Open Access Article
Open Access ArticleMicrobial diversity and metaproteomic analysis of activated sludge responses to naphthalene and anthracene exposure†
Shanshan Li b,
Shaoda Huc,
Sanyuan Shia,
Lu Rena,
Wei Yan
b,
Shaoda Huc,
Sanyuan Shia,
Lu Rena,
Wei Yan b and
Huabing Zhao*a
b and
Huabing Zhao*a
aKey Laboratory of Industrial Fermentation Microbiology, Ministry of Education, Tianjin Key Laboratory of Industrial Microbiology, National Engineering Laboratory for Industrial Enzymes, College of Biotechnology, Tianjin University of Science & Technology, Tianjin 300457, China. E-mail: zhaohuabing@tust.edu.cn; Tel: +86-22-80235816
bDepartment of Environmental Science & Engineering, Xi'an Jiaotong University, Xi'an 710049, China
cTianjin Key Laboratory for Biomarkers of Occupational and Environmental Hazard, Logistics University of Chinese People's Armed Police Forces, Tianjin 300309, China
First published on 24th July 2019
Abstract
The activated sludge process can effectively remove polycyclic aromatic hydrocarbons (PAHs) from wastewater via biodegradation. However, the degradable microorganisms and functional enzymes involved in this process remain unclear. In this study, we successfully employed a laboratory-scale sequential batch reactor to investigate variations in microbial community and protein expression in response to the addition of different PAHs and process time. The analysis of bacterial community structure by 454 pyrosequencing of the 16S rRNA gene indicated that bacteria from Burkholderiales order were dominant in PAHs treated sludge. Mass spectrometry performed with 2D protein profiles of all sludge samples demonstrated that most proteins exhibiting differential expression profiles during the process were derived from Burkholderiales populations; these proteins are involved in DNA replication, fatty acid and glucose metabolism, stress response, protein synthesis, and aromatic hydrocarbon metabolism. Nevertheless, the protein expression profiles indicated that naphthalene, but not anthracene, can induce the expression of PAH-degrading proteins and accelerate its elimination from sludge. Though only naphthalene and anthracene were added into our experimental groups, the differentially expressed enzymes involved in other PAHs (especially biphenyl) metabolism were also detected. This study provides apparent evidence linking the metabolic activities of Burkholderiales populations with the degradation of PAHs in activated sludge processes. Overall, our findings highlighted the successful application of metaproteomics integrated with microbial diversity analysis by high-throughput sequencing technique on the analysis of environmental samples, which could provide a convenience to monitor the changes in proteins expression profiles and their correlation with microbial diversity.
1. Introduction
Activated sludge processes, which are widely applied in biological wastewater treatment processes, are considered a potential and alternative method for the removal of contaminants from wastewater because of their prominent advantages including high efficiency and stability, less secondary pollution, and low cost. The microorganisms in activated sludge can transform or decompose pollutants in wastewater via their metabolic activities. There is an increasing interest in elucidating microbial communities and functions in activated sludge in order to improve degradation range and efficiency. However, the composition of activated sludge is comparatively complex; it contains not only microbial communities with biodegrading activity, but also adsorbed inorganic and organic pollutants from sewage and endogenous respiration residues, which could bring adverse impact on the conventional microbial isolation methods. High-throughput sequencing technology based on 16S rRNA genes has been successfully providing more extensive information regarding the microbial composition in environmental samples; however, the metabolic functions of these environmental samples remain unclear.With the increasing efforts to detect differential protein expression profiles in mixed cultures, transcriptomic and proteomic analysis techniques have been applied in environmental samples including activated sludge. Proteomics employs liquid chromatography (LC) coupled with tandem mass-spectrometry (MS), which can reveal protein expression profiles and provide comprehensive information regarding the metabolic and physiological activities of microbial communities. Wilmes et al. first applied proteomics technique to dephosphorized activated sludge to detect the amount of protein expressed.1 Subsequently, proteomics studies have been carried out with other environmental samples such as soil samples from an estuary,2 heavy metal contaminated freshwater samples,3 and contaminated soil and groundwater.4 Lacerda et al. revealed differences in protein expression of activated sludge prior to and post exposure to the heavy metal cadmium by proteomic analysis.5 In a study on acidic mineral biofilms, Ram et al. identified a large number of proteins using proteomics techniques, one of which is novel and considered to be a key component related to energy conservation in the environment.6 Despite limited reports, proteomics approaches have highlighted the enormous potential of insight into microecological population function.7 However, these studies mentioned above are limited to the proteomics and microbial community structure and macroscopic characteristics of activated sludge in different treatment processes and few studies have examined activated sludge profiles under the stress of certain types of pollutants.
Complex pollutants, including polycyclic aromatic hydrocarbons (PAH), are present in wastewater; sixteen PAHs are listed as preferentially controlled environmental pollutants by the US Environmental Protection Agency (USEPA) due to their broad toxic effects on living organisms. Currently, microbial processes are considered as effective techniques for PAH removal. Some researchers have investigated the removal and fate of PAHs in activated sludge treatment systems.8,9 Trably et al. confirmed high potential of aerobic microorganisms to degrade a wide range of PAHs at trace levels in continuous sludge bioreactors, and the lower bioavailability of PAHs was one of the key restrictive factor on their biodegradation.10 Knightes and Peters found that the PAHs removal efficiency in activated sludge processes is approximately 67% under aerobic conditions.11 Lower molecular weight (2-, 3-, and 4-ring) PAHs are more rapidly degraded than 5- and 6-ring PAHs.12 A laboratory-scale aerobic activated sludge reactor (AASR) system was constructed by Sponza et al. to investigate its removal ability of PAHs. The results indicated that more than 95% of total PAHs (about 55 ng mL−1) were removed by the AASR system, at 25 days SRT.13 However, there are few investigations on the effect of PAHs on the microbial community structure and protein expression profiles in the activated sludge during the biotreatment processes. Some studies illustrated the change of microbial community in the PAHs-contaminated soils. Applying 454 pyrosequencing of the 16S rRNA gene, Laura et al. studied if diverse remediation process would affect the bacterial community structure in anthracene-contaminated soils. The results suggested that the application of the non-ionic surfactant had a strong effect on the bacterial community structure in soils.14 However, the expression profiles of proteins related to xenobiotic degradation and their assignments to microbial taxonomy remain unclear.
The removal and fate of PAHs during activated sludge treatment process have been reported over the years.15–18 However, there is still a space to discover how the addition of PAHs affect the profiles of microbial community structure and the functional protein expression. Here, we utilized microbial diversity analysis by high-throughput sequencing techniques and proteomics to study changes in microbial communities and related proteins expression in activated sludge over time under combined naphthalene and anthracene stress in order to explore the dominant species and PAH-degradation pathway in activated sludge systems.
2. Materials and methods
2.1 Materials
The activated sludge used in this study was obtained from an aeration tank of an industrial wastewater treatment plant in Tianjin Lingang Industrial Zone. The sludge was collected in a 10 L sterile plastic drum, sealed, and sent to the laboratory within 5 h. Following washing and sieving, the activated sludge was stored at −80 °C until further use.Artificial simulated wastewater was comprised of 300 mM glucose, 48 mM sodium acetate, 26.3 mM Na2HPO4, 114.6 mM NH4Cl, 180 mM MgSO4, 10.6 mM CaCl, 125 mM NH4HCO3, and 0.38 mL L−1 trace element solution (containing 3 mM HBO4, 0.5 mM ZnCl2, 0.1 mM CuCl2, 2 mM CoCl2, 0.3 mM MnCl2, 0.3 mM Na2MoO4, and 0.2 mM NiSO4).
2.2 Laboratory-scale sequencing batch reactor (SBR)
Four individual laboratory-scale plexiglass SBRs (volume 8.0 L) were operated with no addition (marked as C group) or the addition of naphthalene (TN group), anthracene (TA group), or naphthalene + anthracene (TC group) in the influent artificial simulated wastewater (Table 1). Briefly, the reactors were closed with tight Teflon-coated caps. Diffusers and magnetic stirrers coated with Teflon were used to aerate and mix the content of the reactors. Air was passed via columns containing activated carbon to supply oxygen to the reactors. The reactors were operated under the following conditions: the initial MLSS in each reactor was controlled at 2.5 ± 0.2 g L−1, dissolved oxygen was controlled at 2–4 mg L−1, and the hydraulic retention time of the influent (artificial simulated wastewater) was 24 h, pH was 6.8–7.2, and the operating temperature was 20 °C.| Time point | 30 days | 90 days | ||||||
|---|---|---|---|---|---|---|---|---|
| a NAPinf: influent naphthalene concentration; ANTinf: influent anthracene concentration. | ||||||||
| Group | C-30 | TN-30 | TA-30 | TC-30 | C-90 | TN-90 | TA-90 | TC-90 |
| PAHs addition | None | Naphthalene | Anthracene | Naphthalene + anthracene | None | Naphthalene | Anthracene | Naphthalene + anthracene |
| NAPinf (mg L−1) | 0 | 50 | 0 | 25 | 0 | 50 | 0 | 25 |
| ANTinf (mg L−1) | 0 | 0 | 50 | 25 | 0 | 0 | 50 | 25 |
At specific points (cultivated for 0, 30 and 90 days), the sampled activated sludge was centrifuged at 2500 × g for 5 min at 4 °C. After discarding the supernatant, the sediments were stored at −80 °C until use.
2.3 Chemical analysis and efficiency calculation
Residual PAHs from sludge pellets and effluent samples were extracted with equivoluminal n-hexane for 20 min, and analyzed on a gas chromatograph (6890, Agilent, USA) coupled with a mass selective detector (5973, Agilent, USA) as previously described.19 Mixed liquor suspended solids (MLSS) was measured as the standard methods.20 The results were presented as mean ± standard deviation (n = 3).The fate of PAHs in a certain period was calculated based on the following mass balance:
| M-PAHbiode = M-PAHinf − M-PAHeff − M-PAHsludge | (1) |
| EPAH% = M-PAHeff/M-PAHinf × 100% | (2) |
| E-BIOPAH% = M-PAHbiode/M-PAHinf × 100% | (3) |
2.4 Microbial community diversity analysis
Total DNA was extracted from the sample, as previously described.21 Following purification, the DNA samples were sent to Biomarker Technologies (Biomarker Technologies Co, LTD, China) for high-throughput sequencing based on the Illumina platform using the following sequencing primers: 338F: 5′-ACTCCTACGGGAGGCAGCA-3′; 806R: 5′-GGACTACHVGGGTWTCTAAT-3′. Triplicate sequencings were generated for each sludge sample. The triplicate pyrosequencing sequences were analyzed using the QIIME (Quantitative Insights into Microbial Ecology) Pipeline.8 Low-quality sequences (<25) and sequences < 200 bp were removed. Sequences with ≥97% similarity were clustered and defined as an operational taxonomic unit (OTU) in order to generate rarefaction curves and calculate the richness and diversity indices. The composition and abundance distribution of the microorganisms in each sample can be performed at the phylum, class, and genus level based on the OTU classification results.2.5 Proteomics
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min at 4 °C, the protein sediment was washed once with methanol and acetone and then stored at −80 °C until used. Protein concentration in the samples was determined using the Bradford method with bovine serum albumin as the standard.23
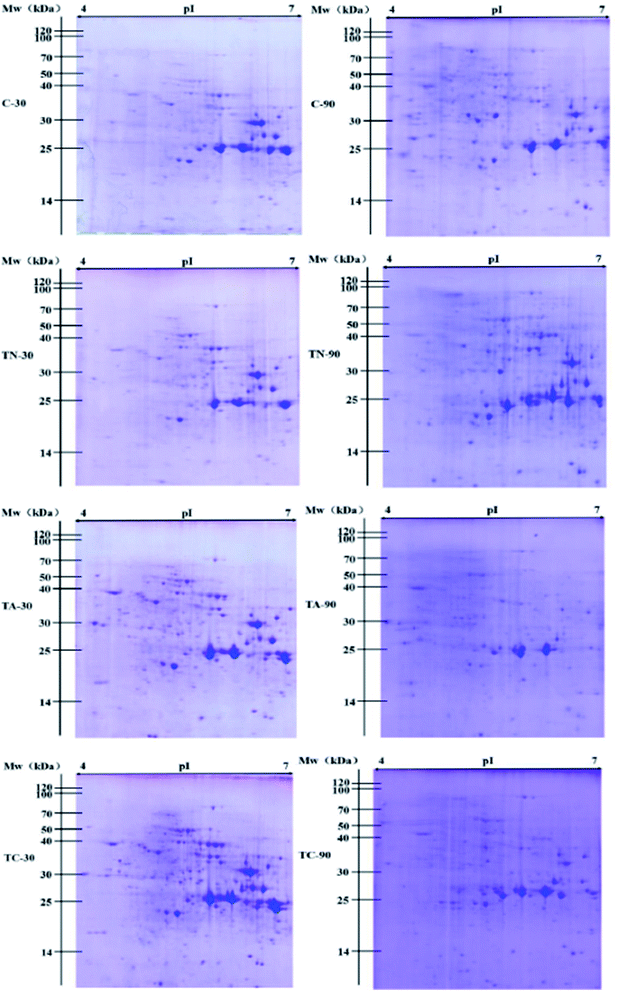
000g for 20 min at 4 °C, the protein sediment was washed once with methanol and acetone and then stored at −80 °C until used. Protein concentration in the samples was determined using the Bradford method with bovine serum albumin as the standard.23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 V h for 6 h. After balancing the strips, a second dimension separation was performed in a 12.5% polyacrylamide at 5 W for 30 min; 17 W to bromophenol blue labeling end at the bottom edge of the glass plate.24 The gel was stained overnight with Coomassie Brilliant Blue R-250 and visualized. Triplicate 2D-PAGE separations were generated for each sludge sample.
000 V h for 6 h. After balancing the strips, a second dimension separation was performed in a 12.5% polyacrylamide at 5 W for 30 min; 17 W to bromophenol blue labeling end at the bottom edge of the glass plate.24 The gel was stained overnight with Coomassie Brilliant Blue R-250 and visualized. Triplicate 2D-PAGE separations were generated for each sludge sample.The obtained gel images were processed and analyzed using PDQuest software (Bio-Rad, Hercules, CA, USA). The gel images were cropped to the same size, automated and manual spot detection and matching were performed, and spot densities were determined for quantification. If the spots densities in the treatment group changed by more than 1.5 folds than the corresponding one in the control group, the protein had been considered differentially expressed.
The data given in all tables are the mean of triplicate measurement with standard deviations (SD) while the data given in figures are the mean of triplicate measurements.
3. Results and discussion
3.1 Performance of SBR reactor
Among different PAHs, naphthalene and anthracene are used as signature compounds for the detection of PAHs contamination. Due to the hydrophobicity and potential bioaccumulation, they are commonly considered as model compounds in the study of bioavailability and degradation rate of PAHs in environment. In this study, four SBR reactors were operated for PAH removal by activated sludge process feeding with naphthalene and/or anthracene. To monitor the performance of SBR reactor, the removal efficiencies of PAHs in the effluent were determined at specific time point (Fig. S1†). The removal efficiencies of naphthalene and anthracene in the effluent of each group were 38–78% and 37–39% respectively at 10 day, and decreased during 10–30 day, then increased until day 70. The results indicated that adsorption may play a dominant role in first 30 days' period, and most cell division had been inhibited by the PAH addition resulting in the weak biodegradation. With the adaption and increasing activity of PAH-degradable microbes, the microbial degradation of PAHs was improving after 30 day. Thus, the sludge samples from each reactor respectively at 30 day and 90 day were used for the furthermore analysis including MLSS, biodegradation efficiency (Table 2), microbial community and proteomics analysis.| Parameter | C-30 | TN-30 | TA-30 | TC-30 | C-90 | TN-90 | TA-90 | TC-90 |
|---|---|---|---|---|---|---|---|---|
| a NAPeff: average effluent naphthalene concentration; ANTeff: average effluent anthracene concentration; NAPsludge: naphthalene in sludge; ANTsludge: anthracene in sludge; ENAP: naphthalene removal efficiency; EANT: anthracene removal efficiency; E-BIONAP: biodegradation efficiency of naphthalene; E-BIOANT: biodegradation efficiency of anthracene.b n = 3, mean ± SD. | ||||||||
| MLSS (g L−1) | 2.3 ± 0.1 | 2.6 ± 0.2 | 2.4 ± 0.1 | 2.4 ± 0.2 | 2.2 ± 0.1 | 3.2 ± 0.3 | 2.5 ± 0.1 | 2.5 ± 0.1 |
| NAPeff (mg L−1) | — | 23.5 ± 0.5 | — | 18.8 ± 0.7 | — | 5.4 ± 0.2 | — | 15.5 ± 0.4 |
| NAPsludge (mg L−1) | — | 38.6 ± 1.5 | — | 25.8 ± 1.0 | — | 23.4 ± 0.7 | — | 12.3 ± 0.2 |
| ENAP (%) | — | 53.0 ± 0.8 | — | 24.8 ± 0.2 | — | 89.2 ± 1.9 | — | 38.2 ± 1.0 |
| E-BIONAP (%) | — | 50.4 ± 0.5 | — | 21.4 ± 0.6 | — | 88.7 ± 1.4 | — | 37.4 ± 0.9 |
| ANTeff (mg L−1) | — | — | 40.3 ± 1.2 | 20.7 ± 1.0 | — | — | 32.2 ± 0.8 | 18.6 ± 1.1 |
| ANTsludge (mg L−1) | — | — | 45.3 ± 2.1 | 34.9 ± 0.8 | — | — | 62.8 ± 1.6 | 52.4 ± 1.7 |
| EANT (%) | — | — | 19.4 ± 0.8 | 17.2 ± 0.4 | — | — | 35.6 ± 0.5 | 25.4 ± 0.9 |
| E-BIOANT (%) | — | — | 16.4 ± 0.3 | 12.5 ± 0.2 | — | — | 34.2 ± 0.6 | 23.3 ± 0.6 |
The potential fates of PAHs in the activated sludge system mainly include being adsorbed in the sludge particles, being degraded by sludge microorganisms, and discharged with the final effluent.13 In this study, the biodegradation rates of PAHs were only a bit lower than the overall PAHs removal efficiency, which indicated that adsorption contributed a little to the PAHs elimination, and most PAHs had been degraded by microbes. For example, naphthalene removal efficiency in TA-30, TC-30, TN-90 and TC-90 was 53.0%, 24.8%, 89.2% and 38.2%, was only 2.6%, 3.4%, 0.5% and 0.8% higher than the corresponding biodegradation efficiency, respectively. These results suggested that the PAHs were commonly removed by biodegradation in aerobic sludge reactors, which was consistent with the previous reports. Sponza et al. investigated the removal efficiencies of 15 PAHs in an aerobic completely stirred tank reactor, and confirmed that adsorption was not an important mechanism controlling the fate of PAHs, and 94% of the PAHs were removed via biodegradation.19
Moreover, the activated sludge process exhibited preferable performance in the removal of naphthalene than anthracene. The removal efficiency of naphthalene (TN group) increased from 53.0% at 30 day to 89.2% at 90 day. However, low level of anthracene removal efficiency (19.4%) was observed in TA group when culturing for 30 days, and it increased to 35.6% at 90 day. When feeding with both naphthalene and anthracene (TC), the biodegradation efficiency of naphthalene decreased drastically compared with that of the naphthalene group (TA). The decreasing naphthalene-biodegrading efficiency in TC group suggested the potential toxicity of anthracene to the microorganisms resulting in the inactivation of degrading microbes in the sludge.
3.2 Microbial structure changes in activated sludge under naphthalene and strontium stress
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 804 raw sequence reads were generated using the high-throughput pyrosequencing technique. After trimming, length, and quality control, 365
804 raw sequence reads were generated using the high-throughput pyrosequencing technique. After trimming, length, and quality control, 365![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 269 (44.07%) of the sequences were used in the analysis, indicating that sufficient sequences were obtained for further analysis.
269 (44.07%) of the sequences were used in the analysis, indicating that sufficient sequences were obtained for further analysis.Based on a similarity threshold of 97%, the 365![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 269 sequences were clustered into 738 different OTUs using the RDP classifier (Table 3). Alpha diversity indices which reflect the community diversity in a sample was calculated by Mothur (version v.1.30), and Shannon index taking into account both community richness and evenness was discussed as the dominant alpha diversity indices.26 The TC-30 sample had the highest number of OTUs, while the C-90 sample exhibited the fewest OTUs. When the same PAH was fed in the influent, the number of OTUs and Shannon diversity index of each group tended to decrease with time, indicating that the PAHs are toxic to microorganisms in activated sludge. Microorganisms that could not adapt to the PAHs in sewage were gradually eliminated; thus, microbial diversity in biosolids decreases with continuous PAH stress. Furthermore, molecular microbial analysis using phylogenetic profiling tools revealed significant shifts in microbial community in response to feeding time in the different PAH exposure groups. The number of OTUs in the C-90 and TN-90 groups (with no addition of PAH and naphthalene in the influent, respectively) was only 33 and 14 lower than in the C-30 and TN-30 groups, respectively. The OTU numbers of the TA-90 and TC-90 groups, fed with anthracene and naphthalene + anthracene, respectively, were reduced by 150 and 114, respectively, compared with the TA-30 and TC-30 groups. Similar profile shifts were observed for the alpha diversity index. For example, the Shannon index decreased from 3.88 and 4.09 in TA-30 and TC-30 to 3.74 and 3.60 in TA-90 and TC-90 group, respectively. These diverse profile changes in alpha diversity index in the experimental groups with or without anthracene may be due to the higher degree of toxicity of anthracene. Anthracene is a tricyclic aromatic compound less able to be utilized and degraded by microorganisms compared with two-ring naphthalene, resulting in a drastic reduction in microbial community population and diversity in the presence of anthracene.27
269 sequences were clustered into 738 different OTUs using the RDP classifier (Table 3). Alpha diversity indices which reflect the community diversity in a sample was calculated by Mothur (version v.1.30), and Shannon index taking into account both community richness and evenness was discussed as the dominant alpha diversity indices.26 The TC-30 sample had the highest number of OTUs, while the C-90 sample exhibited the fewest OTUs. When the same PAH was fed in the influent, the number of OTUs and Shannon diversity index of each group tended to decrease with time, indicating that the PAHs are toxic to microorganisms in activated sludge. Microorganisms that could not adapt to the PAHs in sewage were gradually eliminated; thus, microbial diversity in biosolids decreases with continuous PAH stress. Furthermore, molecular microbial analysis using phylogenetic profiling tools revealed significant shifts in microbial community in response to feeding time in the different PAH exposure groups. The number of OTUs in the C-90 and TN-90 groups (with no addition of PAH and naphthalene in the influent, respectively) was only 33 and 14 lower than in the C-30 and TN-30 groups, respectively. The OTU numbers of the TA-90 and TC-90 groups, fed with anthracene and naphthalene + anthracene, respectively, were reduced by 150 and 114, respectively, compared with the TA-30 and TC-30 groups. Similar profile shifts were observed for the alpha diversity index. For example, the Shannon index decreased from 3.88 and 4.09 in TA-30 and TC-30 to 3.74 and 3.60 in TA-90 and TC-90 group, respectively. These diverse profile changes in alpha diversity index in the experimental groups with or without anthracene may be due to the higher degree of toxicity of anthracene. Anthracene is a tricyclic aromatic compound less able to be utilized and degraded by microorganisms compared with two-ring naphthalene, resulting in a drastic reduction in microbial community population and diversity in the presence of anthracene.27
| Sample ID | Clean reads | OTU number | ACE | Chao1 | Shannon | Simpson |
|---|---|---|---|---|---|---|
| C-30 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 260 |
505 | 550.55 | 561.72 | 3.54 | 0.13 |
| TN-30 | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 719 719 |
576 | 607.96 | 611.66 | 3.95 | 0.05 |
| TA-30 | 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 652 652 |
604 | 618.92 | 631.77 | 3.88 | 0.06 |
| TC-30 | 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 134 134 |
627 | 637.75 | 647.31 | 4.09 | 0.04 |
| C-90 | 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 060 060 |
471 | 516.87 | 520.58 | 3.61 | 0.10 |
| TN-90 | 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 991 991 |
562 | 607.85 | 615.20 | 3.83 | 0.07 |
| TA-90 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 887 887 |
454 | 524.84 | 521.91 | 3.74 | 0.04 |
| TC-90 | 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 566 566 |
513 | 566.00 | 587.17 | 3.60 | 0.11 |
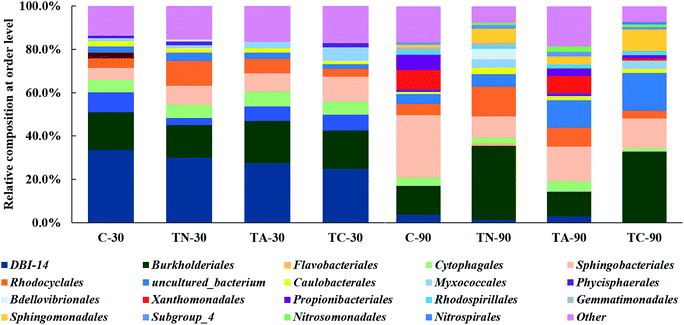
High-throughput sequencing results were assigned at the order level using the RDP classifier (Fig. 1). Following feeding with PAHs for 30 days, the biosolids samples were dominated by DB1-14 and the order Burkholderiales. When feeding was prolonged to 90 days, the dominant microorganisms shifted to Burkholderiales and Sphingobacteriales. These data indicate that the microbial community compositions of activated sludge in the different groups were constantly changing in response to PAH feeding time. The relative abundance of the strains with effective PAH-degrading activity may increase, while microorganisms that cannot adapt were gradually eliminated. For example, in the naphthalene + anthracene group, the dominant DB1-14 populations decreased from 29.91% in TC-30 to <1% when treated for 90 days, indicating the lower adaptability of DB1-14 populations to PAHs especially naphthalene and anthracene.
The relative abundance of the Burkholderiales population increased in response to naphthalene feeding time in the presence or absence of anthracene, from 34.16 and 32.83% in the TN-90 and TC-90 groups to 15.12 and 17.81% in the TC-30 and TN-30 group, respectively. In contrast, the relative abundance of Burkholderiales in the control and anthracene feeding group decreased by 4.15 and 8.06%, respectively at 90 day compared with 30 day. These results indicate that Burkholderiales may possess higher degrading activity for naphthalene than for anthracene.
There have been abundant reports of PAH-degrading bacteria belonging to Burkholderiales order, which is wide distribution in the soil environment around the world.28–30 Liu et al. observed that Betaproteobacteria populations predominated in aerobic activated sludge from an oilfield sewage treatment plant; the relative abundance of Burkholderiales within the Betaproteobacteria was as high as 25%. Furthermore, all the 41 PAH-ring hydroxylating dioxygenase (PAH-RHD) genes retrieved in the experiment were related (95–98%) to those of Burkholderiales order, indicating its dominant role in full-scale process of PAH degradation.31 The PAH-RHD gene that encodes the enzyme responsible for the incorporation of molecular oxygen into the aromatic nucleus during the initial step of PAH metabolism, plays a crucial role in PAH biodegradation in polluted sites.32 The abundance and diversity of PAH-RHD genes have been investigated in various PAH polluted sites.21 PAH degradation-related genes are widely distributed in bacteria belonging to over 80 classes in the order Burkholderiales, including genes involved in the catechol ortho-cleavage, protocatechuic acid ortho-cleavage, homogentisic acid cleavage pathway, and phenylacetyl coenzyme A cleavage pathways. All these crucial genes involved in PAH metabolism could provide Burkholderiales populations with strong and extensive PAH biodegradability in diverse pathways.33
The Sphingomonadales order increased to various levels (1.2–23.49%) over time in all treatment groups. The increase pattern of Sphingomonadales populations indicated their potential PAH-(especially naphthalene and anthracene) degrading activity. Sphingomonadales species have been isolated and studied in recent years for their potential removal of PAH, especially for individual physiological characteristics and highly efficient metabolic activity.34–37 However, current research remains mainly focused on the isolation and identification of Sphingomonadales populations and the related research on the PAH degradation pathway, degrading enzyme system, and associated encoding genes in Sphingomonadales remains limited.
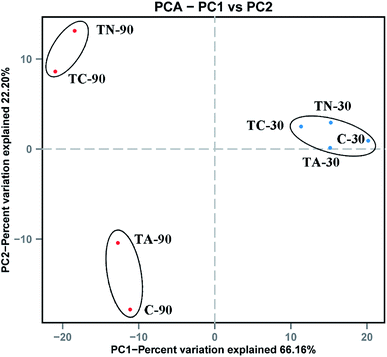 | ||
| Fig. 2 Principal Coordinate Analysis (PCoA) of sludge microbial community. Blue point denote the samples cultured for 30 day, red point denote the samples cultured for 90 day. | ||
The microbial composition diversity among the PAH-fed activated sludge samples detected in this study is consistent with previous biosolids surveys that demonstrated a greater variability in plant-to-plant PAH indicator concentrations. Burkholderiales populations with PAH degradation ability predominated in all experimental groups in this study, while the abundance of the order Sphingomonadales increased in the PAH feeding groups, which may suggest better adaptation and degradation to PAH. The increase in the relative abundance of these two orders has enhanced naphthalene and anthracene degradation ability of activated sludge.
3.3 Metaproteomic analysis of PAH-fed sludge
| Group | Total number of proteins detected | Proteins with typical functions | |||
|---|---|---|---|---|---|
| Carbohydrate metabolism | Protein synthesis | Lipid metabolism | PAH metabolism | ||
| a The results were the average value of triplicate tests. | |||||
| C-30 | 122 | 12 | 17 | 3 | 10 |
| TN-30 | 129 | 14 | 20 | 2 | 11 |
| TA-30 | 157 | 14 | 18 | 3 | 9 |
| TC-30 | 163 | 13 | 15 | 2 | 10 |
| C-90 | 135 | 11 | 16 | 2 | 9 |
| TN-90 | 141 | 15 | 16 | 2 | 13 |
| TA-90 | 63 | 8 | 14 | 1 | 6 |
| TC-90 | 83 | 9 | 13 | 1 | 7 |
| Function | Description | Accession number | Assigned order | Groups | |||||
|---|---|---|---|---|---|---|---|---|---|
| TN-30 | TA-30 | TC-30 | TN-90 | TA-90 | TC-90 | ||||
| Protein synthesis | Elongation factor Tu (EF-Tu) | A2SLF9 | Burkholderiales | 1.16 | 1.50 | 1.88 | 1.15 | −1.22 | — |
| Elongation factor Tu (EF-Tu) | Q46WC7 | Burkholderiales | 1.42 | 1.63 | 1.84 | 1.35 | — | — | |
| Elongation factor Tu (EF-Tu) | B3EU83 | Bacteroidetes | — | — | — | 1.11 | — | — | |
| Diaminopimelate epimerase (DapF) | Q0S1N7 | Actinomycetales | — | — | — | 3.14 | 1.05 | −1.10 | |
| Chorismate synthase (CS) | Q3MFM3 | Nostocales | — | — | — | 1.59 | — | −1.81 | |
| ATP-dependent zinc metalloprotease (FtsH) | C5CES8 | Asterales | 1.06 | 1.53 | 1.34 | 1.34 | — | — | |
| Alanyl-tRNA synthetase (AlaRS) | A8YTJ0 | Actinomycetales | — | — | — | 1.24 | — | — | |
| 50S ribosomal protein | P0A5W1 | Actinomycetales | 1.56 | 1.68 | 1.20 | −1.70 | −3.29 | −1.43 | |
| DNA replication | Chromosomal replication initiator protein (DnaA) | A6W3V4 | Actinomycetales | — | — | — | 1.84 | — | −2.03 |
| Energy metabolism | ATP synthase subunit beta | A1TJ41 | Burkholderiales | — | — | — | −1.75 | −3.40 | −1.12 |
| ATP synthase subunit alpha | A7HIX9 | Myxococcales | 1.41 | 1.18 | 2.21 | −1.16 | — | −1.05 | |
| Carbohydrate metabolism | Isocitrate dehydrogenase kinase/phosphatase (IDH kinase) | Q8Y2Q7 | Burkholderiales | — | — | — | −1.30 | — | — |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | P54226 | Actinomycetales | — | — | — | −1.53 | −7.81 | −3.52 | |
| Enolase | C5CSV6 | Burkholderiales | −1.84 | 1.05 | 1.26 | ||||
| 2-Dehydro-3-deoxyphosphooctonate aldolase (KDO 8-P synthase) | A1K7F7 | Rhodocyclales | — | — | — | −1.05 | — | −1.24 | |
| Stress response | Chaperone protein DnaK | A0L4Z2 | Magnetococcales | — | — | — | −1.16 | — | — |
| 60 kDa chaperonin (GroEL) | A2SCV1 | Burkholderiales | 1.68 | 1.87 | 1.43 | — | — | — | |
| PAH metabolism | Naphthalene 1,2-dioxygenase subunit alpha (NahA3) | P0A110 | Pseudomonadales | — | — | — | 9.12 | — | — |
| 2-Dihydroxy-1,2-dihydronaphthalene dehydrogenase (NagB) | O52384 | Burkholderiales | — | — | — | 8.43 | — | — | |
| Biphenyl 2,3-dioxygenase (BphA1) | Q52438 | Pseudomonadales | 3.08 | 2.93 | 3.35 | 4.22 | −1.12 | −1.17 | |
| 2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (BphD) | Q2VLB9 | Burkholderiales | 3.21 | 1.59 | 1.78 | 1.26 | — | −2.85 | |
| 4-Hydroxy-2-oxovalerate aldolase 1 (BphF) | Q9S152 | Burkholderiales | 1.60 | 1.74 | 1.25 | 1.40 | — | −1.10 | |
| 4-Hydroxy-2-oxovalerate aldolase 1 (BphF) | P51018 | Pseudomonadales | 1.60 | 1.74 | 1.25 | 1.40 | — | −1.10 | |
| Acetaldehyde dehydrogenase (BphG) | Q2VLC6 | Burkholderiales | 1.77 | 1.66 | 1.52 | 1.38 | −4.28 | −1.13 | |
The other interesting consequence which should be highlighted was that more than 40% of up- or down-regulated proteins had been identified associated with Burkholderiales, which was consistent with the result of microbial diversity analysis that the bacteria from Burkholderiales order with PAH degradation ability were dominant in all experimental groups in this study. Therefore, the predominance of Burkholderiales population in the sludge at the stress of naphthalene or (and) anthracene had been confirmed at both microbial diversity and protein level. These large amount of Burkholderiales like strains may have increased the functional significance in the metabolism of PAHs in the activated sludge.
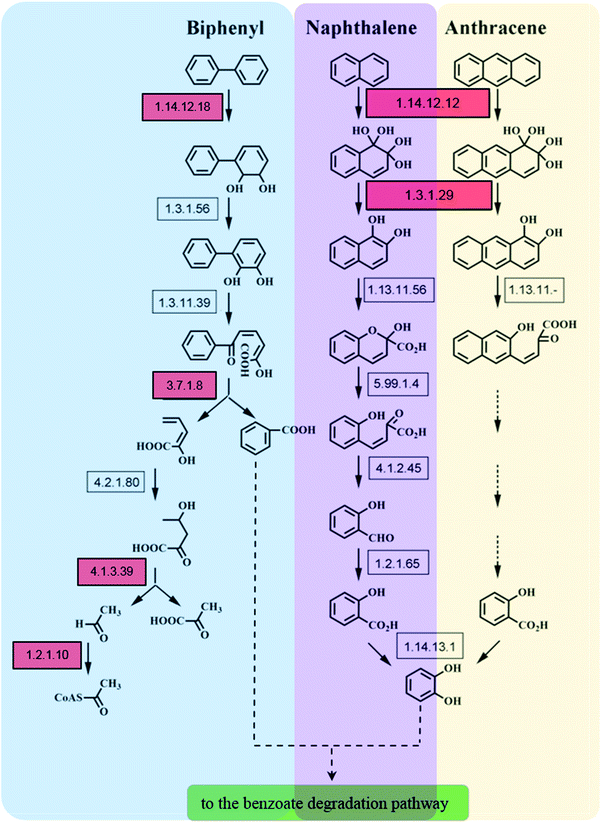
Microorganisms can transform biphenyl into acetyl-CoA, which subsequently undergoes complete degradation upon entering the TCA cycle. Biphenyl 2,3-dioxygenase (BDO, EC 1.14.12.18), which catalyzes the initial step of biphenyl oxidation to form 2,3-dihydrodihydroxybiphenyl, consists of four subunits: BphA1, BphA2, BphA3, and BphA4. The intermediate is further dehydrogenated to generate 2,3-dihydroxybiphenyl by catalysis of 2,3-dihydrobiphenyl-2,3-diol dehydrogenase (BphB, EC 1.3.1.56). This is further catalyzed by 2,3-dihydrodihydroxybiphenyl 1,2-dioxygenase (BphC, EC 1.13.11.39) to form 2-hydroxy-6-keto-6-phenyl-2,4-hexadienoic acid (HOPDA). Finally, HOPDA is catalytically converted to benzoic acid and 2-hydroxy-2,4-pentadienoate by the hydrolase BphD (EC 3.7.1.8), which is sequentially degraded by BphE, BphF, and BphG, eventually entering the TCA cycle for final metabolism.39,40
Though only naphthalene and anthracene were added into our experimental group, the enzymes involved in other PAHs metabolism were also detected. The expression levels of BphA1, BphD, BphF, and BphG related to biphenyl degradation were up-regulated following the addition of PAH (either naphthalene, anthracene, or both) in the sludge for 30 days. Following PAH exposure for 90 days, these four enzymes remained up-regulated in the naphthalene treatment group (TN-90), while the expression levels of the biphenyl degradation-related enzymes were significantly down-regulated in the TA-90 and TC-90 groups. The identification of enzymes associated with biphenyl degradation suggests that the addition of naphthalene and anthracene had induced the expression of enzyme responsible for other PAHs. Furthermore, the major differences in protein expression indicate that the biphenyl degradation-related enzymes may play a role in the degradation of bicyclic aromatic hydrocarbons (naphthalene), but not in anthracene degradation, resulting in the cumulative toxicity effect of anthracene increasing over time. Barriault et al. investigated the functionality of BDO components of Comamonas testosteroni B-356 in naphthalene 1,2-dioxygenase (N1,2-DO, EC 1.14.12.12) of Pseudomonas putida G7. Data showed that there was an efficient electron transfer between the BDO reductase and the N1,2-DO ferredoxin, the BDO ferredoxin could resubstitute N1,2-DO ferredoxin in part (22%), which indicated that BDO might participate in naphthalene degradation.41 Furthermore, Yu et al. investigated the substrate range of E. coli co-expressing the bphA3A4A1A2 genes from Sphingobium yanoikuyae B1, the results demonstrated that recombinant E. coli strain could oxidize both biphenyl and naphthalene to their respective cis-2,3- and 1,2-dihydrodiols. All the above reports supported our proposal that the enzymes in the biphenyl degradation pathway especially for BphA may play a role in the degradation of other PAHs including naphthalene.42 Thus, the addition of naphthalene may result in the differential expression of BphA in this study. Moreover, expression of BphA1 was significantly up-regulated in the TN-90 group, but decreased in both the TA-90 and TC-90 groups. It was further confirmed that anthracene exposure inhibited enzymes related to biphenyl degradation. The cumulative toxic effect on bacteria in the activated sludge was increased with increasing number of benzocyclic compounds among the polycyclic aromatic hydrocarbons and the expression of the biphenyl degradation-related enzyme system was inhibited.
The other differentially expressed proteins related to PAH metabolism in the various experimental groups were identified as enzymes involved in naphthalene degradation. According to the reported degradation mechanism of naphthalene, the initial step of naphthalene oxidation is catalyzed by N1,2-DO, which consists of three α subunits (including NahA3) and three β subunits and is the crucial enzyme in the entire naphthalene degradation pathway. Subsequently, the resulting product can be converted to naphthalene 1,2-diol by naphthalene dihydrodiol dehydrogenase (NagB, EC 1.3.1.29).43 Importantly, based on proteomics analysis, NahA3 and NagB showed consistent trends in the different groups. The expression of NahA3 and NagB was significantly up-regulated in the TN-90 group with naphthalene treatment, indicating the presence of naphthalene-degrading microorganisms in the sludge and that the addition of naphthalene had induced the expression of enzymes responsible for naphthalene degradation.
With the development of protein purification and isolation techniques, metaproteomics has been employed in analyzing environmental samples, such as marine, groundwater, and soil samples.2,6,44 However, reports on the analysis of sludge samples for the degradation of trace pollutants, especially PAH, are rare. In this study, we applied a high-throughput sequencing technique for both 16S rRNA and proteomics to elucidate the relation between microbial community and protein expression profiles in response of PAH (naphthalene and anthracene) stress for different durations. The present research highlights the possibility of applying metaproteomics to environmental samples like activated sludge for the analysis of functional protein expression profiles. The application of 2D-PAGE coupled with high throughput and resolution MALDI-TOF-MS enabled us to obtain more information about the identification and quantification of the proteins and to monitor the changes in proteins expression. The significantly differentially expressed proteins detected belong to several functional categories including protein synthesis, DNA replication, lipid metabolism, glucose metabolism, stress response, and aromatic hydrocarbon metabolism. The expression profiles of the functional enzymes involved in PAH metabolism help illustrate the biochemical process of PAH in sludge (Fig. 4). The initial of the metabolic steps of naphthalene and anthracene were catalyzed by the ring hydroxylating dioxygenase, and the products of which would subsequently be dehydrogenated and ring-cleavage, and finally entering TCA cycle via benzoate pathway. It is important to note that the naphthalene oxidation enzyme N1,2-DO (EC 1.14.12.12), could also be responsible for the initial oxidation of anthracene. N1,2-DO possesses a relaxed substrate specificity, which could also catalyze the hydroxylation of two-ring and three-ring aromatic compounds, such as naphthalene, 1-methylnaphthalene, 2-methylnaphthalene, fluorene, anthracene and benzo[a]pyrene.45 Furthermore, dioxygenases including N1,2-DO (EC 1.14.12.12) and BDO (EC 1.14.12.18) which catalyze the initial oxidation step of corresponding PAHs play an important role in the metabolic process of PAHs in the activated sludge. These evidence support our proposal that the addition of a certain inducer could finally stimulate the co-expression of enzyme associated with the metabolism of a wide range of PAHs, and resulting in the co-degradation of various PAHs.
In addition, this study provides an important direction for future research; subsequent studies should focus on the physiological and biochemical characterization of the PAH degradation-related enzymes that were significantly up-regulated in this study, verify their functions, and elucidate their underlying expression and regulation mechanisms.
4. Conclusions
Analysis of microbial community indicates the diverse dominant bacteria profiles in the activated sludge from the various treatment groups. Furthermore, the diversity in all groups changes with treatment time; Burkholderiales populations were dominant in the process and their abundance increased with treatment time. These findings indicate that the related Burkholderiales species have better degradation ability for polycyclic aromatic hydrocarbons and their increase in activated sludge helps improve the degradation performance of naphthalene.Proteomic analysis showed that the proteins that significantly changed during PAHs stress are involved in DNA replication, fatty acid metabolism, glucose metabolism, stress, protein synthesis, and aromatic hydrocarbon metabolism. Another striking finding is that most functional protein with differential expression profile at PAHs stress were associated with Burkholderiales, which indicated its dominance in the PAHs metabolism consistent with the results from microbial community analysis. The changes in the amount of these polycyclic aromatic hydrocarbon degrading functional enzymes confirmed that activated sludge possesses certain degradation properties for different types of polycyclic aromatic hydrocarbons.
The present study verifies the application of proteomic methodologies along with microbial diversity analysis in the monitoring the level of differential functional proteins and microbial community during activated sludge process of PAHs. More work should be conducted on the other PAHs.
Funding
This work was supported by National Natural Science Foundation of China (Grant number 21477163 and 31670512), and Natural Science Basic Research Plan in Shaanxi Province of China (Grant number 2018JM3039).Conflicts of interest
The authors declare that they have no conflict of interest.References
- P. Wilmes, A. F. Andersson, M. G. Lefsrud, M. Wexler, M. Shah, B. Zhang, R. L. Hettich, P. L. Bond, N. C. VerBerkmoes and J. F. Banfield, ISME J., 2008, 2, 853–864 CrossRef CAS PubMed.
- J. Kan, T. E. Hanson, J. M. Ginter, K. Wang and F. Chen, Saline Syst., 2005, 1, 7 CrossRef PubMed.
- P. A. Maron, L. Ranjard, C. Mougel and P. Lemanceau, Microb. Ecol., 2007, 53, 486–493 CrossRef CAS PubMed.
- D. Benndorf, G. U. Balcke, H. Harms and M. von Bergen, ISME J., 2007, 1, 224–234 CrossRef CAS PubMed.
- C. M. Lacerda, L. H. Choe and K. F. Reardon, J. Proteome Res., 2007, 6, 1145–1152 CrossRef CAS PubMed.
- R. J. Ram, N. C. Verberkmoes, M. P. Thelen, G. W. Tyson, B. J. Baker, R. C. Blake II, M. Shah, R. L. Hettich and J. F. Banfield, Science, 2005, 308, 1915–1920 CrossRef CAS PubMed.
- P. Wilmes and P. L. Bond, Trends Microbiol., 2006, 14, 92–97 CrossRef CAS PubMed.
- B. Clark, G. L. Henry and D. Mackay, Environ. Sci. Technol., 1995, 29, 1488–1494 CrossRef CAS PubMed.
- K. Sexton, S. J. Mongin, J. L. Adgate, G. C. Pratt, G. Ramachandran, T. H. Stock and M. T. Morandi, J. Toxicol. Environ. Health, Part A, 2007, 70, 465–476 CrossRef CAS PubMed.
- E. Trably and D. Patureau, Environ. Sci. Pollut. Res. Int., 2006, 13, 170–176 CrossRef CAS PubMed.
- C. D. Knightes and C. A. Peters, Environ. Eng. Sci., 2003, 20, 207–218 CrossRef CAS.
- A. Chauhan, Fazlurrahman, J. G. Oakeshott and R. K. Jain, Indian J. Microbiol., 2008, 48, 95–113 CrossRef CAS PubMed.
- D. T. Sponza and O. Gok, Water Sci. Technol., 2011, 64, 2282–2292 CrossRef CAS PubMed.
- L. Delgado-Balbuena, J. M. Bello-Lopez, Y. E. Navarro-Noya, A. Rodriguez-Valentin, M. L. Luna-Guido and L. Dendooven, PLoS One, 2016, 11, e0160991 CrossRef PubMed.
- M. D. Ferrari, Rev. Argent. Microbiol., 1996, 28, 83–98 CAS.
- S. Baran and P. Oleszezuk, Pol. J. Environ. Stud., 2003, 12, 523–529 CAS.
- I. Siebielska, Water Sci. Technol., 2014, 70, 1617–1624 CrossRef CAS PubMed.
- S. Khadhar, T. Higashi, H. Hamdi, S. Matsuyama and A. Charef, J. Hazard. Mater., 2010, 183, 98–102 CrossRef CAS PubMed.
- D. T. Sponza and O. Gok, Bioresour. Technol., 2010, 101, 914–924 CrossRef CAS PubMed.
- J. C. Young, L. S. Clesceri and S. M. Kamhawy, Water Environ. Res., 2007, 79, 455–456 Search PubMed.
- M. X. Guo, Z. Q. Gong, R. H. Miao, C. Y. Jia, J. Rookes, D. Cahill and J. Zhuang, Chemosphere, 2018, 212, 15–23 CrossRef CAS PubMed.
- S. S. Li, X. Hu, H. Zhao, Y. X. Li, L. Zhang, L. J. Gong, J. Guo and H. B. Zhao, Biotechnol. Lett., 2015, 37, 1645–1654 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- A. R. Hesketh, G. Chandra, A. D. Shaw, J. J. Rowland, D. B. Kell, M. J. Bibb and K. F. Chater, Mol. Microbiol., 2002, 46, 917–932 CrossRef CAS PubMed.
- P. Wilmes, M. Wexler and P. L. Bond, PLoS One, 2008, 3, e1778 CrossRef PubMed.
- P. D. Schloss, S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn and C. F. Weber, Appl. Environ. Microbiol., 2009, 75, 7537–7541 CrossRef CAS PubMed.
- C. N. Owabor, S. E. Ogbeide and A. A. Susu, Environ. Monit. Assess., 2010, 169, 607–617 CrossRef CAS PubMed.
- M. Afzal, S. Khan, S. Iqbal, M. S. Mirza and Q. M. Khan, Int. Biodeterior. Biodegrad., 2013, 85, 331–336 CrossRef CAS.
- Z. R. Suarez-Moreno, J. Caballero-Mellado, B. G. Coutinho, L. Mendonca-Previato, E. K. James and V. Venturi, Microb. Ecol., 2012, 63, 249–266 CrossRef PubMed.
- N. Gonzalez, R. Simarro, M. C. Molina, L. F. Bautista, L. Delgado and J. A. Villa, Bioresour. Technol., 2011, 102, 9438–9446 CrossRef CAS PubMed.
- R. Y. Liu, Y. X. Gao, Y. F. Ji, Y. Zhang and M. Yang, Water Sci. Technol., 2015, 71, 123–130 CrossRef PubMed.
- M. Song, C. L. Luo, L. F. Jiang, D. Y. Zhang, Y. J. Wang and G. Zhang, Appl. Environ. Microbiol., 2015, 81, 7368–7376 CrossRef CAS PubMed.
- D. Perez-Pantoja, R. Donoso, L. Agullo, M. Cordova, M. Seeger, D. H. Pieper and B. Gonzalez, Environ. Microbiol., 2012, 14, 1091–1117 CrossRef CAS PubMed.
- K. M. L. Hutalle-Schmelzer, E. Zwirnmann, A. Kruger and H. P. Grossart, Microb. Ecol., 2010, 60, 462–475 CrossRef PubMed.
- Z. W. Ma, X. M. Shen, H. B. Hu, W. Wang, H. S. Peng, P. Xu and X. H. Zhang, J. Bacteriol., 2012, 194, 3535–3536 CrossRef PubMed.
- H. P. Bacosa and C. Inoue, J. Hazard. Mater., 2015, 283, 689–697 CrossRef CAS PubMed.
- J. J. A. Dominguez, H. P. Bacosa, M. F. Chien and C. Inoue, Chemosphere, 2019, 234, 789–795 CrossRef CAS PubMed.
- J. Eisenbrand and K. Baumann, Z. Lebensm.-Unters. Forsch., 1969, 140, 210–216 CrossRef CAS.
- L. F. Jiang, C. L. Luo, D. Y. Zhang, M. K. Song, Y. T. Sun and G. Zhang, Environ. Sci. Technol., 2018, 52, 8558–8567 CrossRef CAS PubMed.
- D. H. Pieper, Appl. Microbiol. Biotechnol., 2005, 67, 170–191 CrossRef CAS PubMed.
- D. Barriault and M. Sylvestre, Appl. Microbiol. Biotechnol., 1999, 51, 592–597 CrossRef CAS PubMed.
- C. L. Yu, W. Liu, D. J. Ferraro, E. N. Brown, J. V. Parales, S. Ramaswamy, G. J. Zylstra, D. T. Gibson and R. E. Parales, J. Ind. Microbiol. Biotechnol., 2007, 34, 311–324 CrossRef CAS PubMed.
- S. Li, H. Zhao, Y. Li, S. Niu and B. Cai, J. Bacteriol., 2012, 194, 5154–5155 CrossRef CAS PubMed.
- D. Benndorf, G. U. Balcke, H. Harms and M. von Bergen, ISME J., 2007, 1, 224–234 CrossRef CAS PubMed.
- D. J. Ferraro, A. Okerlund, E. Brown and S. Ramaswamy, IUCrJ, 2017, 4, 648–656 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra04674g |
| This journal is © The Royal Society of Chemistry 2019 |