Open Access Article
Open Access ArticlePreparation and characterization of solid DNA silver nanoclusters with superior aerobic and thermal stability†
Yang-Xiu Lin and
Chih-Wei Chang *
*
Department of Chemistry, National Changhua University of Education, Taiwan. E-mail: cwchang@cc.ncue.edu.tw
First published on 20th August 2019
Abstract
In the current study, we present a universal method to preserve DNA-templated silver nanoclusters (DNA AgNCs) in the solid-state. Our results show that DNA AgNCs must be precipitated before drying. By drying the ethanol precipitated DNA AgNCs, we have successfully prepared solid DNA AgNCs with superior stability in aerobic and high-temperature environments. Although the fluorescence lifetime measurements show that the emission of DNA AgNCs is drastically quenched in the solid-state, the emission can be fully recovered in solution. To our knowledge, this is the first attempt to prepare DNA AgNCs in solid-state, and this finding provides an ideal solution for the transportation and long-term preservation of DNA AgNCs.
Introduction
In past decades, many efforts have been devoted to the synthesis of noble metal nanoclusters (NCs), the small particles that are composed of ten to a hundred atoms.1–8 In solution, the surface of the metal clusters needs to be protected by ligands,9 and various ligands such as thiols, amines, polymers, DNA, proteins, and dendrimers have been employed to synthesize noble metal NCs.9–11 Nowadays, different noble metal (such as Ag, Au, Pt, and Cu) NCs have been reported.6,12 Among these metal clusters, DNA-stabilized silver nanoclusters (DNA AgNCs) have received particular attention because they are highly biocompatible and have been applied in the detection of DNA/RNA,13–18 sensing of small molecules19 and bioimaging20–22 studies. Recent studies have indicated that the emission color of AgNCs is controlled by multiple factors, including the number of silver atoms,23 the shape of silver clusters,24 the oxidization states of the silver atom,25,26 the conformation of deoxyribose27 and the silver–base interaction.28 In the presence of oxygen, the oxidization of Ag0 atoms changes the ratio of the Ag0/Ag+ and ultimately alters the emission properties of AgNCs.29–33 Although certain DNA AgNCs can be stored in aqueous solution for several months or even a year,34 there is no direct correlation between the stability of DNA AgNCs and the sequence of the DNA template. In some cases, the stability of DNA AgNCs is too low, and hence the DNA AgNCs solution must be used fresh, or preserved in the anaerobic environment.31 This limitation explains why finding commercial DNA AgNCs products in the market is difficult, and why the development of a universal long-term preservation method for DNA AgNCs in an aerobic environment is crucial for their application.In contrast with the DNA in the solution phase, the dried DNA is more stable and suitable for long-term storage. However, it is surprising that solid DNA AgNCs have never been reported. In this work, we have employed the cytosine-rich DNA sequence d[T2C3AC3AC4G2C3] (r-AgNCs),35 as our model system and prepared the solid r-AgNCs by different instruments. The result shows that the dried r-AgNCs will lose their fluorescence (FL) due to the oxidation of the AgNCs, and it is essential to precipitate the r-AgNCs prior to the drying process. By drying the ethanol precipitated r-AgNCs, we successfully prepared the solid DNA AgNCs that the emission can be recovered in solution phase. Besides, we also found that the solid DNA AgNCs exhibit superior stability in aerobic or high-temperature environments, which is beneficial for the long-term storage and the transportation of the DNA AgNCs, and the details will be discussed in the following paragraph.
Experimental
Materials
The AgNO3 (≥99.0%), NaBH4 (≥99%), cresyl violet perchlorate (>96.5%), ammonium acetate (≥99%), and sodium acetate (≥99%) were purchased from Sigma Aldrich and used without further purification. The ethanol was purchased from ECHO Chemical, and the HPLC purified d[T2C3AC3AC4G2C3], d[CGC6T2G2CGT] and d[AG3(T2AG3)3] DNA were obtained from MDBio. The TEMED (N,N,N′,N′-tetramethylethylenediamine), ammonium persulfate, loading buffer (0.03% Xylene cyanol FF, 0.03% Bromophenol blue, 10 mM Tris–HCl (pH8.0), 60% glycerol, 60 mM EDTA) and the 10× TBE pH = 8.0 buffer were obtained from Biomate. The 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) acrylamide/bisacrylamide was obtained from Bio Basic. Inc. The SYBR gold was obtained from Fisher scientific. Deionized water (resistivity >18.2 MΩ cm) was obtained from Barnstead™ EasyPure™ II water purification system (Fisher Scientific).
1 (w/w) acrylamide/bisacrylamide was obtained from Bio Basic. Inc. The SYBR gold was obtained from Fisher scientific. Deionized water (resistivity >18.2 MΩ cm) was obtained from Barnstead™ EasyPure™ II water purification system (Fisher Scientific).
Synthesis of r-AgNCs
The 50 μM of d[T2C3AC3AC4G2C3] sequence was dissolved in 10 mM ammonium acetate buffer (pH = 7.0) and annealed at 90 °C for 10 minutes. After cooling, the DNA solution was mixed with the AgNO3 for 15 minutes and then the NaBH4 (DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AgNO3
AgNO3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaBH4 = 1
NaBH4 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). The mixed solution was stored in a 4 °C refrigerator 3 hours prior to further experiment.
5). The mixed solution was stored in a 4 °C refrigerator 3 hours prior to further experiment.
Synthesis of g-AgNCs
The 50 μM of d[CGC6T2G2CGT] sequence was dissolved in 10 mM ammonium acetate buffer (pH = 7.0) and annealed at 90 °C for 10 minutes. After cooling, the DNA solution was mixed with the AgNO3 for 15 minutes, and then the NaBH4 solution (DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AgNO3
AgNO3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaBH4 = 1
NaBH4 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.4). The mixed solution was stored in a 4 °C refrigerator 3 hours prior to further experiment.
2.4). The mixed solution was stored in a 4 °C refrigerator 3 hours prior to further experiment.
Synthesis of Hum 22 AgNCs
The 30 μM of d[AG3(T2AG3)3] sequence was dissolved in de-ionized water and annealed at 90 °C for 10 minutes. After cooling, the DNA solution was mixed with the AgNO3 for 60 minutes and then the NaBH4 (DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AgNO3
AgNO3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaBH4 = 1
NaBH4 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6). The mixed solution was stored in a 4 °C refrigerator 12 hours prior to further experiment.
6). The mixed solution was stored in a 4 °C refrigerator 12 hours prior to further experiment.
Preparation of ethanol precipitated DNA AgNCs
The high concentration sodium acetate solution (3 M, pH = 7.0) was added to the DNA AgNCs solution (50 μM) until the concentration of the sodium acetate reached 300 mM. The ethanol was added to the DNA solution until the final volume percentage of ethanol exceeded 90% and refrigerated in a −20 °C refrigerator for 1 hour to improve the formation of precipitation. The DNA AgNCs ethanol solution was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 684g (12
684g (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, Mikro 200, Hettich) for 30 minutes, at which time the gel-like precipitate was visible at the bottom of the Eppendorf tube. After removing the supernatant, the DNA AgNCs precipitate was dried overnight in the vacuum oven at room temperature.
000 rpm, Mikro 200, Hettich) for 30 minutes, at which time the gel-like precipitate was visible at the bottom of the Eppendorf tube. After removing the supernatant, the DNA AgNCs precipitate was dried overnight in the vacuum oven at room temperature.
The characterization of the DNA AgNCs
The UV-Vis absorption spectra were measured using a Cary 100 UV-Vis spectrophotometer, and the emission spectra were obtained on a Cary Eclipse FL spectrophotometer. The FL quantum yields were estimated by using a cresyl violet perchlorate methanol solution as the reference. The time-resolved FL spectra were recorded using a time-correlated single-photon counting system. The excitation light source was provided by pulsed LED (PLS-600, PicoQuant) in combination with a 580 nm bandpass filter. The polarization of the excitation an emission light is selected through a polarizer, and the FL decay dynamics were measured at the magic angle condition. The instrument response function of the single-photon counting system was recorded by measuring the scattered light of the diluted colloidal silica solution. For the DNA AgNCs solution, the sample was loaded in a 1 cm quartz cuvette for spectroscopy measurement. For the solid DNA AgNCs, the sample was deposited between two glass plates. The transmission electron microscopy (TEM) images were taken using an HT-7700 TEM (Hitachi), and the X-ray photoelectron spectroscopy (XPS) analysis was carried out by a ULVAC-PHI, PHI 5000 VersaProbe/Scanning ESCA Microprobe. The FL microscope image was acquired using an Olympus IX 73 inverted microscope with the Sony SLT-A33 digital camera. The filter set used for the FL microscope: excitation filter = ET550/15× (chroma), the dichroic beam splitter = T570LP (chroma), and the emission filter = ET610/75 M (chroma).Native polyacrylamide gel electrophoresis (PAGE)
The 10 μl (6 μM) DNA and DNA AgNCs solutions were mixed with 2 μl load buffer (0.03% Xylene cyanol FF, 0.03% Bromophenol blue, 10 mM Tris–HCl (pH = 8.0), 60% glycerol, 60 mM EDTA, purchased from Biomate) and then analyzed on 20% native polyacrylamide gel. The electrophoresis was conducted in 1× Tris–borate–EDTA (TBE, 89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.0) at a constant voltage of 120 V for 2.5 hours (Mini-Protean Tetra cell + PowerPac™ basic power supply, Biorad). After finishing the electrophoresis, the gel was stained in SYBR-gold solution for 10 minutes and washing by water and 1× TBE buffer. The gel image was taken by a visible light (470 nm) gel scanner system (Glite 900 BW, Pacific Image). For the PAGE imaged in fluorescence mode, the DNA and DNA AgNCs concentration was increased to 50 uM, and the loading buffer was changed to 60% glycerol + 10 mM Tris–HCl (pH = 8.0). After finishing the electrophoresis, the gel was exposed to 375 nm light and imaged by Sony α33 camera without staining.Results and discussion
Properties of the as-prepared r-AgNCs
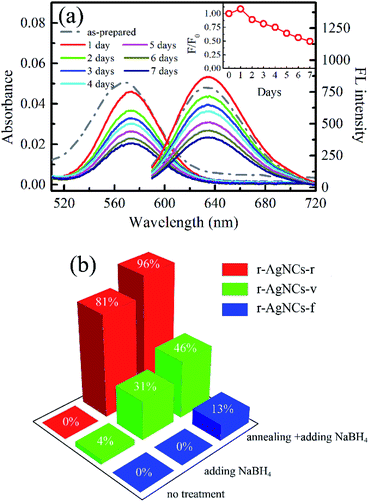
Fig. 1a and b indicate the TEM image and the XPS spectrum of the AgNCs labeled on the cytosine-rich DNA sequence d[T2C3AC3AC4G2C3] (r-AgNCs),35 respectively. As illustrated, the size of the r-AgNCs is ∼3.7 nm, and the XPS spectrum shows typical Ag 3d5/2 and Ag 3d3/2 transition bands for the silver atom.36 Fig. 2a indicates the steady-state spectra of the r-AgNCs solution. Upon 580 nm excitation, the r-AgNCs demonstrate an emission maximum around 630 nm, and the FL quantum yield is estimated to be 0.73 (Fig. S1, ESI†). Like many DNA AgNCs, the r-AgNCs solution is unstable in the aerobic environment, and the FL intensity drops about 50% within 7 days (Fig. 2a, inset). Our controlled experiments indicated that the stability of the r-AgNCs can be significantly improved in anaerobic or refrigerated (4 °C) environments (Fig. S2, ESI†), and hence the FL quenching is associated with the oxidization of the Ag atoms.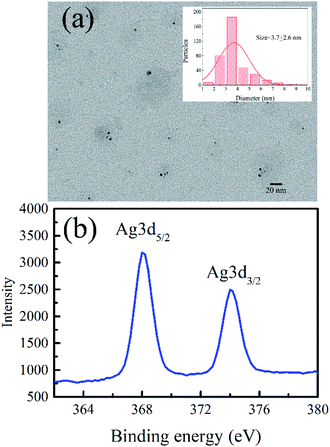 | ||
| Fig. 1 (a) The TEM image, the size distribution (inset) and (b) the XPS spectra of the as-prepared r-AgNCs. | ||
The preparation and the FL recovery yield of the solid AgNCs dried by different instruments
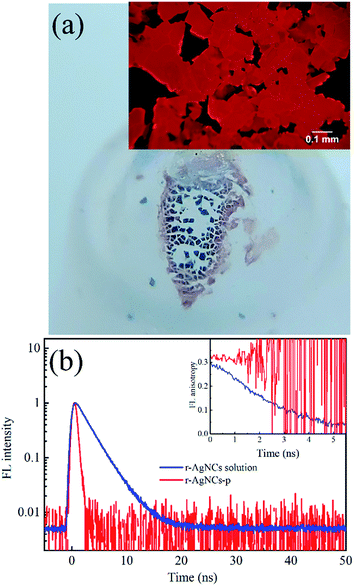
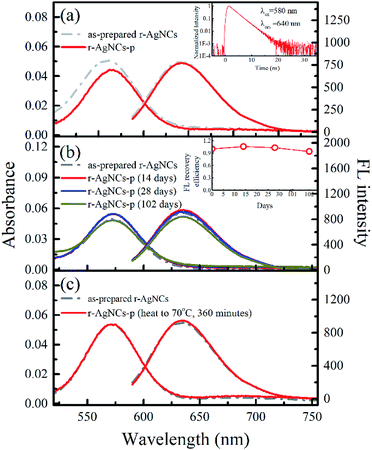
To test the feasibility of preserving DNA AgNCs in their dried state, we have dried the r-AgNCs using a rotary evaporator (r-AgNCs-r), a vacuum oven (r-AgNCs-v), and a freeze dryer (r-AgNCs-f). To estimate the recovery efficiency of the dried r-AgNCs, we have dissolved the dried r-AgNCs in an ammonium acetate buffer and calculated the FL recovery yield according to the ratio between the integrated FL intensity of the re-dissolved and the as-prepared r-AgNCs solutions. The results are illustrated in Fig. 2b (Fig. S3a, ESI†). As delineated, the FL recovery yield for the dried r-AgNCs is close to zero. It should be noted that the FL of the r-AgNCs-r and r-AgNCs-v can be partially recovered by adding the NaBH4 in solution (Fig. 2b and S3b, ESI†), hence the FL quenching is partially associated with the oxidation of Ag atoms. We also found that annealing the solution at 90 °C before the addition of NaBH4 could further improve the FL recovery efficiency (Fig. 2b and S3c, ESI†). Therefore, the conformation change during the drying process is also responsible for the FL quenching of the DNA AgNCs. More importantly, these findings suggest that AgNCs must be protected prior to drying processes.To solve this problem, we precipitate the r-AgNCs by the ethanol precipitation method. After removing the supernatant, we dried the precipitate in the vacuum oven at room temperature, after which the light purple solid was clearly visible at the bottom of the Eppendrof (r-AgNCs-p, Fig. 3a). Upon 550 nm excitation, the r-AgNCs-p appeared red emission under the FL microscope (Fig. 3a, inset). Besides the emission band central at 630 nm, the r-AgNCs-p also exhibits an additional emission band central at ∼690 nm (Fig. S4, ESI†) due to the aggregation of the DNA template. The FL lifetime measurement shows that the averaged FL lifetime decreases from 2.51 ns for the r-AgNCs solution to less than 0.3 ns for the r-AgNCs-p, which implies that the FL of the AgNCs is drastically quenched in the r-AgNCs-p. We also compared the FL anisotropy decay dynamics of the r-AgNCs solution and the r-AgNCs-p. In the FL time window between 0–2.5 ns, the FL anisotropy value of the r-AgNCs solution decreases due to the local structural fluctuation and rotational dynamics of the DNA backbone.31 However, the r-AgNCs-p exhibit constant FL anisotropy value in this range (Fig. 3b, inset). This finding confirms that the AgNCs was confined in a restricted environment in the r-AgNCs-p. Unlike the r-AgNCs dried by the other methods, the r-AgNCs-p shows high FL recovery yield in solution. Fig. 4a illustrates the steady-state spectra of the r-AgNCs-p dissolved in the ammonium acetate buffer. As delineated, the spectra closely resemble that of the as-prepared r-AgNCs solution, and the FL recovery yield is close to 1 (0.968). The FL decay dynamics of the r-AgNCs-p solution is also similar to that of the as-prepared r-AgNCs solution (Fig. 4a, inset). The TEM images show that the size of the recovered r-AgNCs (4.1 nm) is ∼4.1 nm, which is consistent with the size of the as-prepared r-AgNCs (Fig. S5, ESI†).
The superior stability of the ethanol precipitated DNA AgNCs
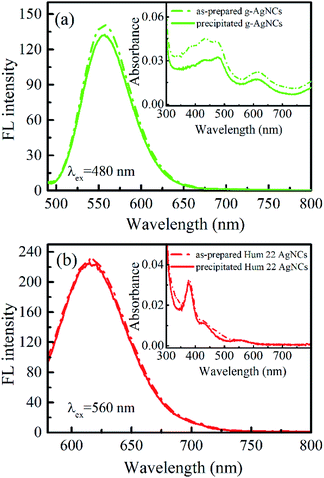
Another unique feature of the r-AgNCs-p is its superior stability in the aerobic environment. Our test shows that the r-AgNCs-p can be stored in laboratory drawers for more than 100 days, and the FL recovery yield still exceeds 90% (Fig. 4b). The native PAGE (Fig. S6 and S7, ESI†) and the fluorescence anisotropy dynamics (Fig. S8, ESI†) also show that the r-AgNCs remain the same after being transformed into the solid-state and long-term storage. The crystal structure and fluorescence anisotropy studies suggested that the AgNCs is surrounded by multiple DNA backbone and close contact with the DNA bases.31,37 Therefore, it is reasonable for us to expect that the precipitated DNA can form a more effective protecting layer than the free DNA molecule to prevent the oxidization of the AgNCs. This feature also leads to the outstanding thermal stability of the r-AgNCs-p. At 70 °C, the emission of the r-AgNCs solution is completely diminished within 360 minutes (Fig. S9, ESI†); however, the FL of r-AgNCs-p stored in the same condition can be completely recovered in the buffer solution (Fig. 4c). To test the versatility of ethanol precipitation method for other DNA AgNCs, we also prepared the precipitated DNA AgNCs for the AgNCs labeled on the other two templates: the d[CGC6T2G2CGT] (g-AgNCs) and d[AG3(T2AG3)3] (Hum 22 AgNCs) sequences. Like the r-AgNCs, the ethanol precipitated g-AgNCs and Hum 22 AgNCs also exhibits superior stability in the aerobic environment, and the FL can be recovered in solution with a high recovery yield (Fig. 5). Although the Hum 22 AgNCs is unstable in the aerobic solution,30 the precipitated Hum 22 AgNCs still shows excellent stability in the aerobic condition (Fig. S10, ESI†). All this evidence indicates that drying the precipitated DNA AgNCs is an ideal long-term storage method for DNA AgNCs.Conclusions
In summary, we have successfully prepared solid DNA AgNCs with superior anaerobic and thermal stabilities. Instead of directly drying the DNA AgNCs solution, we found that precipitating DNA AgNCs prior to the drying processes can effectively prevent the oxidization of AgNCs. Moreover, we also demonstrated that this method is versatile for preparing the solid DNA AgNCs labeled on different templates. Since the most water-soluble contaminants (including free Ag+ ions and the oxidization products of the NaBH4) were removed during the drying process, the precipitated DNA AgNCs are more suitable for applications in biological studies. The preparation of solid DNA AgNCs also provides the opportunity to investigate the excited state dynamics of the AgNCs in highly crowded and dehydrated conditions. Recently, Reveguk et al. have reported the ultrafast spectral relaxation dynamics of DNA AgNCs in the solution phase and concluded that spectral relaxation dynamics are associated with the geometric change of AgNCs in the excited state.38 It will be interesting to examine their model using solid DNA AgNCs. In contrast to DNA AgNCs solution, precipitated DNA AgNCs are more suitable for transportation and long-term storage, and this finding will be important for future applications of DNA AgNCs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We sincerely appreciate the financial support from the Ministry of Science and Technology, Taiwan (project contracts: MOST 107-2113-M-018-004).References
- L.-Y. Chen, C.-W. Wang, Z. Yuan and H.-T. Chang, Anal. Chem., 2014, 87, 216–229 CrossRef PubMed.
- S. Choi, R. M. Dickson and J. Yu, Chem. Soc. Rev., 2012, 41, 1867–1891 RSC.
- J. M. Obliosca, C. Liu and H.-C. Yeh, Nanoscale, 2013, 5, 8443–8461 RSC.
- J. T. Petty, S. P. Story, J.-C. Hsiang and R. M. Dickson, J. Phys. Chem. Lett., 2013, 4, 1148–1155 CrossRef CAS PubMed.
- L. Shang and S. Dong, Chem. Commun., 2008, 1088–1090 RSC.
- L. Zhang and E. Wang, Nano Today, 2014, 9, 132–157 CrossRef CAS.
- X. Yuan, Z. Luo, Q. Zhang, X. Zhang, Y. Zheng, J. Y. Lee and J. Xie, ACS Nano, 2011, 5, 8800–8808 CrossRef CAS PubMed.
- I. Díez and R. H. A. Ras, Nanoscale, 2011, 3, 1963–1970 RSC.
- I. Chakraborty and T. Pradeep, Chem. Rev., 2017, 117, 8208–8271 CrossRef CAS PubMed.
- R. Jin, C. Zeng, M. Zhou and Y. Chen, Chem. Rev., 2016, 116, 10346–10413 CrossRef CAS PubMed.
- X. Wen, P. Yu, Y.-R. Toh and J. Tang, J. Phys. Chem. C, 2013, 116, 11830–11836 CrossRef.
- J. Zheng, P. R. Nicovich and R. M. Dickson, Annu. Rev. Phys. Chem., 2007, 58, 409–431 CrossRef CAS PubMed.
- Z. Huang, F. Pu, Y. Lin, J. Ren and X. Qu, Chem. Commun., 2011, 47, 3487–3489 RSC.
- J. Li, X. Zhong, H. Zhang, X. C. Le and J.-J. Zhu, Anal. Chem., 2012, 84, 5170–5174 CrossRef CAS PubMed.
- S. W. Yang and T. Vosch, Anal. Chem., 2011, 83, 6935–6939 CrossRef CAS PubMed.
- L. Ge, X. Sun, W. Hong and F. Li, ACS Appl. Mater. Interfaces, 2017, 9, 32089–32096 CrossRef CAS PubMed.
- L. Liu, Q. Yang, J. Lei, N. Xu and H. Ju, Chem. Commun., 2014, 50, 13698–13701 RSC.
- P. Shah, S. W. Choi, H.-J. Kim, S. K. Cho, Y.-J. Bhang, M. Y. Ryu, P. W. Thulstrup, M. J. Bjerrum and S. W. Yang, Nucleic Acids Res., 2016, 44, e57 CrossRef PubMed.
- Y. Fu, H. Jin, X. Bu and R. Gui, J. Agric. Food Chem., 2018, 66, 9819–9827 CrossRef CAS PubMed.
- J. Zhu, L. Zhang, Y. Teng, B. Lou, X. Jia, X. Gu and E. Wang, Nanoscale, 2015, 7, 13224–13229 RSC.
- S. Choi and J. Yu, Chem.–Eur. J., 2016, 22, 12660–12664 CrossRef CAS PubMed.
- J. Liu, Trends Anal. Chem., 2014, 58, 99–111 CrossRef CAS.
- S. M. Copp, D. Schultz, S. Swasey, J. Pavlovich, M. Debord, A. Chiu, K. Olsson and E. Gwinn, J. Phys. Chem. Lett., 2014, 5, 959–963 CrossRef CAS PubMed.
- R. R. Ramazanov and A. I. Kononov, J. Phys. Chem. C, 2013, 117, 18681–18687 CrossRef CAS.
- J. Sharma, H.-C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2010, 46, 3280–3282 RSC.
- C. M. Ritchie, K. R. Johnsen, J. R. Kiser, A. Antoku, R. M. Dickson and J. T. Petty, J. Phys. Chem. C, 2007, 111, 175–181 CrossRef CAS PubMed.
- D. Schultz and E. Gwinn, Chem. Commun., 2011, 47, 4715–4717 RSC.
- D. Schultz and E. G. Gwinn, Chem. Commun., 2012, 48, 5748–5750 RSC.
- C. M. Ritchie, K. R. Johnsen, J. R. Kiser, Y. Antoku, R. M. Dickson and J. T. Petty, J. Phys. Chem. C, 2007, 111, 175–181 CrossRef CAS PubMed.
- H.-C. Hsu, M.-C. Ho, K.-H. Wang, Y.-F. Hsu and C.-W. Chang, New J. Chem., 2015, 39, 2140–2145 RSC.
- H.-C. Hsu, Y.-X. Lin and C.-W. Chang, Dyes Pigments, 2017, 146, 420–424 CrossRef CAS.
- D. Schultz, K. Gardner, S. S. R. Oemrawsing, N. Markešević, K. Olsson, M. Debord, D. Bouwmeester and E. Gwinn, Adv. Mater., 2013, 25, 2797–2803 CrossRef CAS PubMed.
- C. Cerretani and T. Vosch, ACS Omega, 2019, 4, 7895–7902 CrossRef CAS.
- J. Sharma, R. C. Rocha, M. L. Phipps, H.-C. Yeh, K. A. Balatsky, D. M. Vu, A. P. Shreve, J. H. Werner and J. S. Martinez, Nanoscale, 2012, 4, 4107–4110 RSC.
- C. Cerretani, M. R. Carro-Temboury, S. Krause, S. A. Bogh and T. Vosch, Chem. Commun., 2017, 53, 12556–12559 RSC.
- I. L. Volkov, A. Smirnova, A. A. Makarova, Z. V. Reveguk, R. R. Ramazanov, D. Y. Usachov, V. K. Adamchuk and A. I. Kononov, J. Phys. Chem. B, 2017, 121, 2400–2406 CrossRef CAS PubMed.
- D. J. E. Huard, A. Demissie, D. Kim, D. Lewis, R. M. Dickson, J. T. Peety and R. L. Lieberman, J. Am. Chem. Soc., 2019, 141, 11465–11470 CrossRef CAS PubMed.
- Z. Reveguk, R. Lysenkon, R. Ramazanov and A. Kononov, Phys. Chem. Chem. Phys., 2018, 20, 28205–28210 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, instruments, methods and other data. See DOI: 10.1039/c9ra04533c |
| This journal is © The Royal Society of Chemistry 2019 |