Open Access Article
Open Access ArticleRetracted Article: Knockdown of long non-coding RNA OIP5-AS1 suppresses cell proliferation and migration in ox-LDL-induced human vascular smooth muscle cells (hVMSCs) through targeting miR-152-3p/PAPPA axis
Xiangya
Yang†
a,
Zhongrui
Li†
b,
Lei
Zhang
a,
Xiaoshan
Wu
a,
Qixin
Kang
a and
Li
Li
 *b
*b
aDepartment of Cardial Surgery, Luoyang Central Hospital Affiliated to Zhengzhou University, Luoyang, China
bDepartment of Nursing, Luoyang Central Hospital Affiliated to Zhengzhou University, No. 288 Middle Zhongzhou Road, Luoyang 471000, China. E-mail: jladsa@163.com; Tel: +86-379-6389201
First published on 11th October 2019
Abstract
Emerging evidence has demonstrated that long non-coding RNA Opa-interacting protein 5 antisense RNA 1 (OIP5-AS1) is associated with cellular behaviors among malignant tumors. However, the role of OIP5-AS1 in atherosclerosis remains largely undefined. The aim of this study was to explore the expression and role of OIP5-AS1 in a cell model of atherosclerosis, as well as the underlying mechanism. We found that expression of OIP5-AS1 was upregulated in human vascular smooth muscle cells (hVMSCs) under oxidized low density lipoprotein (ox-LDL) administration, and knockdown of OIP5-AS1 suppressed cell viability (CCK-8) and proliferating cell nuclear antigen (PCNA) protein level in ox-LDL-treated hVMSCs, as well as inhibited cell migration rate (wound healing assay) and protein expression of matrix metalloproteinase (MMP)-2 and MMP-9. Mechanically, OIP5-AS1 functioned as competing endogenous RNA (ceRNA) to positively regulate PAPPA expression through sponging miRNA-152-3p (miR-152), and pregnancy-associated plasma protein A (PAPPA) was identified as a downstream target gene for miR-152. Moreover, expression of miR-152 was downregulated and PAPPA was upregulated in ox-LDL-treated hVMSCs. Similarly to OIP5-AS1 knockdown, miR-215 overexpression could inhibit cell proliferation and migration of hVMSCs administrated by ox-LDL, which was abated by PAPPA upregulation. Moreover, miR-215 downregulation partially reversed the suppressive role of OIP5-AS1 knockdown as well. In conclusion, knockdown of OIP5-AS1 suppressed ox-LDL-treated hVMSC proliferation and migration presumably through targeting miR-152/PAPPA axis, suggesting a novel OIP5-AS1/miR-152/PAPPA pathway in atherogenesis.
1. Introduction
Atherosclerosis is the most renowned culprit of varying cardiovascular diseases (CVD) including coronary heart disease, cerebral infarction and peripheral vascular disease.1 The pathogenesis of atherosclerosis is generally attributed to the accumulation of fibrous elements and lipids in large and medium arteries.2 The arterial walls are primarily comprised of endothelial cells and vascular smooth muscle cells (VSMCs). Therefore, the role in these cells has been well-documented in the pathological progression of atherogenesis.3 VSMCs serve as the majority of arterial walls and atherosclerotic plaques, contributing to the development of atherosclerosis. Moreover, functions of VSMCs in turn are mainly affected by atherogenesis.2,4 Since atherosclerosis has been the top cause of death globally nowadays,5 it is highly needed to develop an effective therapeutic treatment. Oxidation of low-density lipoprotein (LDL) cholesterol is a key causal risk for the development of atherosclerosis, and LDL reduction can lower the risk of major CVD events.6,7 Thereby, searching the profound molecular mechanisms in oxidized LDL (ox-LDL)-induced VMSCs is one avenue to treat atherosclerosis.Abnormal microRNAs (miRNAs) expression has been shown to be associated with cardiovascular diseases including atherosclerosis.8 miRNAs are best studied family of endogenous non-coding RNAs (ncRNAs) with 20–25 nucleotides in length. Generally, miRNAs regulate generous protein-coding genes through targeting, and thus inhibiting the translation of messenger RNAs (mRNAs). It has been suggested that miRNAs play a role in VSMCs, including phenotypic regulation, cell behaviors and cell interactions.3,9,10 A miRNA expression signature including miRNA-152-3p (miR-152) was co-expressed in plaque tissue and classical monocytes. Whereas, the expression of miR-152 in VMSCs was undetermined, as well as its cellular role.
Long non-coding RNAs (lncRNAs) are another type of ncRNA longer than 200 nucleotides. Unluckily, lncRNAs are much less well characterized than miRNAs in atherosclerosis to date, even though their importance in gene expression has been increasingly recognized. Recent studies have unraveled that emerging lncRNAs, such as lncRNA MALAT1,11 UCA1 (ref. 12) and H19,13 could control biological functions of endothelial cells, macrophages and VMSCs. lncRNA Opa-interacting protein 5 antisense RNA 1 (OIP5-AS1) is always deregulated in human cancers, and then participates in tumor cell progression including proliferation and migration.14–16 Very recently, OIP5-AS1 has been annotated to be higher expressed in human umbilical vein endothelial cells, and the association between OIP5-AS1 and vascular endothelial cells functions has also been addressed.17,18 Nevertheless, the expression and role of OIP5-AS1 in VMSCs remain to be disclosed.
It is wildly reported that ncRNAs play essential roles in regulating atherosclerosis through various mechanisms. Among them, the lncRNA/miRNA/mRNA network has becoming as a promising approach to understand the pathophysiological processes. Pregnancy-associated plasma protein A (PAPPA) is a large glycoprotein derived from placenta, but interestingly plays a significant role in cardiovascular diseases.19 Together with serum lipids, PAPPA is suggested to be one risk factor of atherosclerosis.20 However, the interaction among OIP5-AS1, miR-152 and PAPPA is left to be illuminated.
In this study, we investigated expression of OIP5-AS1, miR-152 and PAPPA in human VMSCs (hVMSCs) under ox-LDL administration, as well as the effects of OIP5-AS1 and miR-152 on cell proliferation and migration. The interaction among OIP5-AS1, miR-152 and PAPPA was further confirmed in both gene expression and biological role.
2. Materials and methods
2.1. Cell culture
Cell line hVMSCs (hVMSCs; CRL-1999) was acquired from American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in ATCC-formulated F-12K medium complemented with components according to the officially recommended method. The cells were incubated in a humidified incubator with 5% CO2 at 37 °C.2.2. ox-LDL-induced cell model of atherosclerosis
At 80% confluence, hVMSCs were treated with ox-LDL (Kalen Biomedical; Montgomery Village, MD, USA) to simulate the high blood lipid environment. In brief, different concentrations of ox-LDL (25–200 μg mL−1) were added into medium for 24 h, and 100 μg mL−1 of ox-LDL was used to stimulate hVMSCs for 0–72 h before measurement.2.3. Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated from ox-LDL-treated hVMSCs using miRNeasy Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's protocols. The cDNA was reverse-transcribed, and then RT-qPCR analysis was operated by the MiScript SYBR Green PCR Kit (Qiagen) on ABI Prism 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The reactions were performed in quadruplicate for each sample at least three independent runs. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA (U6) acted as the endogenous controls. All primers were described in Table 1. The expression of OIP5-AS1, miR-152 and PAPPA was calculated according to comparative threshold (Ct, 2−ΔΔCt) method with fold changes.| Sequence | |
|---|---|
| a OIP5-AS1, lncRNA Opa-interacting protein 5 antisense RNA 1; miR-152, hsa-miRNA-152-3p; PAPPA, pregnancy-associated plasma protein A; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; U6, U6 small nuclear RNA. | |
| si-OIP5-AS1 | Sense: CACCAAACAGGCUUUGUGUUCCUTA |
| Antisense: UAAGGAACACAAAGCCUGUUUGGUGGU | |
| si-con | Sense: AAUUCUCCGAACGTGTCACGT |
| miR-152 mimic | Sense: TCAGTGCATGACAGAACUUGG |
| miR-con mimic | Sense: CUAAGGCAACGAUCAGCAUUACGA |
| miR-152 inhibitor | Sense: CCAAGUUCUGUCAUGCACUGA- |
| miR-con inhibitor | Sense: GAACGGAGCGAGCAGACCUUU |
| OIP5-AS1 | Forward: TGCGAAGATGGCGGAGTAAG |
| Reverse: TAGTTCCTCTCCTCTGGCCG | |
| hsa-miR-152 | Forward: TCAGTGCATGACAGAACTTGG |
| Reverse: GCGAGCACAGAATTAATACGAC | |
| PAPPA | Forward: ATATCTCACGTGACCGAGGA |
| Reverse: AGATGATGGTGCTGGAAGTC | |
| GAPDH | Forward: AATGGGCAGCCGTTAGGAAA |
| Reverse: TGAAGGGGTCATTGATGGCA | |
| U6 | Forward: CGCTTCGGCAGCACATATACTA |
| Reverse: CGCTTCACGAATTTGCGTGTCA | |
2.4. Cell transfection
VMSCs were seeded into 6-well plate or 24-well plate (Corning, NY, USA) for 24 h. For overexpression, OIP5-AS1 and the coding domain sequence (CDS) of PAPPA (NM_002581.5) were separately cloned and amplified into pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA); miR-152 mimic and its negative control (miR-con mimic) was chemically synthesized by Gene Pharma (Shanghai, China). For silencing, siRNA against OIP5-AS1 (si-OIP5-AS1), miR-152 inhibitor (anti-miR-152) and their negative controls (si-con and anti-miR-con) were from Gene Pharma as well. The sequences of these oligonucleotides were shown in Table 1. Then, these vectors (2 μg) or oligonucleotides (50 nM) were transfected into hVMSCs in the logarithmic growth phase with Lipofectamine 2000 (Thermo Scientific, Waltham, MA, USA). After transfection for 24 h, cells were collected for RT-qPCR analysis or treated with 100 μg mL−1 of ox-LDL for further investigation.2.5. Cell counting kit-8 (CCK-8)
Cell proliferative capacity of VSMCs was analyzed using CCK-8 assay (Dojindo, Kyushu Island, Japan). hVSMCs (5 × 103 cells per well) were seeded in 96-well culture plate (Corning) and treated with 100 μg mL−1 of ox-LDL. At 0 h, 24 h, 48 h and 72 h, CCK-8 reagent (10 μL, 5 g L−1) was added into each well for another 3 h. The reactions were performed in sextuplicate for each group. The optical density (OD) values were measured at 450 nm using microplate reader and recorded in cell growth curve.2.6. Western blotting
Total protein was harvested from ox-LDL-treated hVMSCs at 24 h using RIPA lysis buffer (Thermo Scientific). The extraction (20 μg) was loaded for the standard procedures of western blot assay. The primary antibodies including proliferating cell nuclear antigen (PCNA; #2586, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000), matrix metalloproteinase (MMP)-2 (#13132, 1
2000), matrix metalloproteinase (MMP)-2 (#13132, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and MMP-9 (#3852, 1
1000) and MMP-9 (#3852, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) were purchased from Cell Signaling Technology (CST; Danvers, Massachusetts, USA), and β-actin (CST; #4967, 1
1000) were purchased from Cell Signaling Technology (CST; Danvers, Massachusetts, USA), and β-actin (CST; #4967, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) was the loading control. Blots signals were detected using an enhanced chemiluminescence (ECL) system according to the manufacturer's instructions. Image-Pro Plus (Media Cybernetics, Rockville, MD, USA) was used to quantify the densities of the protein signals. Every sample was performed western blotting in triplicate.
1000) was the loading control. Blots signals were detected using an enhanced chemiluminescence (ECL) system according to the manufacturer's instructions. Image-Pro Plus (Media Cybernetics, Rockville, MD, USA) was used to quantify the densities of the protein signals. Every sample was performed western blotting in triplicate.
2.7. Wound healing assay
To confirm cell migratory ability of hVMSCs with and without ox-LDL treatment, wound healing assay was carried out. Generally, hVMSCs were planted in 6-well plate (Corning). At 90% confluence, the medium was then removed and the monolayer cells were manually scraped using a sterile 200 mL pipette tip. The left hVMSCs were cultivated with complete medium added 100 μg mL−1 of ox-LDL for another 48 h, followed by photographed. The width of the wounding gaps was measured using Image-Pro Plus and migration rate was fold change of migration distance versus original distance.2.8. Luciferase reporter assay
hVMSCs were pre-transferred in 24-well plate (Corning) for 24 h. The fragment of OIP5-AS1 (1500 nucleotides) and 3′ UTR of PAPPA (1000 nucleotides) containing the predicted miR-152 binding site were cloned into pGL4 plasmid (Promega, Madison, Wisconsin, USA) respectively, as well as their mismatched sequence. The mutation of wild type of OIP5-AS1 and PAPPA 3′UTR (OIP5-AS1-WT and PAPPA-WT) was performed by site-directed mutagenesis. 50 nM of miR-152 mimic or miR-con mimic was co-transfected in hVMSCs with 100 ng of plasmid expressing OIP5-AS1-WT, PAPPA-WT or their mutant (OIP5-AS1-MUT and PAPPA-MUT). After transfection for 24 h, luciferase activity was detected using dual-luciferase reporter assay system and GloMax Luminometer (Promega). For each transfection, relative luciferase activity was averaged from three replicates. Relative luciferase activity of pGL4 plasmids was presented as Firefly normalized to Renilla.2.9. RNA immunoprecipitation (RIP)
hVMSCs transfected with miR-152 mimic were collected and lysed with RIP buffer in Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA). Then the cell extract was harvested and incubated with magnetic beads conjugated with human Ago2 or IgG antibody (as negative control) in RIP buffer. After incubation of proteinase K, the immunoprecipitated RNAs including miR-152, OIP5-AS1 and PAPPA were detected by RT-qPCR.2.10. Statistical analysis
The data were presented as mean ± standard error of the mean, and the statistical analysis was performed on Graphpad Prism 5 (GraphPad, San Diego, CA, USA). Two-tailed Student t-test and one-way analysis of variance were performed for comparisons between groups. The significant differences were acknowledged as P value < 0.05. P < 0.01 was very significant, and P < 0.001 was extremely significant.3. Results
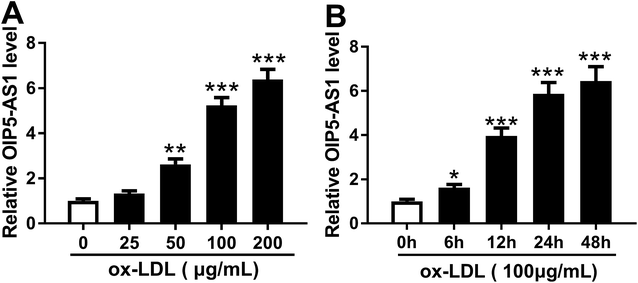
3.1. ox-LDL induced OIP5-AS1 higher expression in hVSMCs in vitro
In order to confirm the expression of OIP5-AS1 in atherosclerosis, we primarily explored its level in cell model of atherosclerosis. hVSMCs were subjected with gradient concentrations of ox-LDL (0–200 μg mL−1), and RT-qPCR analyzed that OIP5-AS1 expression level was gradually increased with ox-LDL treatment (Fig. 1A). Moreover, OIP5-AS1 expression was also significantly upregulated after 100 μg mL−1 of ox-LDL administration for 6–48 h (Fig. 1B). Together, we proposed OIP5-AS1 was highly expressed in hVSMCs induced by ox-LDL in a dose- and time-dependent manner.3.2. Knockdown of OIP5-AS1 inhibited cell proliferation and migration in ox-LDL-treated hVSMCs
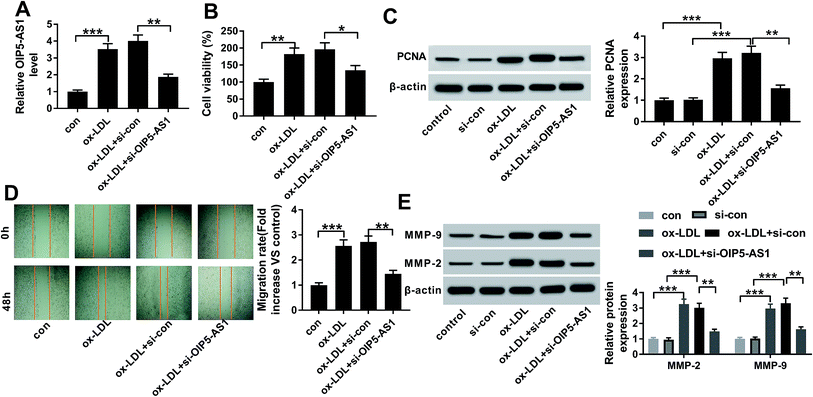
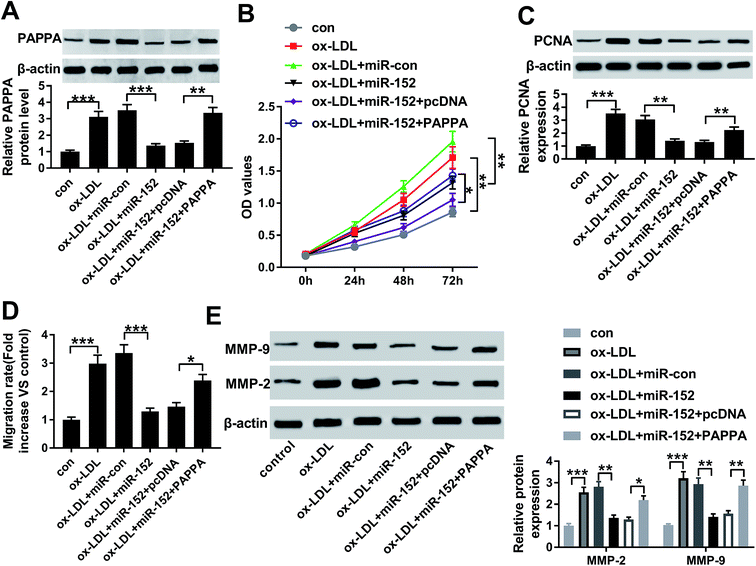
Therefore, we wondered the biological role of OIP5-AS1 dysregulation in hVSMCs. First of all, si-OIP5-AS1 was transfected into hVSMCs to forcedly downregulate OIP5-AS1 expression in ox-LDL-treated hVSMCs. RT-qPCR detected abundantly decreased level of OIP5-AS1 after transfection for 24 h (Fig. 2A), which confirmed a high transfection efficiency. Then, cell proliferation and migration were investigated in vitro. CCK-8 assay monitored the proliferative capacity after 100 μg mL−1 of ox-LDL treatment till 2 days. As a result, ox-LDL could distinctively promote the absorbance value of OD450 in hVSMCs, accompanied with elevated PCNA protein expression according to western blotting (Fig. 2B and C); whereas OIP5-AS1 silencing suppressed the promotion of ox-LDL on cell proliferation of hVSMCs. Wound healing assay demonstrated that migration rate was higher in hVSMCs under ox-LDL administration, which was lowered by si-OIP5-AS1 transfection (Fig. 2D); moreover, ox-LDL-induced upregulation of MMP-2 and MMP-9 was attenuated when OIP5-AS1 was knocked down (Fig. 2E). By the way, si-con transfection affected little on expression of PCNA, MMP-2 and MMP-9; while, PCNA, MMP-2 and MMP-9 levels were significantly induced under ox-LDL treatment with or without si-con transfection (Fig. 2C and E). Collectively, ox-LDL administration could induce hVMSCs cell proliferation and migration, which was suppressed by OIP5-AS1 knockdown.3.3. miR-152 was a target gene of OIP5-AS1
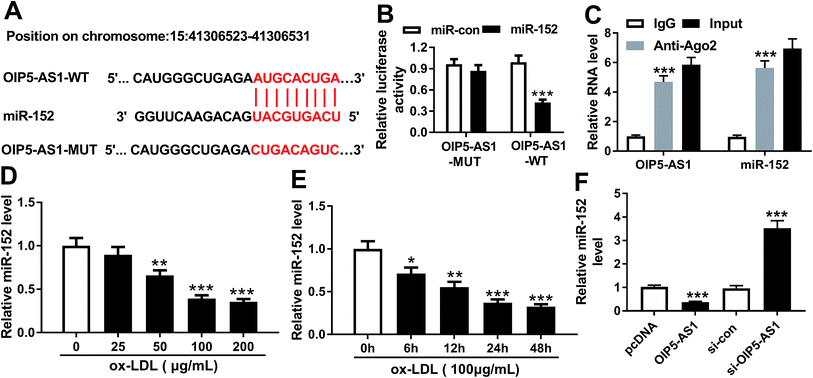
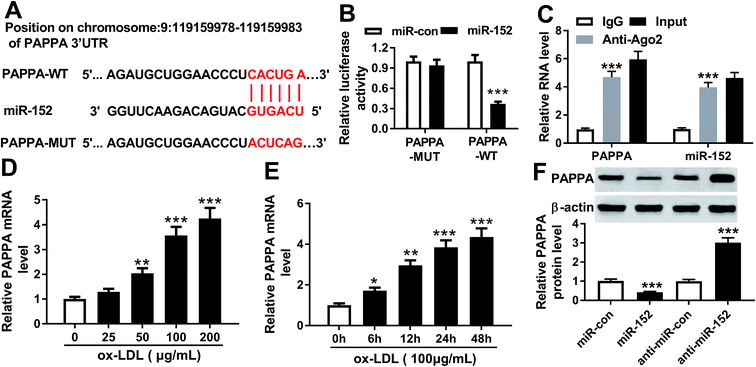
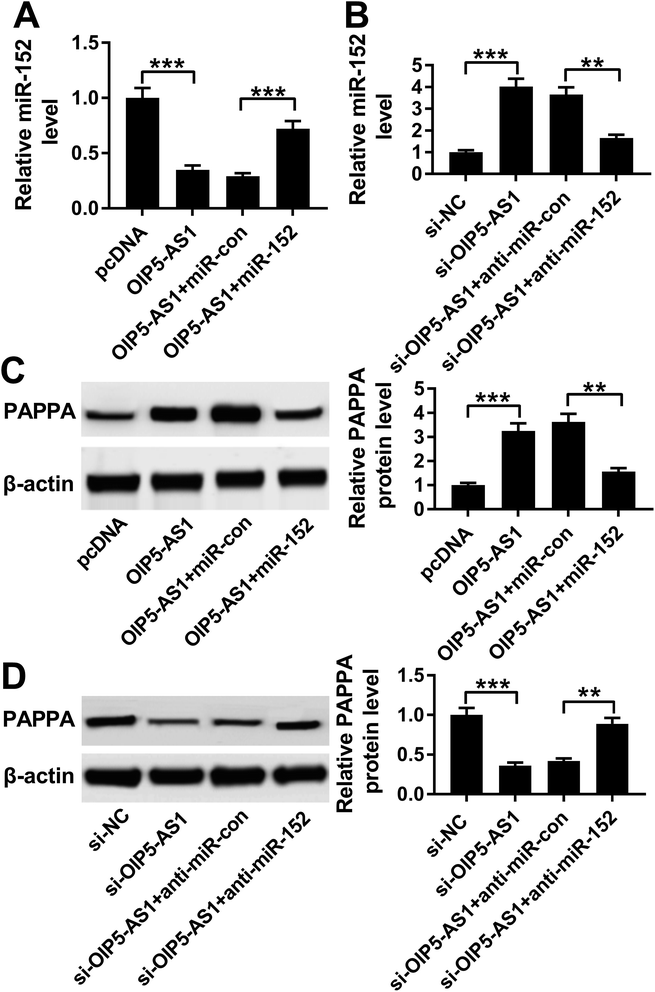
To further explore the in-depth mechanism underlying the anti-proliferation and anti-migration role of OIP5-AS1 knockdown in atherogenesis, we searched the target miRNAs of OIP5-AS1 on LncBase V.2 database (http://diana_tools/lncbasev2-predicted%26miRNAs=ENSG00000247556). In silico data showed a conserved complementary paired site of miR-152 in OIP5-AS1; the wild type of OIP5-AS1 fragment (1500 nucleotides) containing the putative target sites was synthesized and the sequence AUGCACUGA in OIP5-AS1-WT was mutated into CUGACAGUC (Fig. 3A). Luciferase gene reporter analysis and RIP assay were further carried out in hVMSCs to verify this potential target binding. We generated recombinant reporter pGL4 plasmids expressing OIP5-AS1-WT or OIP5-AS1-MUT, and found that luciferase activity of OIP5-AS1-WT was significantly declined when co-transfection with miR-152 mimic (Fig. 3B); meanwhile, there was little difference in OIP5-AS1-MUT groups whether transfected with miR-152 mimic or miR-con mimic. Similarly, expression of OIP5-AS1 and miR-152 was both dramatically enriched in Ago2-RIP instead of IgG-RIP (Fig. 3C). Thus, our data suggested a direct target relationship between OIP5-AS1 and miR-152. On the contrary to OIP5-AS1, miR-152 was downregulated ox-LDL-treated hVMSCs in a certain dose- and time-dependent manner (Fig. 3D and E). Besides, the regulatory effect of OIP5-AS1 on miR-152 was also evaluated. RT-qPCR analysis showed miR-152 level in hVMSCs was extremely lower with pcDNA-OIP5-AS1 transfection, and higher with si-OIP5-AS1 transfection (Fig. 3F). These outcomes indicated miR-152 expression was negatively regulated by OIP5-AS1 in hVMSCs.3.4. Silencing of miR-152 abated the suppressive role of OIP5-AS1 knockdown in ox-LDL-treated hVSMCs
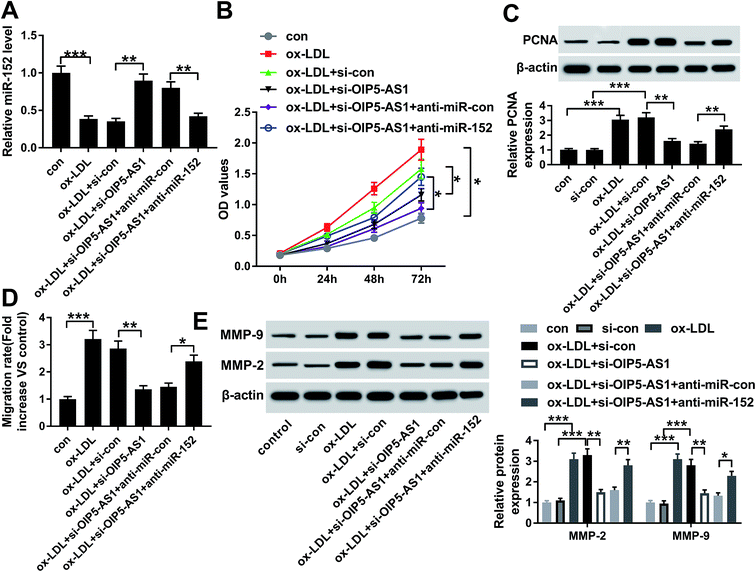
Since the interaction between OIP5-AS1 and miR-152 expression had been confirmed, we intended to further discuss whether miR-152 mediated the role of OIP5-AS1 knockdown in cell model of atherosclerosis. In rescue experiments, ox-LDL-treated hVMSCs were pre-transfected with si-OIP5-AS1 or si-con, and co-transfected with si-OIP5-AS1 and anti-miR-152 or anti-miR-con, and the expression of miR-152 was detected in every group. RT-qPCR analysis depicted si-OIP5-AS1 transfection rescued the inhibition of ox-LDL on miR-152 expression, and this effect was abrogated by co-transfection with anti-miR-152 (Fig. 4A). The suppressive role of OIP5-AS1 knockdown in cell proliferation was partially reversed by miR-152 deletion, as evidenced by improved cell viability and PCNA expression (Fig. 4B and C). Cell migration rate and expression MMP-2/9 were inhibited in hVMSCs transfected with si-OIP5-AS1, which could be mitigated when co-transfection with anti-miR-152 (Fig. 4D and E). These results showed downregulation of miR-152 could abate the suppressive role of OIP5-AS1 knockdown in ox-LDL-induced hVMSCs.3.5. miR-152 negatively regulated PPAPA expression in hVSMCs by target binding
Furthermore, the downstream target genes of miR-152 were sought and identified. Among the putative genes, PAPPA attracted our attention. Online bioinformatics tools (microT-CDS) revealed that there were 1949 targets for hsa-miR-152, and PAPPA 3′UTR showed 8 target sites of miR-152. Besides, the potential binding between miR-152 and PAPPA was also predicted by TargetScan, another database for prediction of miRNA targets. One conserved, potential site of miR-152 in the 3′ UTR of PAPPA at position 1065–1087 (http://dianatools/microT_CDS/hsa-miR-152-3p/threshols=0.57) was further confirmed. As shown in Fig. 5A, the sequence CACUGA was mutated to ACUCAG, which was proposed as the mutant of PAPPA 3′ UTR. Luciferase reporter assay demonstrated that only luciferase activity of vector containing the wild type of PAPPA 3′ UTR fragment (1000 nucleotides) was significantly decreased when miR-152 was overexpressed by transfection (Fig. 5B). The enrichment of miR-152 and PAPPA was obtained by Ago2 antibody, but not IgG antibody (Fig. 5C). Coincident with OIP5-AS1, expression of PAPPA on mRNA level was upregulated in ox-LDL-treated hVMSCs in an extent dose- and time-dependent manner (Fig. 5D and E). The regulatory effect of miR-152 on PAPPA expression was testified by western blotting, and its level was markedly lower in miR-152 mimic-transfected hVMSCs, and higher in anti-miR-152-transfected hVMSCs (Fig. 5F). Collectively, PAPPA expression was inversely regulated by miR-152 via target binding.3.6. Upregulation of PAPPA counteracted the suppressive role of miR-152 overexpression in ox-LDL-treated hVSMCs
The influence of PAPPA dysregulation on biological role of miR-152 was confirmed in cell model of atherosclerosis as well. ox-LDL-treated hVMSCs were pre-transfected with miR-152 mimic alone or together with pcDNA-PAPPA. After ox-LDL administration for 24 h, the expression of PAPPA protein was detected in every group. Western blotting depicted miR-152 mimic transfection inhibited the upregulation of ox-LDL on PAPPA protein expression, and this effect was abrogated by co-transfection with pcDNA-PAPPA (Fig. 6A). Overexpression of miR-152 by transfection impaired cell viability and PCNA expression in hVMSCs, implying anti-proliferation role of miR-152 in ox-LDL-treated hVMSCs (Fig. 6B and C). Consistently, anti-migration activity of miR-152 was supported by declined migration rate and diminished MMP-2/9 expression after miR-152 upregulated (Fig. 6D and E). Notably, the suppressive activities of miR-152 on ox-LDL-treated hVMSCs proliferation and migration were counteracted by pcDNA-PAPPA transfection (Fig. 6B–E). These results showed upregulation of PAPPA could abolish the suppressive role of miR-152 in ox-LDL-induced hVMSCs.3.7. OIP5-AS1 functioned as competing endogenous RNA (ceRNA) for miR-152 to positively modulate PAPPA expression
Considering the target relationship between miR-152 and OIP5-AS1 or PAPPA, we researched the interaction between OIP5-AS1 and PAPPA in hVMSCs. Therefore, hVSMCs were transfected with pcDNA-OIP5-AS1 alone or combined with miR-152 mimic, si-OIP5-AS1 alone or combined with anti-miR-152. OIP5-AS1 overexpression downregulated miR-152 expression, whereas upregulated PAPPA expression, which were both reversed by miR-152 ectopic expression (Fig. 7A and C). Inversely, silencing of OIP5-AS1 induced higher level of miR-152 and lower level of PAPPA, and this regulation was partly overturned by miR-152 deletion (Fig. 7B and D). These data indicated OIP5-AS1 could positively modulate PAPPA expression in hVMSCs through acting as a molecular sponge for miR-152, suggesting a novel OIP5-AS1/miR-152/PAPPA pathway in cell model of atherosclerosis in hVMSCs.4. Discussion
VSMCs are mechanistically implicated in all stages of atherosclerosis, and ncRNAs have been documented to be involved in most cell processes of VMSCs, such as proliferation, apoptosis, autophagy, senescence, migration, and invasion.21–24 Therefore, promising technologies based on epigenetic regulation have been proposed in clinical trials.25 Here, we mainly detected dysregulation of OIP5-AS1, miR-152 and PAPPA in ox-LDL-mediated cell model of atherosclerosis in hVMSCs, as well as their role in cell proliferation and migration.In this present study, we noticed expression of OIP5-AS1 was elevated in ox-LDL-treated hVMSCs in an extent manner of dose and time dependence, which suggested a potential role of OIP5-AS1 dysregulation in VMSCs functions in atherosclerosis. Next, we identified that knockdown of OIP5-AS1 could suppress cell proliferation and migration of hVMSCs with ox-LDL administration as described by decrease of cell viability, migration rate and expression of PCNA, MMP-2 and MMP-9. Therefore, we revealed that OIP5-AS1 might affect VMSCs functions in atherosclerosis. Fortunately, the vital effect of OIP5-AS1 had also been discovered in vascular endothelial cells. Very recently, Wang et al.18 declared higher level of OIP5-AS1 in ox-LDL-mediated cell model of atherosclerosis in human umbilical vein endothelial cells (hUVECs). Silencing of OIP5-AS1 induced G0/G1 arrest, proliferation and apoptosis inhibition in ox-LDL-treated hUVECs. These results together indicate a promising biomarker and target of OIP5-AS1 in atherosclerosis both in endothelial cells and VMSCs. By the way, this may imply that lncRNAs is involved in endothelial-VSMCs interactions in atherosclerosis.3 Besides, the association between OIP5-AS1 and vascular endothelial cells proliferation was also found in hemangioma, which is a benign vascular neoplasm.17
Mechanically, we confirmed OIP5-AS1 functioned as a molecular sponge of miR-152 to regulate PAPPA expression in hVMSCs, as depicted by luciferase reporter assay, RIP and western blotting. Here, expression of miR-152 was detected to be inhibited by ox-LDL and its overexpression by transfection resulted in diminishing cell viability, migration rate and levels of PCNA and MMP-2/9 in ox-LDL-treated hVMSCs. These outcomes suggested that miR-152 upregulation might contribute to eliminate the malignant progression of atherogenesis probably through inhibiting VMSCs proliferation and migration. Furthermore, previous researcher had been demonstrated the functions of miR-152 in cell progression in vascular endothelial cells, and in inflammatory reaction in macrophages in atherogenesis. According to the study from Wu et al.,26 miR-152 exhibited the similar suppressive role in cell proliferation and migration in hUVECs by targeting ADAM17, thus decreasing TNF-α secretion and leading to atherosclerotic protection. They further investigated the involvement of miRNAs in pharmacological effects of atherosclerosis, and observed that, among miRNAs targeting ADAM17, only miR-152 was significantly lower expressed in the serum of atherosclerosis patients than that in normal individuals. This illustrated the potential of circulating miR-152 as biomarker for diagnosis of atherosclerosis. Wang et al.27 noticed reduction of miR-152 level in the aortic tissues of atherosclerotic mice; its upregulation reduced ox-LDL-stimulated high expression of IL-1, IL-6 and TNF-α in RAW264.7 cells, indicating an alleviation of the malignant progression of atherosclerosis. Collectively, miR-152 takes essential part in atherogenesis among endothelial cells, macrophages and VMSCs.
PAPPA is mainly produced by the placental syncytiotrophoblast during pregnancy, as well as VSMCs. Elevated PAPPA levels are dominantly linked to a higher risk of cardiovascular events.19 Circulating PAPPA exerts values of diagnosis and prognosis in patients with CVD including atherosclerosis.20,28 Herein, we discovered higher expression of PAPPA in hVMSCs with ox-LDL stimulation. Functionally, its upregulation could partially eliminate the protective role of miR-152 in ox-LDL-treated hVMSCs proliferation and migration. Moreover, PAPPA was verified to be negatively modulated by OIP5-AS1 via miR-152, suggesting that PAPPA might mediate the effect of OIP5-AS1/miR-152 axis in hVMSCs. It had been reported that PAPPA secreted by VSMCs played a critical role in the development of atherosclerosis through different pathways including miRNAs/PAPPA network. For example, miRNA-490-3p targeting PAPPA inhibited the proliferative capacity of human coronary artery smooth muscle cells induced by ox-LDL.29 Similarly, Zhang et al.30 mentioned that miRNA-141 exhibited a protective effect on VSMC proliferation in response to ox-LDL by directly targeting PAPPA. PAPPA/IGF-binding protein-4 (IGFBP-4)/IGF system participated in cell behaviors including proliferation, migration and inflammation in atherosclerotic VSMCs, endothelial cells and macrophages.19,31 However, there is still limited information on PAPPA-related miRNAs, thus miRNAs/PAPPA network remains largely undisclosed. Meanwhile, the regulatory role of lncRNAs on PAPPA is little uncovered yet. Our research might provide a novel direction in the development and treatment of atherosclerosis through regulating OIP5-AS1/miR-152/PAPPA pathway.
Whereas, there were few limitations of this study. On one hand, we did not explore the expression of OIP5-AS1 and miR-152 in human aortic atherosclerotic lesions in this study, as well as the underlying signaling pathways such as IGF signaling cascade.28,32 On the other hand, the pro-atherosclerosis of OIP5-AS1 should be further confirmed in animal model, such as apolipoprotein E-deficient mice fed a high-fat diet.33 Even that, we confirmed the suppressive activities of OIP5-AS1 knockdown on cell proliferation and migration in ox-LDL-induced cell model of atherosclerosis in hVMSCs depending on OIP5-AS1/miR-152/PAPPA pathway.
5. Conclusion
Collectively, expression of OIP5-AS1 and PAPPA was upregulated and miR-152 was downregulated in cell model of atherosclerosis in hVMSCs induced by ox-LDL. Both OIP5-AS1 knockdown and miR-152 upregulation could inhibit cell proliferation and migration in ox-LDL-induced hVMSCs. Mechanically, miR-152 was confirmed to target OIP5-AS1 and PAPPA. PAPPA expression mediated the expression and role of OIP5-AS1 and miR-215 in the malignant progression of atherogenesis in ox-LDL-treated hVMSCs. Our results provide a novel OIP5-AS1/miR-152/PAPPA pathway in hVMSCs of atherogenesis.Author agreement
All authors have seen and approved the final version of the manuscript and confirm that this manuscript has never been published elsewhere so far.Authors' contribution
Conceptualization and methodology: Zhongrui Li and Lei Zhang; formal analysis and data curation: Xiaoshan Wu and Qixin Kang; validation and investigation: Xiangya Yang and Li Li; writing – original draft preparation and writing – review and editing: Xiangya Yang, Zhongrui Li and Lei Zhang.Conflicts of interest
The authors have no financial conflicts to declare.References
- P. N. Hopkins, Physiol. Rev., 2013, 93, 1317–1542 CrossRef CAS.
- A. J. Lusis, Nature, 2000, 407, 233–241 CrossRef CAS.
- M. Li, M. Qian, K. Kyler and J. Xu, Front. Cardiovasc. Med., 2018, 5, 151, DOI:10.3389/fcvm.2018.00151. eCollection 2018.
- A. C. Doran, N. Meller and C. A. McNamara, Arterioscler., Thromb., Vasc. Biol., 2008, 28, 812–819 CrossRef CAS.
- G. L. Basatemur, H. F. Jorgensen, M. C. H. Clarke, M. R. Bennett and Z. Mallat, Nat. Rev. Cardiol., 2019 DOI:10.1038/s41569-019-0227-9.
- W. Herrington, B. Lacey, P. Sherliker, J. Armitage and S. Lewington, Circ. Res., 2016, 118, 535–546 CrossRef CAS.
- A. J. Kattoor, S. H. Kanuri and J. L. Mehta, Curr. Med. Chem., 2019, 26, 1693–1700 CrossRef CAS.
- Y. Lu, T. Thavarajah, W. Gu, J. Cai and Q. Xu, Arterioscler., Thromb., Vasc. Biol., 2018, 38, e159–e170 CAS.
- L. Li, Y. Li and C. Tang, Cell Biol. Int., 2019, 43, 1102–1112 CrossRef CAS.
- B. N. Davis, A. C. Hilyard, P. H. Nguyen, G. Lagna and A. Hata, J. Biol. Chem., 2009, 284, 3728–3738 CrossRef CAS.
- K. Wang, C. Yang, J. Shi and T. Gao, Eur. J. Pharmacol., 2019, 858, 172338, DOI:10.1016/j.ejphar.2019.04.019.
- X. Hu, R. Ma, W. Fu, C. Zhang and X. Du, J. Cell. Physiol., 2019, 234, 14154–14160 CrossRef CAS.
- L. Zhang, H. Cheng, Y. Yue, S. Li, D. Zhang and R. He, J. Biomed. Sci., 2018, 25, 11, DOI:10.1186/s12929-018-0418-4.
- J. Deng, H. Deng, C. Liu, Y. Liang and S. Wang, Biomed. Pharmacother., 2018, 98, 102–110 CrossRef CAS PubMed.
- N. Yang, J. Chen, H. Zhang, X. Wang, H. Yao, Y. Peng and W. Zhang, Cell Death Dis., 2017, 8, e2975, DOI:10.1038/cddis.2016.145.
- Z. Zhang, F. Liu, F. Yang and Y. Liu, Biomed. Pharmacother., 2018, 101, 14–23 CrossRef CAS.
- J. Zhang, T. Zhao, L. Tian and Y. Li, Front. Pharmacol., 2019, 10, 449, DOI:10.3389/fphar.2019.00449..
- M. Wang, Y. Liu, C. Li, Y. Zhang, X. Zhou and C. Lu, Am. J. Transl. Res., 2019, 11, 1827–1834 Search PubMed.
- L. Consuegra-Sanchez, S. Fredericks and J. C. Kaski, Atherosclerosis, 2009, 203, 346–352 CrossRef CAS.
- J. Woelfle, C. L. Roth, R. Wunsch and T. Reinehr, Eur. J. Endocrinol., 2011, 165, 613–622 CAS.
- N. J. Leeper and L. Maegdefessel, Cardiovasc. Res., 2018, 114, 611–621 CrossRef CAS PubMed.
- Z. Song, D. Wei, Y. Chen, L. Chen, Y. Bian, Y. Shen, J. Chen and Y. Pan, Toxicol. Appl. Pharmacol., 2019, 364, 45–54 CrossRef CAS.
- B. Zhang, Y. Dong and Z. Zhao, Biochem. Biophys. Res. Commun., 2019, 510, 171–176 CrossRef CAS.
- L. Zhang, C. Zhou, Q. Qin, Z. Liu and P. Li, J. Cell. Biochem., 2019, 120, 14670–14678 CrossRef CAS.
- D. Skuratovskaia, M. Vulf, A. Komar, E. Kirienkova and L. Litvinova, Biomolecules, 2019, 9, 226 CrossRef CAS PubMed.
- Y. Wu, A. Huang, T. Li, X. Su, H. Ding, H. Li, X. Qin, L. Hou, Q. Zhao, X. Ge, T. Fang, R. Wang, C. Gao, J. Li and N. Shao, FEBS Lett., 2014, 588, 2063–2069 CrossRef CAS.
- W. Wang, Y. Zhang, L. Wang, J. Li, Y. Li, X. Yang and Y. Wu, Biomed. Pharmacother., 2019, 109, 2409–2414 CrossRef CAS.
- X. H. Yu, L. H. He, J. H. Gao, D. W. Zhang, X. L. Zheng and C. K. Tang, Atherosclerosis, 2018, 278, 250–258 CrossRef CAS.
- Y. Sun, D. Chen, L. Cao, R. Zhang, J. Zhou, H. Chen, Y. Li, M. Li, J. Cao and Z. Wang, Cardiovasc. Res., 2013, 100, 272–279 CrossRef CAS.
- Y. Zhang, B. Chen, L. Ming, H. Qin, L. Zheng, Z. Yue, Z. Cheng, Y. Wang, D. Zhang, C. Liu, W. Bin, Q. Hao, F. Song and B. Ji, Int. J. Clin. Exp. Pathol., 2015, 8, 14401–14408 CAS.
- F. Andreotti, T. Rio and E. Conti, Atherosclerosis, 2009, 203, 353–354 CrossRef CAS.
- K. Shan, Q. Jiang, X. Q. Wang, Y. N. Wang, H. Yang, M. D. Yao, C. Liu, X. M. Li, J. Yao, B. Liu, Y. Y. Zhang, Y. J. and B. Yan, Cell Death Dis., 2016, 7, e2248, DOI:10.1038/cddis.2016.145.
- L. Zhang, H. Cheng, Y. Yue, S. Li, D. Zhang and R. He, Cardiovasc. Pathol., 2018, 33, 6–15 CrossRef CAS PubMed.
Footnote |
| † The two authors contributed to this work equally as co-first authors. |
| This journal is © The Royal Society of Chemistry 2019 |