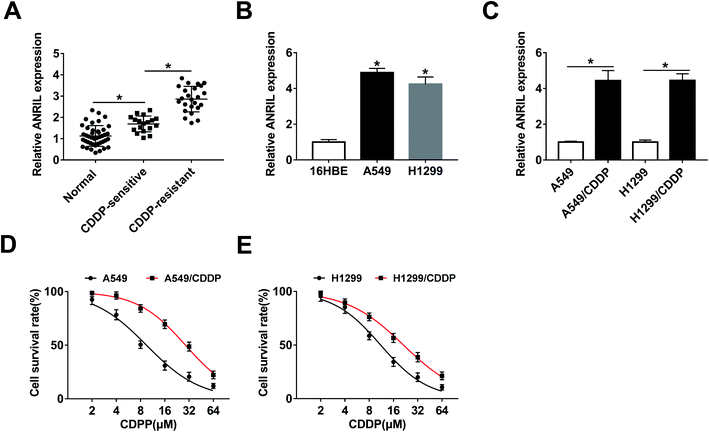
To explore the lncRNA level in NSCLC tissues, 42 patients were recruited and divided into CDDP-sensitive (n = 18) and -resistant groups (n = 24). As shown in Fig. 1A, the expression of ANRIL was significantly increased in NSCLC tissues compared with that in normal samples. Meanwhile, higher level of ANRIL was shown in CDDP-resistant group than that in sensitive group. Moreover, the expression of ANRIL was abnormally enhanced in A549 and H1299 cells when compared with 16HBE cells (Fig. 1B). In addition, the abundance of this lncRNA was obviously elevated in A549/CDDP and H1299/CDDP cells in comparison to the corresponding CDDP-sensitive cells (Fig. 1C). Besides, MTT assay displayed that CDDP-resistant cells had higher survival rate than sensitive cells upon treatment of CDDP (IC50: A549, 9.47 ± 0.53 μM; A549/CDDP, 28.86 ± 0.36 μM; H1299, 11.09 ± 0.46 μM; H1299/CDDP, 21.27 ± 0.41 μM) (Fig. 1D and E).Knockdown of ANRIL suppresses CDDP resistance in CDDP-resistant NSCLC cells
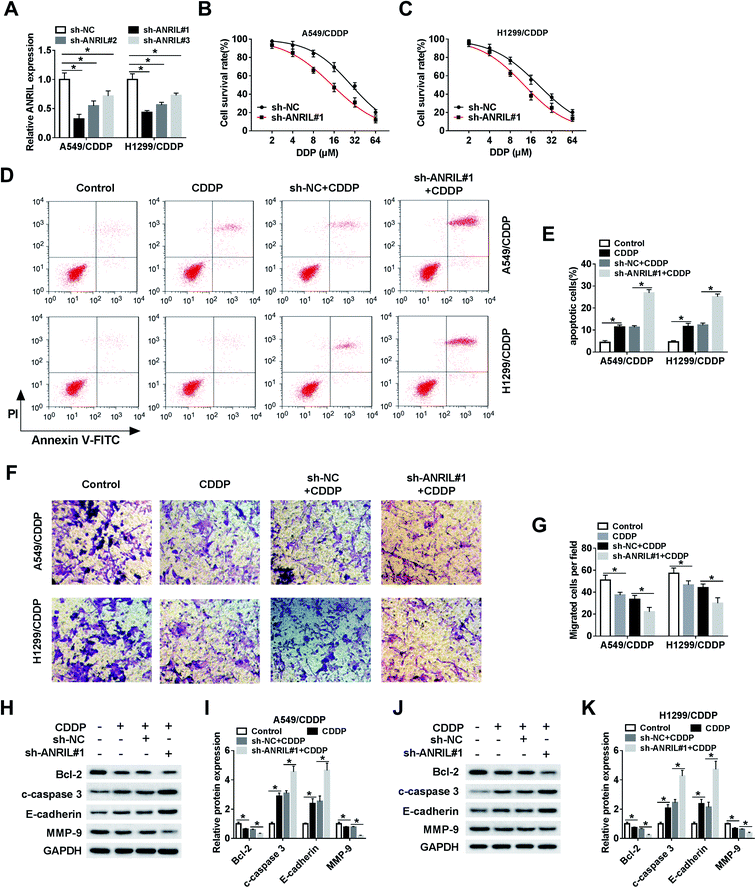
To investigate the role of ANRIL in CDDP resistance, CDDP-resistant cells were transfected with sh-NC or sh-ANRIL#1–#3. The transfection efficacy was confirmed with lowest level of ANRIL in sh-ANRIL#1 group (Fig. 2A). Hence, the loss-of-function experiments were performed by using sh-ANRIL#1. MTT assay showed that knockdown of ANRIL remarkably decreased cell survival in A549/CDDP and H1299/CDDP cells after challenge of CDDP (Fig. 2B and C). Moreover, silence of ANRIL significantly promoted cell apoptosis induced by CDDP in the two CDDP-resistant cell lines (Fig. 2D and E). In addition, the data of trans-well assay described that treatment of CDDP evidently reduced the migrated ability of A549/CDDP and H1299/CDDP cells, which was exacerbated by interference of ANRIL (Fig. 2F and G). What's more, the levels of proteins involved in apoptosis and EMT were measured and results showed that exposure of CDDP led to obviously increased c-caspase 3 and E-cadherin and reduced Bcl-2 and MMP-9 levels in A549/CDDP and H1299/CDDP cells, and absence of ANRIL aggravated this effect (Fig. 2H–K).Silence of ANRIL increases CDDP sensitivity in NSCLC xenograft model
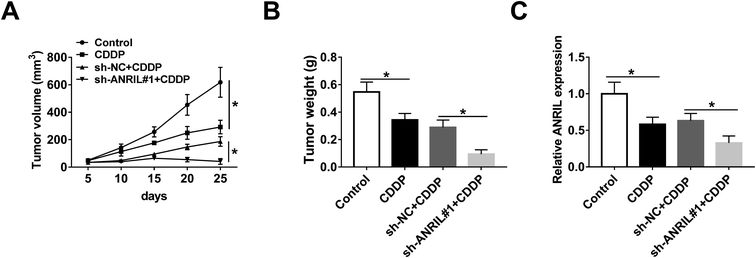
To further evaluate the regulatory effect of ANRIL on CDDP resistance in vivo, A549/CDDP cells were used for xenograft model and then the mice were treated by CDDP. As shown in Fig. 3A and B, tumor volume and weight were significantly decreased in CDDP-treated group compared with those in control group, and these events were further deteriorated by silence of ANRIL. Moreover, qRT-PCR assay revealed that the expression of ANRIL was specially decreased by CDDP treatment, which was aggravated in knockdown of ANRIL group (Fig. 3C).miR-656-3p is a target of ANRIL in CDDP-resistant NSCLC cells
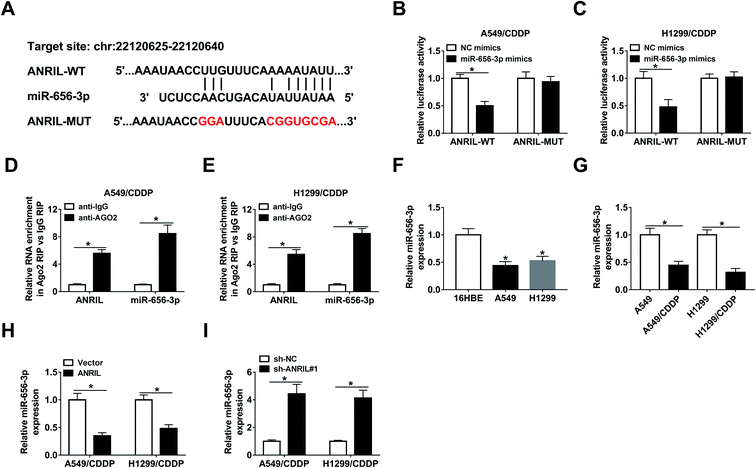
In order to explore the mechanism mediated by ANRIL, LncBase predicted database online assessed the targets of this lncRNA, in which miR-656-3p has the potential binding sites of ANRIL (Fig. 4A). Through generating and transfecting luciferase reporter vectors ANRIL-WT and ANRIL-MUT into A549/CDDP and H1299/CDDP cells, luciferase activity was remarkably decreased by miR-656-3p overexpression in ANRIL-WT group, while there was not significant change in ANRIL-MUT group (Fig. 4B and C). Furthermore, RIP analysis confirmed there were great enrichment of ANRIL and miR-656-3p in the complex of Ago2 RIP group compared with IgG group (Fig. 4D and E). Moreover, the abundance of miR-656-3p was significantly decreased in NSCLC cells (Fig. 4F), especially in CDDP-resistant cells (Fig. 4G). Additionally, the expression of miR-656-3p in A549/CDDP and H1299/CDDP cells was inhibited by ANRIL overexpression but increased by ANRIL knockdown (Fig. 4H and I).Deficiency of miR-656-3p reverses the promoting effect of ANRIL silence on CDDP sensitivity in CDDP-resistant NSCLC cells
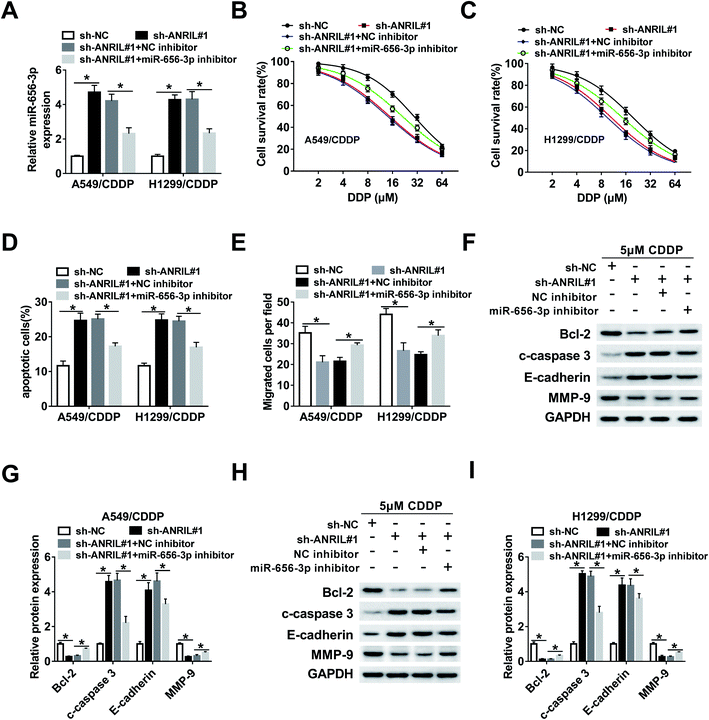
To explore whether miR-656-3p was required for the regulation of ANRIL on CDDP resistance in NSCLC, A549/CDDP and H1299/CDDP cells were transfected with sh-NC, sh-ANRIL#1, sh-ANRIL#1 and NC inhibitor or miR-656-3p inhibitor. The results of qRT-PCR showed that the abundance of miR-656-3p increased by ANRIL silence was effectively reduced by introduction of miR-656-3p inhibitor (Fig. 5A). Moreover, deficiency of miR-656-3p attenuated the survival inhibition and apoptosis production mediated by ANRIL knockdown in A549/CDDP and H1299/CDDP cells treated by CDDP (Fig. 5B–D). Additionally, silence of ANRIL-induced inhibition of migration was reversed by miR-656-3p knockdown in the two CDDP-resistant cell lines (Fig. 5E). Besides, the effect of ANRIL silence on protein levels of Bcl-2, c-caspase 3, E-cadherin and MMP-9 was mitigated by down-regulation of miR-656-3p in A549/CDDP and H1299/CDDP cells treated by CDDP (Fig. 5F–I).SOX4 is a target of miR-656-3p in CDDP-resistant NSCLC cells
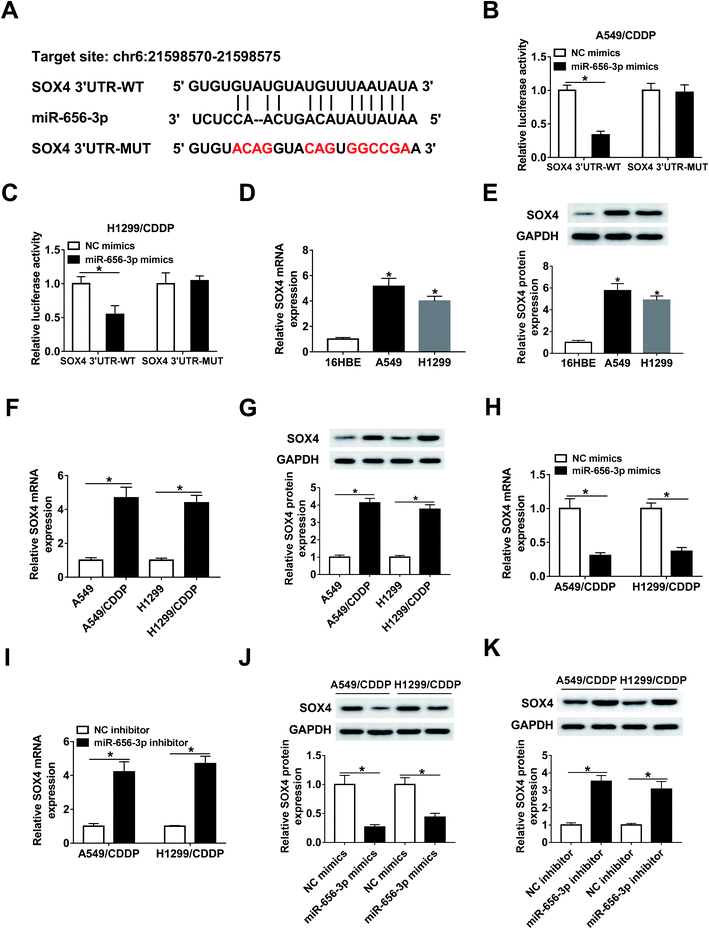
The targets of miR-656-3p were searched by starBase, which displayed the putative binding sites between miR-656-3p and SOX4 (Fig. 6A). Luciferase reporter assay was used to validate this association with the results of the decreased luciferase activity in A549/CDDP and H1299/CDDP cells co-transfected with miR-656-3p mimics and SOX4 3′UTR-WT when compared to corresponding control group (Fig. 6B and C). Furthermore, increased expression of SOX4 at mRNA and protein levels was shown in A549 and H1299 cells (Fig. 6D and E), and the corresponding CDDP-resistant cells exhibited higher levels of SOX4 than sensitive cells (Fig. 6F and G). Besides, the mRNA and protein levels of SOX4 were negatively regulated by miR-656-3p in A549/CDDP and H1299/CDDP cells (Fig. 6H–K).ANRIL regulates SOX4 expression by miR-656-3p
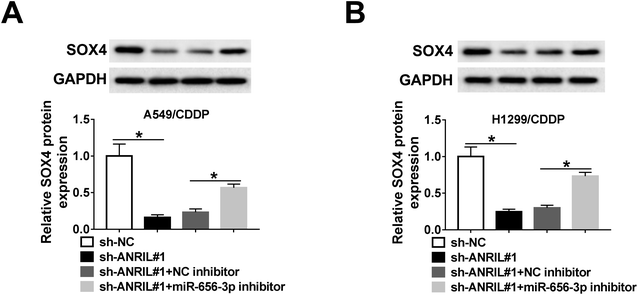
To explore whether ANRIL could mediate SOX4 expression in NSCLC, A549/CDDP and H1299/CDDP cells were transfected with sh-NC, sh-ANRIL#1, sh-ANRIL#1 and NC inhibitor or miR-656-3p inhibitor. The data of western bolt revealed that the protein level of SOX4 was significantly decreased by knockdown of ANRIL in A549/CDDP and H1299/CDDP cells, which was restored by inhibition of miR-656-3p (Fig. 7A and B).Discussion
Development of drug resistance limits the efficacy of chemotherapy in NSCLC and lncRNAs could participate in the regulation of CDDP sensitivity.26,27 Previous efforts have displayed that ANRIL could promote NSCLC progression by increasing cell proliferation.28,29 Nevertheless, little is known about the regulatory effect of ANRIL on CDDP sensitivity in NSCLC. In the present work, we disclosed the suppressive effect of ANRIL silence on CDDP resistance to NSCLC. Furthermore, this research was the first to confirm the ceRNA network of ANRIL/miR-656-3p/SOX4 in NSCLC.Here we first collected the CDDP-resistant and -sensitive tissues and generated the resistant cells. By comparing to sensitive group, we found that ANRIL was highly expressed in CDDP-resistant tissues and cells, implying that high expression of ANRIL might promote CDDP resistance. Next, loss-of-function experiments were performed to confirm that ANRIL knockdown inhibited CDDP resistance in CDDP-resistant cells, indicating that ANRIL inhibitor might be regarded as promising target for improving chemotherapeutic effect, which is consistent with the results as previous studies in other cancers.10–12 Furthermore, Chen et al. reported that autophagy and apoptosis were implicated in the regulation of CDDP sensitivity in NSCLC.30 Similarly, this study also confirmed that ANRIL knockdown enhanced CDDP sensitivity by inducing apoptosis through detecting apoptotic rate and levels of anti-apoptotic protein Bcl-2 and pro-apoptotic protein c-caspase 3. However, whether autophagy was also involved in the mechanism mediated by ANRIL would be explored in future. Furthermore, EMT has been regarded as an important pathway associated with metastasis and drug resistance in cancers.31–33 By detecting migration and protein levels of E-cadherin and MMP-9, this study showed that ANRIL silence suppressed CDDP resistance by decreasing EMT. Collectively, ANRIL knockdown could facilitate apoptosis and repress EMT to decrease CDDP resistance in NSCLC. Meanwhile, we used xenograft model to mimic the microenvironment in vivo to further validate the sensitizing effect of ANRIL silence.
Previous studies have suggested that the ceRNA network mediated by ANRIL might be responsible for the tumorigenesis and development of drug resistance in human cancers.7,12 Hence, to figure out the potential ceRNA network involved in CDDP resistance, the targets of ANRIL were explored and this study confirmed the interaction between ANRIL and miR-656-3p, which has been reported as a suppressor in progression of NSCLC.17 However, there is no evidence disclosing the effect of this miRNA on CDDP resistance. The rescue experiments showed that inhibition of miR-656-3p decreased the suppressive effect of ANRIL knockdown on CDDP resistance, indicating that ANRIL contributed to CDDP resistance by sponging miR-656-3p. To further uncover the ceRNA network, the targets of miR-656-3p were expected to be explored. SOX4 as an inducer of EMT could promote metastasis and development of cancers.19,34 Furthermore, a number of evidences have been reported on the promoting role of SOX4 in chemoresistance in multiple cancers, including NSCLC.20,35,36 Here we confirmed SOX4 as a target of miR-656-3p in NSCLC cells and further showed that ANRIL could regulate SOX4 expression by competitively binding miR-656-3p. Moreover, we found that SOX4 expression was increased in CDDP-resistant cells, indicating that high expression of SOX4 might be associated with CDDP resistant in NSCLC, which is also in agreement with a former work.20 Meanwhile, that study has validated that SOX4 could increase CDDP resistance in NSCLC cells. Hence, although the function of SOX4 was not investigated, we also concluded that ANRIL regulated CDDP resistance in NSCLC by miR-656-3p/SOX4 axis in vitro. Previous studies have disclosed the importance of Wnt and mitogen activated protein kinase signaling pathways in development of CDDP resistance in NSCLC.37,38 Moreover, these two pathways could be mediated by ANRIL in retinoblastoma and head and neck squamous cell carcinoma.7,39 Hence, we hypothesized that Wnt and mitogen activated protein kinase signaling pathways might be also involved in the regulation of ANRIL on CDDP resistance in NSCLC, which should be explored in future.
Conclusion
To summarize, this research demonstrated the sensitizing effect of ANRIL knockdown on NSCLC in response to CDDP through inducing apoptosis and inhibiting migration and EMT, possibly by up-regulating miR-656-3p and down-regulating SOX4, providing a new target for improving chemotherapeutic efficacy of patients with NSCLC.Conflicts of interest
The authors declare that they have no financial conflicts of interest.References
- R. S. Herbst, D. Morgensztern and C. Boshoff, Nature, 2018, 553, 446–454 CrossRef CAS PubMed.
- N. Duma, R. Santana-Davila and J. R. Molina, Mayo Clin. Proc., 2019, 94, 1623–1640 CrossRef CAS PubMed.
- A. Rossi, M. Di Maio, P. Chiodini, R. M. Rudd, H. Okamoto, D. V. Skarlos, M. Fruh, W. Qian, T. Tamura, E. Samantas, T. Shibata, F. Perrone, C. Gallo, C. Gridelli, O. Martelli and S. M. Lee, J. Clin. Oncol., 2012, 30, 1692–1698 CrossRef CAS PubMed.
- F. Corra, C. Agnoletto, L. Minotti, F. Baldassari and S. Volinia, Front. Oncol., 2018, 8, 327 CrossRef PubMed.
- M. A. Osielska and P. P. Jagodzinski, Biomed. Pharmacother., 2018, 101, 322–333 CrossRef CAS PubMed.
- L. Wang, L. Ma, F. Xu, W. Zhai, S. Dong, L. Yin, J. Liu and Z. Yu, Thorac. Cancer, 2018, 9, 761–768 CrossRef CAS PubMed.
- L. M. Zhang, H. Y. Ju, Y. T. Wu, W. Guo, L. Mao, H. L. Ma, W. Y. Xia, J. Z. Hu and G. X. Ren, Am. J. Cancer Res., 2018, 8, 2296–2310 CAS.
- X. Wang, X. Zhang, Y. Han, Q. Wang, Y. Ren, B. Wang and J. Hu, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 2265–2273 CrossRef CAS PubMed.
- L. Liu, S. B. Ning, S. Fu, Y. Mao, M. Xiao and B. Guo, Eur. Rev. Med. Pharmacol. Sci., 2019, 23, 6194–6201 CAS.
- W. G. Lan, D. H. Xu, C. Xu, C. L. Ding, F. L. Ning, Y. L. Zhou, L. B. Ma, C. M. Liu and X. Han, Oncol. Rep., 2016, 36, 263–270 CrossRef CAS PubMed.
- G. Li and Y. Zhu, Pathol., Res. Pract., 2019, 215, 931–938 CrossRef CAS PubMed.
- J. T. Miao, J. H. Gao, Y. Q. Chen, H. Chen, H. Y. Meng and G. Lou, Biosci. Rep., 2019, 39, BSR20182101 CrossRef CAS PubMed.
- L. Lin, Z. T. Gu, W. H. Chen and K. J. Cao, Diagn. Pathol., 2015, 10, 14 CrossRef PubMed.
- M. Naemura, C. Murasaki, Y. Inoue, H. Okamoto and Y. Kotake, Anticancer Res., 2015, 35, 5377–5382 CAS.
- H. Zang, J. Peng, W. Wang and S. Fan, J. Cancer, 2017, 8, 3856–3861 CrossRef PubMed.
- M. Guo, Z. Jiang, X. Zhang, D. Lu, A. D. Ha, J. Sun, W. Du, Z. Wu, L. Hu, K. Khadarian, J. Shen and Z. Lin, Carcinogenesis, 2014, 35, 1698–1706 CrossRef CAS PubMed.
- T. Chen, S. Qin, Y. Gu, H. Pan and D. Bian, Mol. Genet. Genomic Med., 2019, 7, e757 Search PubMed.
- D. Grimm, J. Bauer, P. Wise, M. Kruger, U. Simonsen, M. Wehland, M. Infanger and T. J. Corydon, Semin. Cancer Biol., 2019 DOI:10.1016/j.semcancer.2019.03.004.
- X. Wu, Z. Xin, Z. Zou, C. Lu, Z. Yu, S. Feng, P. Pan, G. Hao, Y. Dong and Y. Yang, Semin. Cancer Biol., 2019 DOI:10.1016/j.semcancer.2019.06.008.
- B. Hu, H. Zhang, Z. Wang, F. Zhang, H. Wei and L. Li, Cancer Biol. Ther., 2017, 18, 974–983 CrossRef CAS PubMed.
- J. Chang, F. Gao, H. Chu, L. Lou, H. Wang and Y. Chen, J. Cell. Physiol., 2019 DOI:10.1002/jcp.29099.
- A. Renganathan and E. Felley-Bosco, Adv. Exp. Med. Biol., 2017, 1008, 199–222 CrossRef CAS PubMed.
- G. J. Hao, Y. H. Ding, H. Wen, X. F. Li, W. Zhang, H. Y. Su, D. M. Liu and N. L. Xie, Biochem. Biophys. Res. Commun., 2017, 488, 501–508 CrossRef CAS PubMed.
- R. S. Huang, Y. L. Zheng, J. Zhao and X. Chun, Biomed. Pharmacother., 2018, 98, 538–544 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- K. A. Olaussen and S. Postel-Vinay, Ann. Oncol., 2016, 27, 2004–2016 CrossRef CAS PubMed.
- Y. Hu, Q. N. Zhu, J. L. Deng, Z. X. Li, G. Wang and Y. S. Zhu, OncoTargets Ther., 2018, 11, 3185–3194 CrossRef PubMed.
- F. Q. Nie, M. Sun, J. S. Yang, M. Xie, T. P. Xu, R. Xia, Y. W. Liu, X. H. Liu, E. B. Zhang, K. H. Lu and Y. Q. Shu, Mol. Cancer Ther., 2015, 14, 268–277 CrossRef CAS PubMed.
- Y. Lu, X. Zhou, L. Xu, C. Rong, C. Shen and W. Bian, OncoTargets Ther., 2016, 9, 3077–3084 CAS.
- L. Chen, X. Han, Z. Hu and L. Chen, Cancer Chemother. Pharmacol., 2019, 83, 921–931 CrossRef CAS PubMed.
- F. M. Davis, T. A. Stewart, E. W. Thompson and G. R. Monteith, Trends Pharmacol. Sci., 2014, 35, 479–488 CrossRef CAS PubMed.
- T. Shibue and R. A. Weinberg, Nat. Rev. Clin. Oncol., 2017, 14, 611–629 CrossRef PubMed.
- J. Roche, R. M. Gemmill and H. A. Drabkin, Cancers, 2017, 9, E72, DOI:10.3390/cancers9070072.
- A. Sasaki, H. Abe, S. Mochizuki, M. Shimoda and Y. Okada, Pathol. Int., 2018 DOI:10.1111/pin.12685.
- T. M. Yoon, S. A. Kim, W. S. Cho, D. H. Lee, J. K. Lee, Y. L. Park, K. H. Lee, J. H. Lee, S. S. Kweon, I. J. Chung, S. C. Lim and Y. E. Joo, BMC Cancer, 2015, 15, 888 CrossRef PubMed.
- R. Sun, B. Jiang, H. Qi, X. Zhang, J. Yang, J. Duan, Y. Li and G. Li, Cell Death Dis., 2015, 6, e1990 CrossRef CAS PubMed.
- D. J. Stewart, J. Natl. Cancer Inst., 2014, 106, djt356 CrossRef PubMed.
- I. W. Achkar, N. Abdulrahman, H. Al-Sulaiti, J. M. Joseph, S. Uddin and F. Mraiche, J. Transl. Med., 2018, 16, 96 CrossRef CAS PubMed.
- F. Yu, G. Pang and G. Zhao, Int. J. Biol. Macromol., 2019, 128, 583–592 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |