 Open Access Article
Open Access ArticleMolecular imprinting on PtPd nanoflowers for selective recognition and determination of hydrogen peroxide and glucose†
Caini Fana,
Junjia Liub,
Haiying Zhaoa,
Ling Lia,
Min Liu*a,
Jing Gaob and
Li Ma *b
*b
aDepartment of Hypertension, Henan Provincial People's Hospital, Zheng Zhou, 450003, China. E-mail: liumin136@126.com
bSchool of Chemical Engineering and Technology, Hebei University of Technology, Tianjin 300130, China. E-mail: mali0502@hebut.edu.cn
First published on 21st October 2019
Abstract
PtPd nanoflowers (PtPd NFs) exhibit intrinsic peroxidase-like activity as nanozymes, but the nanozymes lack substrate specificity and have low catalytic activity. Herein, a molecularly imprinted nanogel on PtPd NFs was prepared by using 3,3′,5,5′-tetramethylbenzidine (TMB) as the template through the aqueous precipitation polymerization method. After the TMB was washed out, many substrate binding pockets were retained in the PtPd NFs. Scanning electron microscopy (SEM), transmission electron microscopy (TEM) and powder X-ray diffraction (XRD) were employed to characterize the molecularly imprinted polymer (MIP) PtPd nanoflowers (T-MIP-PtPd NFs). The obtained T-MIP-PtPd NFs exhibited enhanced catalytic activity and specific recognition for TMB. Compared with PtPd NFs, T-MIP-PtPd NFs showed a linear range from 0.01–5000 μM and a detection limit of 0.005 μM toward the detection of H2O2. Glucose can also be sensitively detected through cascade reaction by the T-MIP-PtPd NFs and glucose oxidase. Therefore, molecular imprinting on nanozymes technology shows promising application in biocatalysis and sensing fields.
Introduction
Nanozymes, a nanomaterial with enzyme-like activity, are attracting more and more attention thanks to their outstanding properties.1–3 To date, various nanozymes, including metal–organic frameworks (MOFs),4–7 metal nanoparticles,8 oxidized metal nanoparticles9,10 and some other nanomaterials,11 have been successfully prepared and used as enzyme mimics for catalytic applications.2 As one of the enzyme mimics, Pt nanoparticles have been demonstrated to exhibit versatile catalytic activity like peroxidase, catalase and superoxide dismutase.2,12 Recent studies reported that the Pt-based bimetallic nanoparticles (i.e. PtPd nanodendrites) can exhibit peroxidase-like activity.13,14 For this kind of nanozyme, the creation of mesoporous nanostructures is considered to be an effective method to improve the enzyme mimetic activity.14 With the presence of a mesoporous structure, specific surface area and accessible pores of the Pt-based nanozymes will be increased, and the activity will be improved. Recently, a surfactant-directing method for synthesizing multimetallic spheres with ultra-large mesoporous was reported15,16 and the obtained mesoporous multimetallic spheres exhibited enhanced electrochemical activity thanks to the high surface area and mesoporous structure. Despite the progress made in the synthesis of multi-metal nanospheres, their enzyme mimetic properties are rarely reported and the mechanism remains elusive. In addition, there are still disadvantages for these nanozymes, such as complicated synthesis process, low specificity and low catalytic activity.1 Thus, the synthesis of the PtPd bimetallic nanomaterials with high peroxidase-mimic activity and substrate specificity are still challenges for further investigation.Compared to natural enzymes, nanozymes are stable to denaturation, low in cost, highly resistant to high concentrations of substrates, flexible storage conditions and easy to manipulate catalytic activity. However, most nanozymes have no substrate specificity and can catalyze a lot of substrates leading to the lack of molecular recognition function. Molecularly imprinted polymers (MIPs) could create functional polymer materials with selective recognition properties.17 MIPs are usually prepared by bulk polymerization, photopolymerization, surface grafting polymerization and sol–gel mechanisms to generate imprinting specific cavities.18,19 Thus, combining MIPs with nanozymes provides a new class of nanozymes with improved selectivity and activity.20–22 For example, Zhang et al.23 imprinted substrate binding pockets gels on three different nanozymes and the experimental results showed that the molecular imprinting on nanozymes remarkably increased the selectivity and catalytic activity.
Thus, in this study, PtPd nanoflowers (PtPd NFs) were synthesized by a surfactant-directing method. Then, the MIP layer was created on the surface of PtPd NFs and the obtained nanocomposites were named as T-MIP-PtPd NFs. The structure and composition of PtPd and T-MIP-PtPd were analyzed by scanning electron microscopy (SEM), transmission electron microscope (TEM), energy dispersive X-ray spectroscopy (EDX) and powder X-ray diffraction (XRD). Their intrinsic peroxidase-like activities were investigated. The T-MIP-PtPd NFs was used to detect H2O2 and glucose through a sensitive colorimetric method. To the best of our knowledge, it is the first report regarding the preparation of TMB imprinted PtPd NFs and its application in detecting hydrogen peroxide and glucose.
Materials and methods
Chemicals
Chloroplatinic acid (H2PtCl6) was purchased from Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China). Potassium tetrachloroplatinate(II) (K2PtCl4), sodium tetrachloropalladate(II) (Na2PdCl4), ascorbic acid, 3,3,5,5-tetramethylbenzidine (TMB), 2,2′-azinobis-(3-ethylbenzthiazoline-6-sulphonate) (ABTS), F127 and glucose oxidase (GOD) were purchased from Shanghai Aladdin Bio-Chem Technology Co., Ltd. (Shanghai, China). Glacial acetic acid, anhydrous sodium acetate, and ethanol were purchased from Tianjin Fengchuan Chemical Reagent Technologies Co., Ltd.Preparation of PtPd NFs
Typically, the solutions of H2PtCl6 (20 mM, 1.2 mL), K2PtCl4 (20 mM, 1.8 mL) and Na2PdCl4 (20 mM, 0.6 mL) were mixed uniformity in a 10 mL centrifuge tube. Then F127 (60 mg) and HCl solution (6 M, 60 μL) were added. After F127 was completely dissolved, ascorbic acid (0.1 M, 3 mL) was added with ultrasonic at 40 °C. After 4 hours, the obtained PtPd NFs were centrifuged and washed with Milli-Q water and ethanol, respectively.Molecular imprinting on PtPd NFs
The aqueous precipitation polymerization method is used to prepare TMB imprinted PtPd NFs (T-MIP-PtPd NFs). Typically, PtPd NFs (1 mg mL−1, 0.5 mL) and TMB solution (7.5 mM, 20 μM) were mixed in HEPES buffer solution (20 mM, pH 7.6) that was purged with N2. Acrylamide (2.9 mg, 42 μM), N-isopropylacrylamide (NIPAAm) (4.6 mg, 42 μM), N,N′-methylenebisacrylamide (MBAAm as crosslinker) (2.4 mg, 16 μM) and SDS (0.8 mg) were dissolved in 0.5 mL HEPES buffer solution to prepare a monomer solution. Polymerization of the monomer solutions was initiated by adding ammonium persulfate (0.2 mg) and tetramethyl-ethylenediamine (0.3 μL). The monomer and the PtPd NFs-containing solutions were mixed after 20 min of initiation, and then the mixture was continued to react for 1 h at room temperature. The resulting imprinted gels were collected by centrifugation at 5000 rpm for 5 min. Then 1 mM H2O2 was added to react with the imprinted substrates so they can be easily removed by subsequent washing. As a control experiment, non-imprinted PtPd NFs nanozymes (NIP-PtPd NFs) were also prepared in the same way except that no substrate template was added.Determination of peroxidase-like activity of PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs
Typically, 10 μL (0.1 mg mL−1) nanozymes (PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs) and 50 μL of substrate (0.375 mM, TMB or ABTS) were added to 0.93 mL HAc–NaAc buffer solution (0.05 M, pH 4.0) containing 10 μL H2O2 (0.2 M). The reaction mixture was incubated at 25 °C for 10 min, and the peroxidase-like activity was investigated by detecting the absorbance (652 nm for TMB and 420 nm for ABTS). To measure the steady-state kinetics, various concentrations of substrates (TMB and ABTS) were used, respectively. For colorimetric detection, 0.1 mg mL−1 of nanozymes (PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs) and 0.1 mM of H2O2 were added to 9.3 mL acetate buffer solution (0.1 M, pH 4) containing substrates (TMB and ABTS) with various concentrations for 10 min at 25 °C. The kinetic parameters were calculated by the Michaelis–Menten equation (eqn (1)):
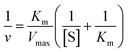 | (1) |
Detection of H2O2 and glucose
The detection of H2O2 by a colorimetric assay was carried out as follows: 10 μL of nanozyme (0.1 mg mL−1) (PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs), 50 μL of TMB (7.5 mM), and 10 μL of H2O2 (0.2 M) were added into 930 μL of HAc–NaAc buffer solution (0.05 M, pH 4.0). Afterward, the absorbance was measured at 652 nm after 15 min of incubation at room temperature. For comparison, ABTS (50 μL 7.5 mM) was also used as colorimetric substrate to detect H2O2, and the absorbance was measured at 420 nm after 15 min of incubation at room temperature.PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs were also used to detect glucose through catalyzing the production of H2O2 from the oxidation of glucose by glucose oxidase. In a typical process, 300 μL of PBS (phosphate buffer solution, pH 7.0) containing a serials concentration of glucose and 100 μL of PBS (pH 7.0) containing 1 mg mL−1 glucose oxidase were mixed at 37 °C for 30 min. Then 540 μL of HAc–NaAc buffer solution, 50 μL of TMB (0.35 mM) and 10 μL of (0.1 mg mL−1) nanozymes (PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs) were added to the above mixture, and incubated at 25 °C for 10 min to oxidize the reaction solution from colorless to blue.
Results and discussion
Characterization of T-MIP-PtPd NFs
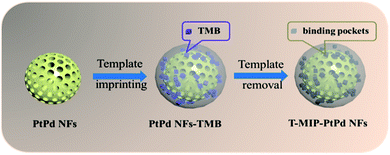
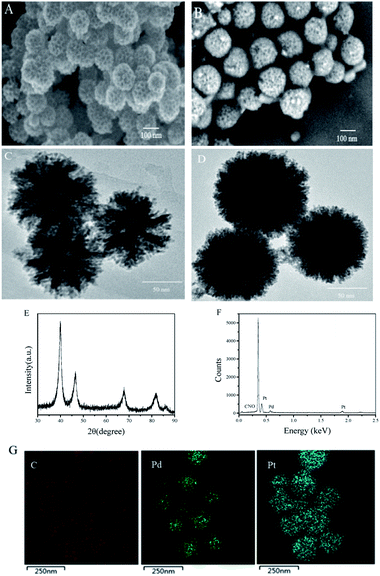
As shown in Fig. 1, PtPd NFs were synthesized firstly and then the monomers including acrylamide, NIPAAm and MBAAm were mixed with PtPd NFs to perform precipitation polymerization. TMB was used as a template for imprinting. After the TMB imprinted polymer layer was formed on the PtPd NFs surface, the TMB molecules were washed away, leaving behind the binding pockets that can selectively rebinding TMB. As shown in Fig. 2A and C, SEM and TEM images of PtPd NFs indicated a spherical nanostructure with flower-like morphology and mesoporous on the surface. The particle size of PtPd NFs was approximately 120 nm (Fig. 2A and C). Compared to PtPd NFs, T-MIP-PtPd NFs had a bigger size due to the polymer coating on the surface of PtPd NFs (Fig. 2B and D). XRD pattern of T-MIP-PtPd NFs showed four strong diffraction peaks at 39.7°, 46.4°, 67.5° and 81.2°, which can be assigned to the (111), (200), (220) and (311) crystalline planes of the standard pattern of Pt and Pd, respectively16,24 (Fig. 2E). EDX analysis of the T-MIP-PtPd NFs showed obvious Pt, Pd, C, N and O peaks (Fig. 2F). Moreover, elemental mapping images of T-MIP-PtPd NFs (Fig. 2G) showed that C, Pt and Pd element were homogeneously distributed through the T-MIP-PtPd NFs structure. The content of C element is much lower than Pt and Pd, and the boundary between C and Pt (and Pd) is unclear, indicating the polymer layer coating on the PtPd NFs is thin.Peroxidase-like activity of T-MIP-PtPd NFs
The peroxidase activity of PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs was investigated at 25 °C using TMB as the substrate (Fig. S1†). Besides, similar reaction was also carried out using ABTS as reductant (Fig. S2†). These results confirmed that PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs were peroxidase mimicking nanozyme (Fig. 3A and B). The mechanism for the oxidation of TMB may be as follows: firstly, the d-band of platinum will become nearly bare when palladium atoms combine with platinum atoms. Then, the lone electron pairs of H2O2 will fill the d-band. After that, H2O2 cleave into –OH, and –OH can oxidize TMB to TMB+.25 Although both TMB and ABTS can be oxidized by T-MIP-PtPd NFs, it showed poor catalytic activity for ABTS. | ||
| Fig. 3 Photographs with color change showing the activity and specificity of T-MIP-PtPd NFs for oxidation of (A) TMB and (B) ABTS with H2O2. | ||
Imprinting enhanced the activity of nanozyme
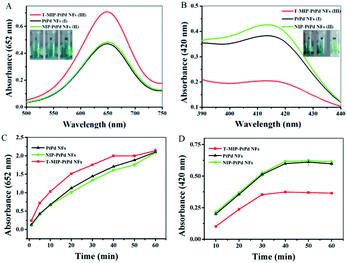
To compare the catalytic activity of PtPd NFs, NIP-PtPd NFs and T-MIP-PtPd NFs, the value changes in absorption spectra during the catalytic process was measured using UV-vis spectrophotometer. When PtPd NFs was used as catalyst, the absorbance at 652 nm reached 0.47 after reaction for 10 min (Fig. 4A, black trace). In contrast, when T-MIP-PtPd NFs were used as catalyst, the absorbance reached 0.71 (Fig. 4A, red trace), indicating the enhanced activity. According to these results, T-MIP-PtPd NFs could enhance the catalytic activity for TMB oxidation. The improved catalytic activity can be attributed to the TMB pockets, which imprinted in the PtPd NFs and had selective recognition for TMB molecules (Fig. 4C). Moreover, the increased local substrate concentration after MIP may also enhance the catalytic rate according to the mass action law.26 The system of NIP-PtPd NFs with TMB and H2O2 exhibited similar absorption value to PtPd NFs system (Fig. 4A, green trace), indicating similar catalytic activity of NIP-PtPd NFs and PtPd NFs. Because the polymer layer around the PtPd NFs was thin, the polymer layer might have no obvious influence on the activity. In contrast, when ABTS was used as another substrate to investigate the selectivity of T-MIP-PtPd NFs, the catalytic activity of T-MIP-PtPd NFs for ABTS was lower than that of PtPd NFs and NIP-PtPd NFs (Fig. 4B and D). Therefore, after TMB imprinting, the selectivity of T-MIP-PtPd NFs was improved for TMB oxidation and inhibited for ABTS oxidation. In order to achieve an optimal colorimetric effect, the effect of pH, temperature, reaction time, H2O2 concentration, and quantity of T-MIP-PtPd NFs on relative activity were investigated (Fig. S4†). The highest absorbance was reached when 0.1 mg mL−1 of T-MIP-PtPd NFs, 0.35 mM TMB, and 2.0 M H2O2 were used. Therefore, the above condition was selected in the following experiments.Kinetic analysis
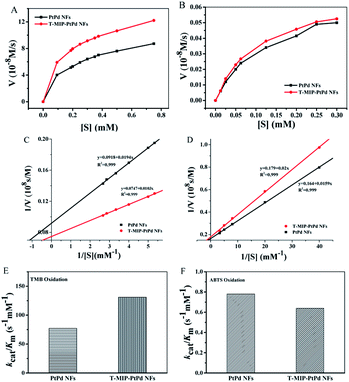
In order to investigate the mechanism of peroxidase-mimetic activity, the reaction kinetics of PtPd NFs and T-MIP-PtPd NFs were measured (Fig. 5A and B). Reaction rates with PtPd NFs and T-MIP-PtPd NFs as catalysts were tested by varying the concentration of TMB while fixing H2O2 concentration. For comparison, Km of free HRP was also determined. The Km value for PtPd NFs and T-MIP-PtPd NFs with TMB as substrate was 0.211 mM and 0.137 mM, respectively (Fig. 5C and D). The Km value for T-MIP-PtPd NFs is much lower than those of PtPd NFs and free HRP (Table 1), indicating that T-MIP-PtPd NFs had stronger affinity to TMB than PtPd NFs and free HRP. However, the Km for T-MIP-PtPd NFs with ABTS as substrate was higher than that of PtPd NFs, implying that the imprinting process only offer accessible site for TMB and then enhance substrate specificity.Using TMB as substrate, the catalytic efficiency (kcat/Km) of PtPd NFs and T-MIP nanozymes were also investigated. Compared to PtPd NFs, the kcat value of the T-MIP-PtPd NFs increased by 0.2-fold, while the Km value dropped by 10% for ABTS oxidation. We plotted kcat/Km of T-MIP-PtPd NFs for TMB and ABTS oxidation to compare PtPd NFs (Fig. 5E and F). The catalytic efficiency of T-MIP-PtPd NFs showed 0.7-fold higher than that of PtPd NFs for oxidizing TMB, but 0.15-fold lower for oxidizing ABTS. The imprinting in PtPd NFs can selectively enrich TMB substrate near the T-MIP-PtPd NFs and then result in enhanced catalytic activity.
Detection of H2O2
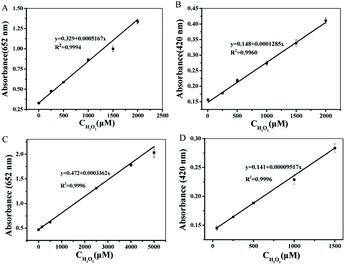
Under the optimized experimental conditions (see ESI†), a series concentration of H2O2 were detected based on the peroxidase-like activity of T-MIP-PtPd NFs through colorimetric method. When TMB was used as substrate, the absorbance of oxidized TMB is linearly correlated to concentration of H2O2 from 0.05–2000 μM (limit of detection (LOD) was 0.02 μM) catalyzed by PtPd NFs (Fig. 6A). The absorbance ranged from 0.01–5000 μM (LOD was 0.005 μM) catalyzed by T-MIP-PtPd NFs. This result proved that molecular imprinted nanozyme enhances its activity thanks to the pockets of TMB molecule (Fig. 6C). When ABTS was used as the substrate, the linear range between the absorbance and concentration of H2O2 was ranged from 0.05–2000 μM by PtPd NFs (Fig. 6B). For T-MIP-PtPd NFs, the linear range for H2O2 sensing was ranged from 0.01–1500 μM (Fig. 6D), indicating that the special pockets of TMB will hinder the approach of ABTS molecule. It can be seen from Table 2 that detection of H2O2 based on the T-MIP-PtPd NFs exhibited good performance, which further confirmed that the molecular imprinted method can enhance the catalytic activity.Detection of glucose
Through combining T-MIP-PtPd NFs and GOx, we further developed a colorimetric method for the determination of glucose. Glucose can be catalyzed by GOx to generate gluconic acid and H2O2. In the presence of H2O2, TMB can be oxidized through the catalytic action of peroxidase-like nanozymes. The combination of two catalytic reactions was used to detect glucose through colorimetric method. With the increasing concentrations of glucose, the absorbance will increase from 0.05 to 1500 μM and a LOD of 0.023 μM for PtPd NFs was obtained (Fig. 7A). The T-MIP-PtPd NFs exhibits a linear detection range from 0.02 to 4000 μM and a LOD of 0.017 μM (Fig. 7B). Compared with the reported glucose sensors, T-MIP-PtPd NFs showed better catalytic performance for detecting glucose owing to the molecularly imprinted process (Table 3). This efficient detection performance was mainly ascribed to: (i) the T-MIP-PtPd NFs had excellent selectivity to TMB; (ii) the T-MIP-PtPd NFs provided an optimal microenvironment to stabilize GOx.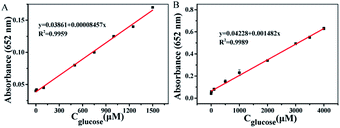 | ||
| Fig. 7 Linear calibration plot for glucose with different concentration of glucose in absorbance of 652 nm with (A) PtPd NFs and (B) T-MIP-PtPd NFs. | ||
To evaluate the selectivity of the established method for glucose detection, the influence of lactose, galactose, fructose, and arabinose were investigated (Fig. S5†). All above interferences showed negligible effect on glucose detection. This phenomenon demonstrates that the reaction system is highly selective for glucose. The reproducibility of the T-MIP-PtPd NFs was investigated by analyzing 0.6 mM glucose for six times in parallel. The relative standard deviation (RSD) was 4.8%, which indicates that the testing results had acceptable reproducibility. To verify the feasibility of the as-prepared colorimetric method, glucose concentration in human serum samples were measured by the standard addition method (ESI†). All the obtained results were listed in Table S1.† It was found that the colorimetric method has a favorable recovery of 92.3–98.7% to detect glucose, which indicates that T-MIP-PtPd NFs, TMB and H2O2 system could be reliable for glucose detection in the biological environment.
Conclusions
In summary, molecular imprinting PtPd nanoflowers were successfully fabricated. The integration of PtPd nanozyme and molecular imprinting can enhance the peroxidase like activity and specificity of T-MIP-PtPd NFs. The obtained T-MIP-PtPd NFs exhibit better catalytic efficiency than that of PtPd NFs and HRP. Based on T-MIP-PtPd NFs, colorimetric sensor was developed for detecting hydrogen peroxidase and glucose. The T-MIP-PtPd showed better catalytic performance as peroxidase mimetic in the optical detection of hydrogen peroxidase and glucose. Thanks to these benefits, T-MIP-PtPd NFs provides a facile, low-cost, sensitive and selective method for colorimetric detection of H2O2 and glucose molecules. With good enzyme-like activity and specificity, the method of combining molecular imprinting with nanozyme can be extended to fabricate other kinds of nanozymes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Science and technology projects in Henan province (182102311235), the National Natural Science Foundation of China (No. 21576068, 21276060 and 21878068), the Natural Science Foundation of Hebei Province, China (B2017202056), the Program for Top 100 Innovative Talents in Colleges and Universities of Hebei Province, China (SLRC2017029) and Hebei High level personnel of support program, China (A2016002027).Notes and references
- D. Jiang, D. Ni and Z. T. Rosenkrans, et al., Chem. Soc. Rev., 2019, 48, 3683–3704 RSC.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- S. Jin, C. Wu and Z. Ye, et al., Sens. Actuators, B, 2019, 283, 18–34 CrossRef CAS.
- F. Cui, Q. Deng and L. Sun, RSC Adv., 2015, 5, 98215–98221 RSC.
- Y. Zhang, C. Dai and W. Liu, et al., Microchim. Acta, 2019, 186, 340–349 CrossRef.
- W. Xu, L. Jiao and H. Yan, et al., ACS Appl. Mater. Interfaces, 2019, 25, 22096–22101 CrossRef.
- Z. Zhao, T. Lin and W. Liu, et al., Spectrochim. Acta, Part B, 2019, 219, 240–247 CrossRef CAS.
- X. Liu, X. Wang and C. Qi, et al., Appl. Surf. Sci., 2019, 479, 532–539 CrossRef CAS.
- X. Cheng, L. Huang and X. Yang, et al., J. Colloid Interface Sci., 2019, 535, 425–435 CrossRef CAS.
- X. Cao and N. Wang, Analyst, 2011, 136, 4241–4246 RSC.
- X. Wang, Y. Hu and H. Wei, Inorg. Chem. Front., 2016, 3, 41–60 RSC.
- T. Yu, W. Wang and J. Chen, et al., J. Phys. Chem. C, 2012, 116, 10516–10521 CrossRef CAS.
- X. Chen, B. Su and Z. Cai, et al., Sens. Actuators, B, 2014, 201, 286–292 CrossRef CAS.
- S. Ge, W. Liu and H. Liu, et al., Biosens. Bioelectron., 2015, 71, 456–462 CrossRef CAS.
- B. Jiang, C. Li and M. Imura, et al., Adv. Sci., 2015, 2, 1500112 CrossRef.
- L. Ma, L. Zhou and Y. He, et al., Electroanalysis, 2018, 30, 1801–1810 CrossRef.
- M. Dinc, C. Esen and B. Mizaikoff, TrAC, Trends Anal. Chem., 2019, 114, 202–217 CrossRef CAS.
- H. Zhang, Adv. Mater., 2019, e1806328 CrossRef.
- M. Zarejousheghani, W. Lorenz and P. Vanninen, et al., Polymers, 2019, 11, 888–905 CrossRef CAS.
- W. Zhao, H. Yang and S. Xu, et al., Colloids Surf., A, 2018, 555, 95–102 CrossRef CAS.
- T. Kajisa and T. Sakata, ACS Appl. Mater. Interfaces, 2018, 10, 34983–34990 CrossRef CAS.
- D. Duan, H. Yang and Y. Ding, et al., Electrochim. Acta, 2018, 261, 160–166 CrossRef CAS.
- Z. Zhang, X. Zhang and B. Liu, et al., J. Am. Chem. Soc., 2017, 139, 5412–5419 CrossRef CAS.
- L. Wang and Y. Yamauchi, J. Am. Chem. Soc., 2013, 135, 16762–16765 CrossRef CAS PubMed.
- R. Mandal, A. Baranwal and A. Srivastava, et al., Biosens. Bioelectron., 2018, 117, 546–561 CrossRef CAS.
- Z. Zhang, Y. Li and X. Zhang, et al., Nanoscale, 2019, 11, 4854–4863 RSC.
- V. K. Singh, P. K. Yadav and S. Chandra, et al., J. Mater. Chem. B, 2018, 6, 5256–5268 RSC.
- N. Lu, M. Zhang and L. Ding, et al., Nanoscale, 2017, 9, 4508–4515 RSC.
- J. Liu and Y. Lu, Angew. Chem., Int. Ed., 2005, 45, 90–94 CrossRef.
- W. Zhang, D. Ma and J. Du, Talanta, 2014, 120, 362–367 CrossRef CAS.
- Y. Jv, B. Li and R. Cao, Chem. Commun., 2010, 46, 8017–8019 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Color evolution of TMB and ABTS in different reaction systems, optimization of experimental parameters, the selectivity of glucose detection. See DOI: 10.1039/c9ra05677g |
| This journal is © The Royal Society of Chemistry 2019 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000